Optimized Protocol for Efficient Transfection of Dendritic Cells without Cell Maturation
Summary
We present our optimized high-throughput nucleofection protocol as an efficient way of transfecting primary human monocyte-derived dendritic cells with either plasmid DNA or siRNA without causing cell maturation. We further provide evidence for successful siRNA silencing of targeted gene RIG-I at both the mRNA and protein levels.
Abstract
Dendritic cells (DCs) can be considered sentinels of the immune system which play a critical role in its initiation and response to infection1. Detection of pathogenic antigen by naïve DCs is through pattern recognition receptors (PRRs) which are able to recognize specific conserved structures referred to as pathogen-associated molecular patterns (PAMPS). Detection of PAMPs by DCs triggers an intracellular signaling cascade resulting in their activation and transformation to mature DCs. This process is typically characterized by production of type 1 interferon along with other proinflammatory cytokines, upregulation of cell surface markers such as MHCII and CD86 and migration of the mature DC to draining lymph nodes, where interaction with T cells initiates the adaptive immune response2,3. Thus, DCs link the innate and adaptive immune systems.
The ability to dissect the molecular networks underlying DC response to various pathogens is crucial to a better understanding of the regulation of these signaling pathways and their induced genes. It should also help facilitate the development of DC-based vaccines against infectious diseases and tumors. However, this line of research has been severely impeded by the difficulty of transfecting primary DCs4.
Virus transduction methods, such as the lentiviral system, are typically used, but carry many limitations such as complexity and bio-hazardous risk (with the associated costs)5,6,7,8. Additionally, the delivery of viral gene products increases the immunogenicity of those transduced DCs9,10,11,12. Electroporation has been used with mixed results13,14,15, but we are the first to report the use of a high-throughput transfection protocol and conclusively demonstrate its utility.
In this report we summarize an optimized commercial protocol for high-throughput transfection of human primary DCs, with limited cell toxicity and an absence of DC maturation16. Transfection efficiency (of GFP plasmid) and cell viability were more than 50% and 70% respectively. FACS analysis established the absence of increase in expression of the maturation markers CD86 and MHCII in transfected cells, while qRT-PCR demonstrated no upregulation of IFNβ. Using this electroporation protocol, we provide evidence for successful transfection of DCs with siRNA and effective knock down of targeted gene RIG-I, a key viral recognition receptor16,17, at both the mRNA and protein levels.
Protocol
1. Program the Amaxa 96 well shuttle Nucleofector
- Open a new parameter file.
- Select the number of wells you will be using for standard transfection by dragging the cursor over the 96 well plate diagram. Use a minimum of 3 wells to pool for each experimental sample.
- Input the program code: in part1 select ‘FF’ and in part2 select ‘168’ from the pull down menus
- From Solution box select ‘Monocyte, human’
- Under Control Option select ‘standard’.
- Click on Apply.
- To include a no-transfection control, select further wells from the diagram as required and then choose ‘No Program Control’ from Control Option and click on Apply.
- Select any remaining unused wells on the plate diagram and click on Undefine.
2. Prepare DCs for transfection
- All work should be done under sterile conditions in a cell culture hood where possible. Prepare the nucleofection solution by adding 96-well supplement to Human Monocyte 96-well Nucleofector Solution in the ratio of 450 to 2025. Mix and allow to warm to room temperature. You will need 20μl of nucleofection solution per well. Make an excess of 10% to allow for pipetting error.
- Place the number of nucleocuvette modules you need into the nucleocuvette plate in the correct orientation ie inserting the first one in to rows 1 and 2.
- Determine the number of DCs needed for your experiment calculating on 500,000 cells per well and pellet by centrifuging at 400g for 10min. Carefully remove the supernatant.
- Resuspend the cells in the nucleofection solution by gently pipetting up and down a few times.
- Divide the correct volume of resuspended cells into eppendorfs labeled for the particular treatment eg GLO and RIG-I.
- Add 0.25μg siRNA per 500,000 cells and mix by pipetting. Use non-targeting siRNA in your no-transfection control sample.
- Pipette 20μl of the above mixtures into the nucleocuvette modules, according to your experimental layout, ensuring the liquid is delivered to the bottom of the well.
- Cover the nucleocuvette plate with the lid and tap the plate on a hard surface a couple of times to help ensure removal of air bubbles.
3. Transfect DCs
- Insert plate into the Nucleofector 96-well shuttle tray, click on Upload and then start.
- Follow progress of transfection process on the lap top display; a black cross on a green background signifies a successful transfection in that well, whereas a black bar on a red background means it was unsuccessful.
- On completion of the transfection process, remove the plate and add 80μl of DC growth medium to each well using a multichannel pipette.
- Incubate plate for 10 min at 37°C and 5% CO2.
- Transfer the 100μl volume from the nucleocuvettes into matrix tubes containing 100μl of pre-warmed DC growth medium, maintaining the correct orientation.
