An Immunofluorescence Technique for Viral Protein Localization in Infected Cells
Abstract
Source: Samrat, S. K. et al., Temporal Analysis of the Nuclear-to-cytoplasmic Translocation of a Herpes Simplex Virus 1 Protein by Immunofluorescent Confocal Microscopy. J. Vis. Exp. (2018)
This video demonstrates a method for monitoring infected cell protein 0, ICP0 trafficking in herpes simplex virus-1 infection. Post-de novo synthesis, ICP0 translocates to the nucleus, later moving to the cytoplasm during infection progression. Immunofluorescence microscopy reveals and analyzes the protein's subcellular localization, offering insights into its trafficking across infection phases.
Protocol
1. Cell Seeding and Virus Infection
- At 20–24 h before the virus infection, seed 5 x 104 of human embryonic lung (HEL) fibroblast cells or other cells to be examined on a 4-well 11 mm staggered slide in growth medium (Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS)). Incubate the cells at 37 °C with 5% carbon dioxide (CO2).
NOTE: Each well should have 70-80% cell confluency at the time of infection. - On the next day, remove the growth medium and infect the cells with viruses in Medium-199 at a range of 4–10 pfu/cell. Incubate virus-infected cells for 1 h at 37 °C. Keep shaking the slide during the incubation period.
- After the 1 h incubation, remove Medium-199 and supplement with growth medium.
NOTE: Drugs that interfere with different infection phases can be added at this step or prior to viral absorption. - Incubate the virus-infected cells at 37 °C with 5% CO2 for various lengths of infection period.
2. Fixation and Permeabilization
- At proper infection time, quickly wash the infected cells with phosphate-buffered saline (PBS) 3 times and add 200 μL of 4% paraformaldehyde freshly prepared in PBS. Incubate the cells with paraformaldehyde for 8–10 min at room temperature to fix the cells in each well.
- Aspirate paraformaldehyde and wash the wells with 200 μL of PBS for 3 times. Completely aspirate PBS after the 3rd wash.
- Add 100 μL of 0.2% non-ionic surfactant to each well to permeabilize the cells for 5–10 min.
- Aspirate the non-ionic surfactant and wash the wells with 200 μL of PBS for 3 times.
3. Immunofluorescent Staining
- Completely aspirate PBS and add 200 μL of blocking buffer (1% bovine serum albumin (BSA) and 5% horse serum in PBS) in each well and incubate at room temperature for 1 h or at 4 °C overnight.
- Add experimentally determined concentration of primary antibody against infected cell protein 0 (rabbit anti-ICP0 polyclonal antibody) in blocking buffer and incubate primary antibody at room temperature for 2 h or at 4 °C overnight.
- Wash with blocking buffer 3 times with 10 min incubation. Add Alexa 594-conjugated goat anti-rabbit secondary antibody (1:400 diluted in blocking buffer) and incubate the slides at room temperature for 1 h. Then wash the slides 3 times with blocking buffer at 10 min intervals.
- Finally wash the slide once with PBS to remove residual BSA and horse serum.
- Add one drop of antifade mounting medium with 4',6-diamidino-2-phenylindole (DAPI) to mount the slide and seal it with coverslip using transparent nail polish.
4. Confocal Imaging
- With a confocal microscope, set the wavelength at 590–650 nm for Alexa 594 and 410–520 nm for DAPI. Select image format at 1024 x 1024 and line average of 8 to acquire high resolution images.
- Analyze each well on the 4-well slide under a confocal microscope. Acquire representative cell images under the 100X objective, as shown in Figure 1 and Figure 2.
- For counting large numbers of cells, take images of consecutive fields under the 40X objective.
NOTE: It requires 5–10 images to accumulate over 200 infected cells from each time point of each infection. - In each experiment, take pictures with constant confocal parameters for all samples that need to be compared.
5. Analyzing Nuclear vs. Cytoplasmic Distribution
- Open project with the confocal application software. Select an image from which cells need to be tabulated for nuclear vs. cytoplasmic distribution of ICP0.
- Click the tab "Quantity" from top menu and select "sort ROIs" from tools menu.
- Draw a longitudinal line across the cell to be analyzed by selecting "Draw line" from top menu.
NOTE: Histogram will appear showing the fluorescence intensity along the line for both ICP0 and DAPI. In the histogram, blue line represents DAPI pixels and marks the boundary of the nucleus whereas the red line represents ICP0 pixels. - Based on background staining, set up a constant threshold for ICP0 intensity to analyze ICP0 subcellular distribution in each experiment.
