Foodborne Pathogen Screening Using Magneto-fluorescent Nanosensor: Rapid Detection of E. Coli O157:H7
Summary
The overall goal of this protocol is to synthesize functional nanosensors for the portable, cost-effective, and rapid detection of specifically targeted pathogenic bacteria through a combination of magnetic relaxation and fluorescence emission modalities.
Abstract
Enterohemorrhagic Escherichia coli O157:H7 has been linked to both waterborne and foodborne illnesses, and remains a threat despite the food- and water-screening methods used currently. While conventional bacterial detection methods, such as polymerase chain reaction (PCR) and enzyme-linked immunosorbent assays (ELISA) can specifically detect pathogenic contaminants, they require extensive sample preparation and lengthy waiting periods. In addition, these practices demand sophisticated laboratory instruments and settings, and must be executed by trained professionals. Herein, a protocol is proposed for a simpler diagnostic technique that features the unique combination of magnetic and fluorescent parameters in a nanoparticle-based platform. The proposed multiparametric magneto-fluorescent nanosensors (MFnS) can detect E. coli O157:H7 contamination with as little as 1 colony-forming unit present in solution within less than 1 h. Furthermore, the ability of MFnS to remain highly functional in complex media such as milk and lake water has been verified. Additional specificity assays were also used to demonstrate the ability of MFnS to only detect the specific target bacteria, even in the presence of similar bacterial species. The pairing of magnetic and fluorescent modalities allows for the detection and quantification of pathogen contamination in a wide range of concentrations, exhibiting its high performance in both early- and late-stage contamination detection. The effectiveness, affordability, and portability of the MFnS make them an ideal candidate for point-of-care screening for bacterial contaminants in a wide range of settings, from aquatic reservoirs to commercially packaged foods.
Introduction
The persistent occurrence of bacterial contamination in both commercially produced food and water sources has created a need for increasingly rapid and specific diagnostic platforms.1,2 Some of the more common bacterial contaminants responsible for food and water contamination are from the Salmonella, Staphylococcus, Listeria, Vibrio, Shigella, Bacillus, and Escherichia genera.3,4 Bacterial contamination by these pathogens often results in symptoms such as fever, cholera, gastroenteritis, and diarrhea.4 Contamination of water sources often has drastic and adverse effects on communities without access to sufficiently filtered water, and food contamination has led to a great number of illnesses and product recall efforts.5,6
In order to reduce the occurrence of illnesses caused by bacterial contamination, there have been a number of efforts to develop methods by which water and food can be efficiently scanned prior to sale or consumption.3 Techniques such as PCR,1,7,8,9,10 ELISA,11,12 loop-mediated isothermal amplification (LAMP),13,14 among others,15,16,17,18,19,20,21,22,23,24 have recently been used for detection of various pathogens. Compared to traditional bacterial culturing methods, these techniques are far more efficient with regards to specificity and time. However, these techniques still struggle with false positives and negatives, complex procedures, and cost.1,3,25 It is for this very reason that multiparametric magneto-fluorescent nanosensors (MFnS) are proposed as an alternative method for bacterial detection.
These nanosensors uniquely pair together magnetic relaxation and fluorescent modalities, allowing for a dual-detection platform that is both rapid and accurate. Using E. coli O157:H7 as a sample contaminant, the ability of MFnS to detect as little as 1 CFU within minutes is demonstrated. Pathogen-specific antibodies are used to increase specificity, and the combination of both magnetic and fluorescent modalities allows for the detection and quantification of bacterial contaminants in both low- and high-contamination ranges.16 In the case of bacterial contamination, the nanosensors will swarm around the bacteria due to the targeting abilities of the pathogen-specific antibodies. The binding between the magnetic nanosensors and bacteria limits the interaction between the magnetic iron core and the surrounding water protons. This causes an increase in the T2 relaxation times, as recorded by a magnetic relaxometer. As the concentration of bacteria in solution rises, the nanosensors disperse with the increased number of bacteria, resulting in lower T2 values. Conversely, fluorescence emission will increase in proportion with the concentration of bacteria, due to the increased number of nanosensors directly bound to pathogen. Centrifugation of the samples, and isolation of the bacterial pellet, will only conserve the nanoparticles directly attached to the bacteria, removing any free-floating nanosensors, and directly correlating the fluorescence emission with the number of bacteria present in solution. A schematic representation of this mechanism is represented in Figure 1.