- Remove and discard those tubes where transfection did not occur.
- Incubate at 37°C and 5% CO2 for 24h or other desired time interval.
4. Infect cells with NDV
- Remove matrix tubes from the incubator to the cell culture hood and pool tubes for each experimental sample into eppendorfs
- Pellet the cells gently by spinning in a desk top centrifuge for 5 min and remove supernatant.
- Resuspend the cells in serum-free growth medium containing NDV at an MOI of 1 and incubate at 37°C and 5% CO2 for 45 min, with eppendorfs loosely covered in a sterile fashion.
- Add 900μl of DC growth medium, and re-incubate for 8-10 h.
5. Harvest cells
- Pellet cells by spinning eppendorfs in a desk top centrifuge and remove supernatant.
- Harvest cells for RNA or protein extraction according to your protocol.
6. Representative results:
Using our optimized protocol we transfected DCs with RIGI-targeting siRNA for 24 h and then infected the cells with NDV (a paramyxovirus detected by RIG-I) to stimulate the interferon response pathway. By qRT-PCR analysis we demonstrated knock down of the gene by 75% at the transcription level. We also observed a similar reduction in the expression of IFNβ, which is a downstream effector of RIG-I in the IFN signaling cascade. Furthermore, we observed that the expression of IFNβ in non-infected, control-transfected cells was not detectable while that of MxA, an IFNβ downstream response gene, was minimal (Figure 1A).
A second transfection of DCs with RIGI-targeting siRNA was performed using enough cells to include Western blot analysis. Where the RT-PCR results were similar to what we saw previously (62% and 66% knock down of RIG-I and IFNβ expression respectively) (Figure 1B), the Western blot probed for RIG-I revealed that the expression of this gene had been completely blocked (Figure 1C).
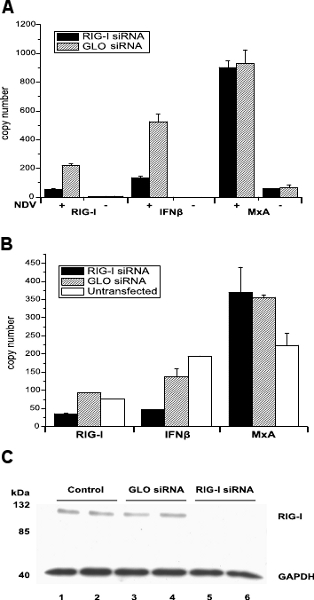
Figure 1. (A) MoDCs were transfected with either siRNA targeting RIG-I or nonspecific GLO siRNA and the effect on the expression of RIG-I, IFNβ and MxA determined by qRT-PCR. The transfection protocol used a monocyte-specific buffer and nucleoporation program FF168 (Lonza Walkersville Inc.) Following incubation for 24 h, transfected cells were either infected with NDV (+) or left uninfected (-). After a further incubation for 10 h, cells were harvested and RNA extracted Transcript levels represent the results of two replicate experiments. Taken from Bowles et al 16.
(B) MoDCs obtained from a different buffy coat as used for Figure 1A were again transfected with either RIG-I-targeting siRNA or nonspecific GLO siRNA using the same transfection protocol as above. An additional control using untransfected MoDCs was incorporated into the experiment. Following a 24 h incubation all cells were infected with NDV and incubated for a further 10 h before being harvested for both RNA and protein extraction. Transcript levels of RIG-I, IFNβ and MxA as determined by qRT-PCR represent the results of two replicate experiments. Taken from Bowles et al 16.
(C) Lysates from cells described in Figure 1B above were analyzed by Western blot. Lanes 1 and 2, lysates from untransfected cells; lanes 3 and 4, lysates from GLO siRNA transfected cells; lanes 5 and 6, lysates from cells transfected with RIG-I-targeting siRNA. Samples were probed for RIG-I and also GAPDH (as a loading control) and were run in duplicate. Taken from Bowles et al 16.
Discussion
Efficient transfection of naïve primary dendritic cells is important for high throughput analysis and reverse engineering of cellular inflammatory pathways in this key cell mediating the innate-adaptive immune transition. However, most investigators find that these cells are difficult to transfect both efficiently and without the transfection procedure inducing cell maturation when using standard transfection techniques. We investigated whether these limitations could be overcome by high throughput protocol optimization using a simultaneous, independent 96-well commercial nuclear transfection system (Lonza).
Using fluorescent activated cell sorting (FACS), we measured a GFP-expressing plasmid as a marker for transfection efficiency and CD86 and MHCII immunoreactivity as a readout of cell maturation. A series of experiments evaluating buffers and electroporation programs ultimately identified conditions showing >50% transfection and an absence of increase in maturation markers.