- As exemplified in Figure 2, if the red signal on average is below the threshold in the nuclear region but is above the threshold beyond the blue boundary, categorize the red signal as predominantly located in the cytoplasm.
- If the red signal is above the threshold throughout the nucleus and beyond the boundary of blue signal, group the red signal as nucleus plus cytoplasmic localization.
- If the red signal is above the threshold in the nucleus but on average is below it outside the boundary of blue signal, group the red signal as nuclear localization.
- Tabulate more than 200 infected cells from each sample at different infection time and plot in bar graph to illustrate ICP0 movement according to time (Figure 3).
Representative Results
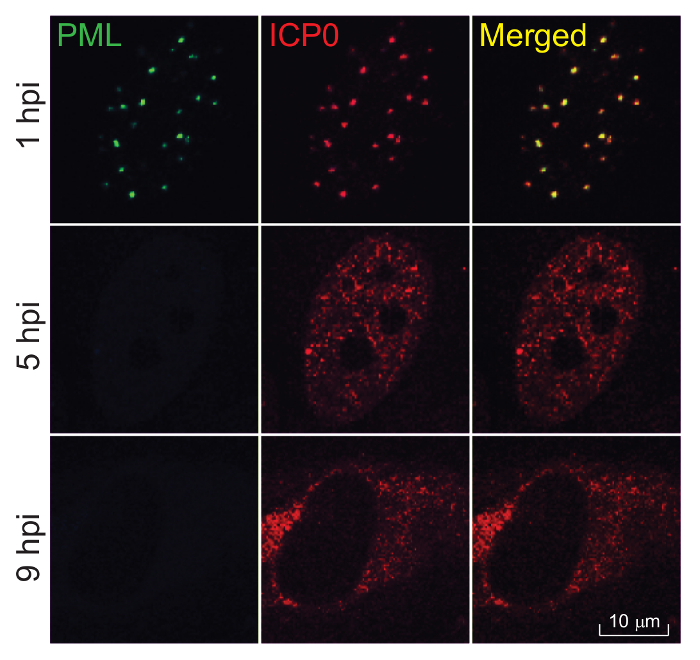
Figure 1: Dynamic trafficking of ICP0 during HSV-1 infection. HEL cells grown on 4-well slides were infected with prototype HSV-1 (strain F) at 10 pfu/cell. At 1, 5, and 9 h post infection (hpi), cells were fixed, permeabilized, and reacted to rabbit anti-ICP0 and mouse anti-PML primary antibodies, and then reacted to Alexa 594-conjugated anti-rabbit and Alexa 488-cojugated anti-mouse secondary antibodies for imaging under 100X objectives. Promyelocytic leukemia (PML) protein serves as a marker protein for ND10 nuclear bodies, which disappears at 5 and 9 hpi due to PML degradation in infection. Scale bar = 10 µm.
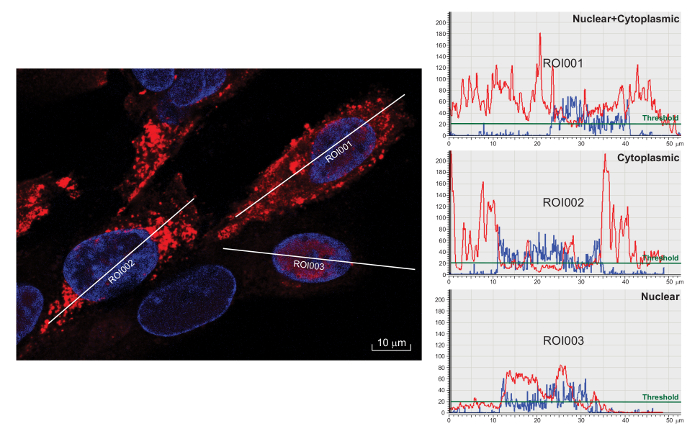
Figure 2: Analysis of ICP0 subcellular distribution. Left panel: With a confocal microscope, representative cells were enlarged to show the longitudinal line drawn across the cell that defines the region of interest (ROI). Right panel: Fluorescence pixel intensities in ROI were quantified for both ICP0 and DAPI in individual cells and illustrated as histograms by the confocal application software. An arbitrary threshold (green line) was set to reflect the background staining. Scale bar = 10 µm.