This MFnS platform has been designed with point-of-care screening in mind, resulting in low-cost and portable characteristics. MFnS are stable at room temperature, and are only required in very low concentrations for accurate detection of bacterial contaminants. Furthermore, after synthesis, use of the MFnS is simple and does not require the use of trained professionals in the field. Lastly, this diagnostic platform allows for highly customizable targeting, providing a means by which this one platform may be used to detect pathogens of all kinds, in many different settings.
Protocol
1.Synthesis and Functionalization of Multi-Parametric Magneto-Fluorescent Nanosensors (MFnS).
- Synthesis of superparamagnetic iron oxide nanoparticles (IONPs)
- To prepare for IONP synthesis, prepare the following 3 solutions: Solution 1: FeCl3 (0.70 g) and FeCl2 in H2O (2 mL), Solution 2: NH4OH (2.0 mL, 13.4 M) in H2O (15 mL), and Solution 3: polyacrylic acid (0.855 g) in H2O (5 mL).
- Add 90 µL of 2 M Hydrochloric acid (HCl) to Solution 1 and then immediately mix with Solution 2 while vortexing at 875 rpm. Then add Solution 3. Continue vortexing for 60 min.
- Centrifuge for 20 min at 1,620 x g. Centrifuge the supernatant for 20 min at 2,880 x g.
- Purify the final supernatant via dialysis. Add the supernatant to a dialysis bag (MWCO 6−8 K) and place it in a beaker containing phosphate buffered saline (PBS) (pH = 7.4) and spin bar. Let it soak for 12 h, replacing with fresh buffer every 2-3 h.
- Conjugation of target-specific antibodies to surface of IONPs
- Prepare the following four solutions: Solution A: 5 mg of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) in 250 µL MES [2-(N-morpholino)ethanesulfonic acid] buffer (0.1 M, pH = 6.8), Solution B: 3 mg of N-Hydroxysuccinimide (NHS) in 250 µL of MES buffer (0.1 M, pH = 6.8), Solution C: 5 µg of IgG1, the E. coli mAb, in 225 µL of phosphate-buffered saline (PBS) (pH 7.4), and Solution D: 4 mL of IONP (5.0 mmol) in 1 mL of PBS (pH 7.4).
- Add 2-(N-Morpholino)ethanesulfonic acid (MES) buffer to Solution A, then immediately add Solution A to Solution D in small increments within 15 s, inverting the solution after each addition for mixing.
- Immediately begin to add Solution B to Solution D in small increments over a three-minute period. Mix by inverting the solution after each addition.
- Add Solution C to solution in increments of 5 µL, inverting the solution after each addition for mixing.
- Allow reaction to continue for 3 h at room temperature and then continue the incubation at 4 °C overnight.
- Purify the resulting Ab-conjugated IONPs via magnetic column using PBS (pH = 7.4, final [Fe] = 3.5 mmol) to remove any unconjugated antibodies.
- Wash the magnetic column with PBS (1 mL). Attach the column to the magnetic plate, and add IONP solution.
- Wash the magnetic column with PBS (1 mL) again. Remove the magnetic column from magnetic plate. Add PBS to the column and collect the solution. Store at 4 °C
- Encapsulation of fluorescent 1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindocarbocyanine Perchlorate (DiI) dye into the polyacrylic acid (PAA) coating of the antibody-conjugated IONPs using a solvent diffusion method.