To further investigate possible activation of DCs by the nucleofection process we analyzed the expression levels of IFNβ and its downstream response gene MxA at the mRNA level. MxA expression is exquisitely sensitive to IFNβ production and can be used as a bioassay to detect very low levels of the cytokine18. Through qRT-PCR analysis we saw no detectable IFNβ expression but did see some upregulation of MxA thus indicating above-baseline IFNβ induction. However this minimal upregulation of MxA in non-infected, control-transfected cells was negligible compared to that resulting from virus-mediated stimulation of the IFNβ signaling cascade (Figure 1A) and confirmed that we could successfully use our nucleoporation protocol to transfect DCs without activating them to any significant level.
The utility of our optimized protocol was confirmed by Western blot analysis. Transfection of DCs with siRNA targeting RIG-I caused a complete loss of detectable RIG-I protein (Figure 1C). We should note that for successful perturbation of other genes, the amount of siRNA used and length of transfection time may have to be optimized.
To our knowledge, this work allows for the first time the design of high-throughput loss-of-function studies in primary human DCs, thus solving a difficult impediment to research in this field. This should provide new opportunities for the study of DC signaling and may contribute to advancement in developing DC-based immunotherapies.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The project was supported by NIH NIAID Contract No. HHSN2662000500021C. We thank Ming Chen for his technical assistance.
Materials
| Equipment/Reagent | Company | Catalogue # | Comments |
|---|---|---|---|
| Amaxa Nucleofector 96-well Shuttle | Lonza | 108S0109 | Serial number |
| Amaxa Human Monocyte 96-well Nucleofector Kit | Lonza | VHPA-2007 | Contains the Human Monocyte 96-well Nucleofector Solution, the 96-well Supplement and the Nucleocuvettes and plates |
| RIG-I siRNA | Dharmacon | L-012511-00 | |
| GLO siRNA | Dharmacon | D-001600-01-20 | |
| RPMI 1640 | Invitrogen | 11875 | Supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin to make DC growth medium |
| DMEM | Invitrogen | 11965 | |
| L-glutamine | Invitrogen | 25030081 | |
| Penicillin/streptomycin | Invitrogen | 15070063 | |
| Fetal Calf Serum | HyClone | 3070.03 | |
| Dendritic Cells | New York Blood center | DCs are purified from buffy coats using a standard procedure |
Riferimenti
- Reis e Sousa, C. Activation of dendritic cells: translating innate into adaptive immunity. Curr. Opin. Immunol. 16, 21-25 (2004).
- Bancherau, J., Steinman, R. M. Dendritic cells and the control of immunity. Nature. 392, 245-252 (1998).
- Clark, G. J. The role of dendritic cells in the innate immune system. Microbes Infect. 2, 257-272 (2000).
- Hamm, A. Efficient transfection method for primary cells. Tissue Eng. 8, 235-245 (2002).
- Henderson, R. A. Human dendritic cells genetically engineered to express high levels of the human epithelial tumor antigen mucin (MUC-1). Cancer Res. 56, 3763-3770 (1996).
- Reeves, M. E. Retroviral transduction of human dendritic cells with a tumor-associated antigen gene. Cancer Res. 56, 5672-5677 (1996).
- Aicher, . Successful retroviral mediated transduction of a reporter gene in human dendritic cells: feasibility of therapy with gene-modified antigen presenting cells. Exp. Hematol. 25, 39-44 (1997).
- Thomas, C. E. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 4, 346-358 (2003).
- Jooss, K. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J. Virol. 72, 4212-4223 (1998).
- Mitchell, D. A. RNA-transfected dendritic cells in cancer immunotherapy. J. Clin. Invest. 106, 1065-1069 (2000).
- Mincheff, M. In vivo transfection and/or cross-priming of dendritic cells following DNA and adenoviral immunizations for immunotherapy of cancer-changes in peripheral mononuclear subsets and intracellular IL-4 and IFN-gamma lymphokine profile. Crit. Rev. Oncol. Hematol. 39, 125-132 (2001).
- Roth, . Helper-dependent adenoviral vectors efficiently express transgenes in human dendritic cells but still stimulate antiviral immune responses. J. Immunol. 169, 4651-4656 (2002).
- Tendeloo, V. F. V. a. n. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 98, 49-56 (2001).
- Lenz, P. Nucleoporation of dendritic cells: efficient gene transfer by electroporation into human monocyte-derived dendritic cells. FEBS Lett. 538, 149-154 (2003).
- Prechtel, A. T. Small interfering RNA (siRNA) delivery into monocyte-derived dendritic cells by electroporation. J. Immunol. Methods. 311, 139-152 (2006).
- Bowles, R. Validation of efficient high-throughput plasmid and siRNA transfection of human monocyte-derived dendritic cells without cell maturation. J. Immunol. Methods. , .
- Kato, H. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 23, 19-28 (2005).
- Kato, H. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441, 101-105 (2006).
- Haller, O. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9, 1636-1643 (2007).