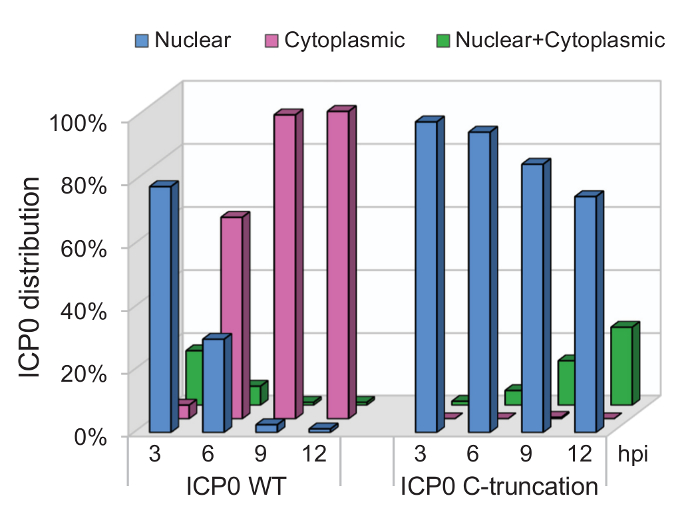
Figure 3: Percentage of subcellular distribution for wild-type and C-terminal truncated ICP0. HEL cells were infected by recombinant viruses containing wild-type ICP0 (ICP0 WT) or C-terminal truncated ICP0 (ICP0 C-truncation) at 4 pfu/cell. At indicated time points, cells were stained and analyzed as described above. Over 200 cells were tabulated for ICP0 location. Percentage of cells containing nuclear, cytoplasmic, or nuclear+cytoplasmic ICP0 were plotted with a spreadsheet computation software. This is an exemplary experiment to show that using this method, we have identified ICP0 C-terminus as a domain required for ICP0 nuclear-to-cytoplasmic translocation.
Divulgazioni
The authors have nothing to disclose.
Materials
| Cells and viruses | |||
| Human Embryonic Lung fibroblasts (HEL Cells) | Dr. Thomas E. Shenk (Princeton University) | HEL cells were grown in DMEM supplemented with 10% FBS | |
| HSV-1 viral Stock (Strain F) | Dr. Bernard Roizman Lab | ||
| Medium | |||
| Dulbecco's modified Eagle's medium (DMEM) | Invitrogen | 11965-092 | |
| Fetal Bovine Serum (FBS) | Sigma | F0926-500ml | |
| Medium-199 (10X) | Gibco | 11825-015 | |
| Reagents | |||
| 4- well 11 mm staggered slide | Cel-Line/Thermofisher Scientific | 30-149H-BLACK | |
| 16% Paraformaldehyde solution(w/v) Methanol free | Thermo Scientific | 28908 | |
| Triton X-100 | Fisher reagents | BP151-1C0 | |
| Bovine Serum Abumin (BSA) | Calbiochem | CAS 9048-46-8 | |
| Horse Serum | Sigma | H1270 | |
| Phosphate Buffered Saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, pH7.4) | Dr. Haidong Gu lab | ||
| NaCl | Fisher Bioreagent | BP358-212 | |
| KH2PO4 | Fisher Bioreagent | BP362-500 | |
| KCl | Fisher Scientific | BP366-500 | |
| Na2HPO4 | Fisher Bioreagent | BP332-500 | |
| Blocking buffer (PBS with 1% BSA and 5% Horse serum ) | Dr. Haidong Gu lab | ||
| Rabiit Anti-ICP0 antibody | Dr. Haidong Gu lab | ||
| PML (PG-M3)-Mouse monoclonal IgG | santa Cruz Biotechnology | SC-966 | |
| Alexa Fluor 594-goat anti-rabbit IgG | invitrogen | A11012 | |
| Alexa Fluor 488-goat anti-mouse IgG | invitrogen | A11001 | |
| Vectashield Mouting medium with DAPI | Vector laboratories | H-1200 | |
| Pasteur pipette | Fisher Brand | 13-678-20D | |
| Nail Polish | Sally Hansen | ||
| Equipment | |||
| Confocal Microscope | Leica SP8 | ||
| Confocal Software | Leica LAS X Application suite | ||
| Excel software | Microsoft Excel | ||
| HERAcell 150i CO2 incubator | Thermo Scientific | Order code 51026282 |