- To 4 mL of the IONPs, add dropwise 2.0 µL of DiI dye (2 mmol) in 100 µL of DMSO with continuous mixing at 1100 rpm.
- Dialyze the resulting solution (MWCO 6-8 KDa, 12 h) against PBS, creating a final MFnS solution with a final iron concentration of [Fe] = 2 mmol.
- Characterization of MFnS
- For spectrophotometric analysis, use a plate reader to detect encapsulated DiI dye with fluorescence emission at 595 nM.
- Dynamic Light Scattering
- Place a sample of the MFnS solution into a zetasizer to determine the average size and surface charge of the MFnS.
Note: Here, the average size and surface charge of functional MFnS were found to be 77.09 nm and -22.3 mV, respectively.
- Place a sample of the MFnS solution into a zetasizer to determine the average size and surface charge of the MFnS.
2. Bacterial Culturing and Stock Solution Preparation
- Bacterial Culturing
- Hydrate a freeze-dried pellet (E. coli O157:H7) in 1 mL of nutrient broth and then add an additional 5 mL of broth.
- Apply 100 µL of this solution to an agar plate and streak using a sterile inoculation loop, followed by incubation at 37 °C for 24 h.
- After 24 h, select an isolated colony from the agar plate and add it to a 15 mL of culture broth. Incubate at 37 °C for 4-6 h, while monitoring the optical density value (absorbance at 600 nm).
- Once an optical density value of 0.1 is obtained, stop culturing, and perform serial dilutions.
- Serial dilutions
- Add 900 µL of nutrient broth to eight sterile microcentrifuge tubes.
- Add 100 µL of the bacterial suspension to the first tube (dilution 10-1).
- Take 100 µL from the first tube and add it to the second, to obtain a dilution of 10-2. Continue this pattern for the remaining tubes, ending with a final dilution of 10-8.
- Transfer 100 µL from each tube to separate agar plates, and incubate for 24 h at 37 °C.
- The next day, count the colony forming units (CFUs) on each plate. Select the plate with ~100 CFUs. Use the corresponding dilutions for further experiments (100 µL = ~100 CFUs).
Note: Here it was the 10-6 dilution that resulted in ~100 CFUs.
3. Rapid Detection of E. coli O157:H7 using MFnS
- Spike various PBS solutions (1X, pH 7.4, 300 µL) with increasing amounts of the 10-6 bacterial stock, resulting in CFU ranges from 1-100. Add a consistent amount of MFnS (100 µL) to these solutions.
- Create one baseline solution that contains only PBS (1X, pH 7.4, 300 µL) and MFnS (100 µL).
- Incubate solutions for 30 min at 37 °C and then allow them to cool to room temperature.
- Transfer individual solutions to the magnetic relaxometer (0.47 Tesla) and record changes in relaxation times (T2) relative to the CFUs in each solution.
- To record changes in T2 values, begin by measuring the T2 value of the baseline solution that contains only PBS and MFnS.
- Place the solution in the magnetic relaxometer. Open the respective software, select the "T2 relaxation" setting and press "measure."
- Following the collection of the baseline T2, measure the T2 values of the additional solutions that were spiked with various concentrations of bacteria.
Note: The change in T2 is equivalent to baseline T2 subtracted from the spiked T2.
- To record changes in T2 values, begin by measuring the T2 value of the baseline solution that contains only PBS and MFnS.
- Remove samples from the magnetic relaxometer and centrifuge the tubes at 2880 x g for 10 min.
- Decant the supernatant and resuspend bacterial pellets in 100 µL of PBS (1X, pH 7.4).
- Add 80 µL of each resuspension to a 96-well plate and record fluorescence intensities at 595 nM.
Note: Testing can be repeated using different solvents, including lake water, milk and others as described in the results section
Representative Results
The mechanism of MFnS action is represented in Figure 1. The clustering of MFnS around the surface of bacterial contaminants interferes with the interactions between the magnetic cores of the MFnS and the surrounding hydrogen nuclei. As a result of this clustering, magnetic relaxation values increase. As the concentration of bacterial contaminants increases, clustering reduces, and the change in T2 values decreases. Therefore, the addition of a fluorescent modality is crucial. As the bacterial concentration increases, the strength of the fluorescent signal produced by MFnS increases, allowing for the sensitive detection of bacterial contaminants in both low and high concentration ranges.
Following the protocol,16 MFnS were synthesized and first tested in solutions of PBS (1X, pH 7.4). The initial modality used for detection, magnetic relaxation, was used to record changes in T2 values. In accordance with the protocol, the samples were then centrifuged, pellets were resuspended, and fluorescence data were collected. The corresponding ΔT2 and fluorescence emission values are presented in Figure 2. As shown, the ΔT2 value was greatest at lower concentrations of bacterial contamination, and reached saturation at roughly 20 CFUs. Fluorescence emission, on the other hand, became more accurate at >20 CFUs, displaying the importance of combining these two modalities. Following these primary assays, similar tests were conducted in complex media, including lake water and milk. This was done to ensure that the nanosensors are still effective in more complex media. The data collected from these assays are presented in Figure 3 and are very similar to the assays conducted in PBS. This verifies that these nanosensors remain effective in complex media.
After confirming that MFnS could detect the presence of bacterial contaminants in both simple and complex media, a number of tests were then conducted to determine the level of specificity maintained by the nanosensors. To this end, MFnS were incubated in solutions containing target bacteria, heat-killed target bacteria, non-target bacteria (non-pathogenic E. coli and S. typhimurium) and a mixture. As this was only a specificity assay, the collection of magnetic relaxation data alone was sufficient, and the corresponding data is presented in Figure 4. As can be seen, the MFnS bound only to live E. coli O157:H7, and not to the other heat-killed or non-target bacteria. This level of specificity is attributed to the conjugated targeting antibodies, and further confirms that MFnS will be effective at detecting specific pathogen contaminants, even when in the presence of non-pathogenic bacterial species.
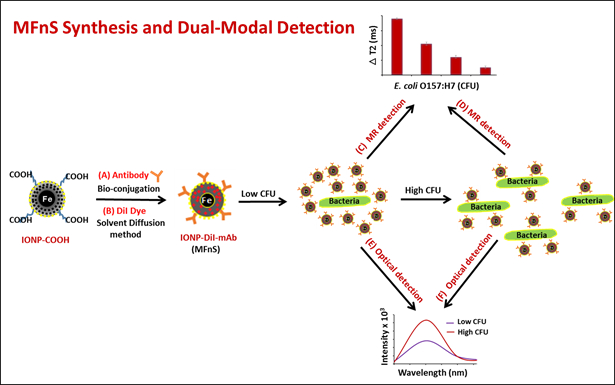
Figure 1: Magneto-Fluorescent Detection Mechanism. Schematic representation of MFnS synthesis and the dual-modal mechanism of bacterial detection. Following a short incubation period (30 min), MFnS are able to sensitively detect the target bacteria via a combination of magnetic resonance and fluorescence emission. Clustering of MFnS around the surface of bacterial contaminants causes changes in magnetic relaxation values, which are detectable by a magnetic relaxometer. In addition, fluorescent signals may be collected from the MFnS directly bound to bacterial contaminants. This provides a dual-modal detection platform capable of detection bacterial contaminants in both low and high concentration ranges.16 This figure has been modified with permission from.16
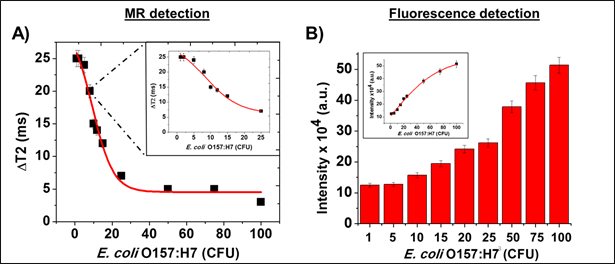
Figure 2: Proof of Concept. A) Magnetic relaxation data (ΔT2) was first collected from dilutions of E. coli O157:H7 (1-100 CFU) in PBS solvent (1X, pH = 7.4). MR detection of bacteria was highly sensitive at lower CFU counts (inset: ranging from 1-20 CFU). However, the ΔT2 readings became saturated at higher bacterial concentrations (>20 CFU), indicating that MR is more valuable for the detection and quantification of early-stage bacterial contamination. B) Fluorescence emission data from the same spiked PBS solutions (inset: linearity plot). The results showed that the fluorescence detection method is more sensitive at higher CFU counts, while it is lacking in sensitivity for low CFU samples. Together, these data demonstrate the ability of both magnetic and fluorescent modalities to detect and quantify bacterial contamination in early- and late-stages of bacterial contamination.16 Average values of three measurements are depicted ± standard error. This figure has been modified with permission from.16
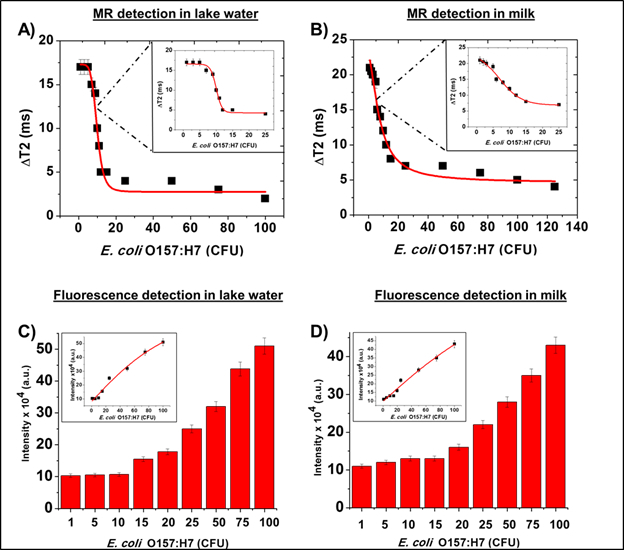
Figure 3: Bacterial Detection in Complex Media. Magnetic relaxation ΔT2 data collected from more complex media, including A) lake water and B) whole milk. Similar to the data collected from the previous PBS solutions, it was noted that detection of bacterial contaminants was more sensitive in the range of 1-20 CFU (insets). Corresponding fluorescence emission data was collected for the C) lake water and D) milk samples (insets: linearity plots), which showed higher sensitivity with increasing CFU counts. Once again, these data demonstrate the effectiveness with which each modality complements the other, and validate their ability to remain functional in complex media.16 Average values of three measurements are depicted ± standard error. This figure has been modified with permission from.16
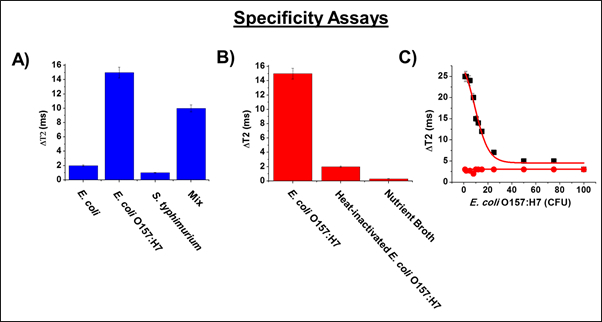
Figure 4: MFnS Specificity. The specificity of MFnS was tested using MR analysis in nutrient broth solutions containing A) a number of bacterial cross-contaminants and a mixture, and B) heat-inactivating E. coli O157:H7. As shown, it is clear that the nanosensors have little to no binding with non-targeted bacteria, and are still capable of detecting the target bacteria in the presence of other contaminants, ensuring that this form of screening would be viable for use in complex media that contain a number of non-pathogenic bacterial species. C) Further specificity testing was conducted by synthesizing MFnS with an isotype antibody (red circles: Anti-E. coli O111), which resulted in little to no binding compared to the O157:H7 antibody-conjugated MFnS (black squares). These data demonstrate that MFnS only react with viable target bacteria, further verifying their effectiveness as a point-of-care diagnostic platform.16 Average values of three measurements are depicted ± standard error. This figure has been modified with permission from.16
Discussion
This protocol has been designed to produce fully functional MFnS as simply as possible. However, there are many key points at which alteration of the protocol may be useful, depending on the user’s end goal. For example, the use of different antibodies would allow for targeting of many other pathogens. In addition, this protocol is not limited to the use of antibodies as targeting molecules. Any molecule which has specific binding affinity for target pathogens, such as host cell receptors, may also be used as targeting molecules. As long as the targeting molecule has a primary amine or alcohol group, or can be functionalized to have one, it may be conjugated to the surface of iron oxide nanoparticles according to the EDC/NHS conjugation chemistry listed in the protocol. Secondly, the amount of MFnS used in each detection solution may be varied. It is important to use enough so that the T2 value for the baseline solution (containing only solvent and nanosensor without target pathogen) is in the range of 100-250 ms. If the baseline value is below 100, then the changes in T2 values will be less sensitive, due to an overabundance of nanosensor. To raise the baseline T2 value, reduce the amount of nanosensor in solution. If, however, the baseline is above 250, it becomes overly sensitive, and false positives may become more likely. For these reasons, it is suggested to aim for a baseline solution T2 value between 100-250 ms.
Current limitations of this protocol, when considering its use as a point-of-care diagnostic, are the reliance on the current benchtop magnetic relaxometer. While effective and stable, this magnetic relaxation platform could be reduced in size to increase its portability. This would increase the ease with which this platform could be used in different environments, such as at water sources or in grocery markets, for effective on-site bacterial screening. This could also reduce the cost of this machine, making it more feasible for point-of-care pathogen detection.
Concerning disease diagnosis and pathogen detection in low-resource settings, this platform has the potential to be more effective than current pathogen diagnosis techniques used today, including PCR and ELISA. While these traditional techniques have proven to be largely efficient, they are highly complex, costly, and slow. As such, there is a need for an accurate diagnostic platform that could be developed for use in low-resource settings for point-of-care detection of pathogens and disease diagnosis. This platform is relatively simple to use, cost effective and rapid. Aside from initial start-up costs of the required machines (relaxometer and plate reader), the actual MFnS are relatively cheap and very stable. In addition, the collection of T2 values is simple and does not require a professional lab technician.
In the future, collaboration with chemical and biological engineers will be pursued to design a more portable, hand-held device that could be used for both the magnetic and fluorescent detection of bacterial contamination using MFnS. The development of a smaller device would help to further reduce the costs associated with this detection platform and increase its effectiveness in the field. Furthermore, this platform has the potential to be used to analyze the effectiveness of developing biocide agents. As shown in our data, our MFnS were only able to bind with live-targeted bacteria, and produced little to no signal when incubated with heat-killed bacteria. Based upon this, our platform could be used to test the effectiveness of biocide candidates, and we plan to explore this further in the future.
Critical steps in this protocol include the initial synthesis of MFnS and collection of T2 data. It is important to properly purify the MFnS to ensure that free-floating antibodies do not interfere with T2 data collection. In addition, the amount of MFnS placed in each test sample must be consistent, as changes in the MFnS concentration will alter T2 values, risking false positives/negatives.
In conclusion, MFnS are a novel step forward in the fight against bacterial contamination, and will be further developed as the targeting of additional pathogens is achieved and the design more portable machinery is pursued. The low cost and low-complexity associated with the use of MFnS makes them a viable candidate to be used for on-site detection of bacterial contamination. While the synthesis of these nanosensors requires careful preparation, their actual use is relatively simple, and gives them the potential to reduce foodborne and waterborne illnesses. With further development, the implementation of these nanosensors may be seen in the screening of water reservoirs and commercially produced foods at packaging sites, points of sale, and perhaps even homes.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work is supported by K-INBRE P20GM103418, Kansas Soybean Commission (KSC/PSU 1663), ACS PRF 56629-UNI7 and PSU polymer chemistry startup fund, all to SS. We thank the university videographer, Mr. Jacob Anselmi, for his outstanding work with the video. We also thank Mr. Roger Heckert and Mrs. Katha Heckert for their generous support for research.
Materials
| Ferrous Chloride Tetrahydrate | Fisher Scientific | I90-500 | |
| Ferric Chloride Hexahydrate | Fisher Scientific | I88-500 | |
| Ammonium Hydroxide | Fisher Scientific | A669S-500 | |
| Hydrochloric Acid | Fisher Scientific | A144S-500 | |
| Polyacryllic Acid | Sigma-Aldrich | 323667-100G | |
| EDC | Thermofisher Scientific | 22980 | |
| NHS | Fisher Scientific | AC157270250 | |
| Anti-E. coli O111 antibody | sera care | 5310-0352 | |
| Anti-E. coli O157:H7 antibody [P3C6] | Abcam | ab75244 | |
| DiI Stain | Fisher Scientific | D282 | |
| Nutrient Broth | Difco | 233000 | |
| Freeze-dried E. coli O157:H7 pellet | ATCC | 700728 | |
| Magnetic Relaxomteter | Bruker | mq20 | |
| Zetasizer | Malvern | NANO-ZS90 | |
| Plate Reader | Tecan | Infinite M200 PRO | |
| Magnetic Column | QuadroMACS | 130-090-976 | |
| Centrifuge | Eppendorf | 5804 Series | |
| Centrifuge (accuSpin Micro 17) | Fisher Scientific | 13-100-676 | |
| Floor Model Shaking Incubator | SHEL LAB | SSI5 | |
| Analytical Balance | Metler Toledo | ME104E | |
| Digital Vortex Mixer | Fisher Scientific | 02-215-370 | |
| Open-Air Rocking Shaker | Fisher Scientific | 02-217-765 |
References
- Law, J. W., Ab Mutalib, N. S., Chan, K. G., Lee, L. H. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 5, 770 (2014).
- Pandey, P. K., Kass, P. H., Soupir, M. L., Biswas, S., Singh, V. P. Contamination of water resources by pathogenic bacteria. AMB Express. 4, 51 (2014).
- Zhao, X., Lin, C. W., Wang, J., Oh, D. H. Advances in rapid detection methods for foodborne pathogens. J Microbiol Biotechnol. 24 (3), 297-312 (2014).
- Heithoff, D. M., et al. Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS Pathog. 8 (4), e1002647 (2012).
- Ishii, S., Sadowsky, M. J. Escherichia coli in the Environment: Implications for Water Quality and Human Health. Microbes Environ. 23 (2), 101-108 (2008).
- Chiou, C. S., Hsu, S. Y., Chiu, S. I., Wang, T. K., Chao, C. S. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J Clin Microbiol. 38 (12), 4621-4625 (2000).
- Zhou, G., et al. PCR methods for the rapid detection and identification of four pathogenic Legionella spp. and two Legionella pneumophila subspecies based on the gene amplification of gyrB. Appl Microbiol Biotechnol. 91 (3), 777-787 (2011).
- Chen, J., Tang, J., Liu, J., Cai, Z., Bai, X. Development and evaluation of a multiplex PCR for simultaneous detection of five foodborne pathogens. J Appl Microbiol. 112 (4), 823-830 (2012).
- LeBlanc, J. J., et al. Switching gears for an influenza pandemic: validation of a duplex reverse transcriptase PCR assay for simultaneous detection and confirmatory identification of pandemic (H1N1) 2009 influenza virus. J Clin Microbiol. 47 (12), 3805-3813 (2009).
- Mahony, J. B., Chong, S., Luinstra, K., Petrich, A., Smieja, M. Development of a novel bead-based multiplex PCR assay for combined subtyping and oseltamivir resistance genotyping (H275Y) of seasonal and pandemic H1N1 influenza A viruses. J Clin Virol. 49 (4), 277-282 (2010).
- Alvarez, M. M., et al. Specific recognition of influenza A/H1N1/2009 antibodies in human serum: a simple virus-free ELISA method. PLoS One. 5 (4), e10176 (2010).
- Huang, C. J., Dostalek, J., Sessitsch, A., Knoll, W. Long-range surface plasmon-enhanced fluorescence spectroscopy biosensor for ultrasensitive detection of E. coli O157:H7. Anal Chem. 83 (3), 674-677 (2011).
- Zhang, J., et al. Rapid visual detection of highly pathogenic Streptococcus suis serotype 2 isolates by use of loop-mediated isothermal amplification. J Clin Microbiol. 51 (10), 3250-3256 (2013).
- Han, F., Wang, F., Ge, B. Detecting potentially virulent Vibrio vulnificus strains in raw oysters by quantitative loop-mediated isothermal amplification. Appl Environ Microbiol. 77 (8), 2589-2595 (2011).
- Wang, J., et al. Rapid detection of pathogenic bacteria and screening of phage-derived peptides using microcantilevers. Anal Chem. 86 (3), 1671-1678 (2014).
- Banerjee, T., et al. Multiparametric Magneto-fluorescent Nanosensors for the Ultrasensitive Detection of Escherichia coli O157:H7. ACS Infect Dis. 2 (10), 667-673 (2016).
- Shelby, T., et al. Novel magnetic relaxation nanosensors: an unparalleled "spin" on influenza diagnosis. Nanoscale. 8, 19605-19613 (2016).
- Bui, M. P., Ahmed, S., Abbas, A. Single-Digit Pathogen and Attomolar Detection with the Naked Eye Using Liposome-Amplified Plasmonic Immunoassay. Nano Lett. 15 (9), 6239-6246 (2015).
- Farnleitner, A. H., et al. Rapid enzymatic detection of Escherichia coli contamination in polluted river water. Lett Appl Microbiol. 33 (3), 246-250 (2001).
- Huh, Y. S., Lowe, A. J., Strickland, A. D., Batt, C. A., Erickson, D. Surface-enhanced Raman scattering based ligase detection reaction. J Am Chem Soc. 131 (6), 2208-2213 (2009).
- Jayamohan, H., et al. Highly sensitive bacteria quantification using immunomagnetic separation and electrochemical detection of guanine-labeled secondary beads. Sensors (Basel). 15 (5), 12034-12052 (2015).
- Kaittanis, C., Naser, S. A., Perez, J. M. One-step, nanoparticle-mediated bacterial detection with magnetic relaxation. Nano Lett. 7 (2), 380-383 (2007).
- Meeker, D. G., et al. Synergistic Photothermal and Antibiotic Killing of Biofilm-Associated Staphylococcus aureus Using Targeted Antibiotic-Loaded Gold Nanoconstructs. ACS Infect Dis. 2 (4), 241-250 (2016).
- Wang, Y., Ye, Z., Si, C., Ying, Y. Subtractive inhibition assay for the detection of E. coli O157:H7 using surface plasmon resonance. Sensors (Basel). 11 (3), 2728-2739 (2011).
- Zhao, X., et al. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc Natl Acad Sci U S A. 101 (42), 15027-15032 (2004).

