Intracranial Implantation with Subsequent 3D In Vivo Bioluminescent Imaging of Murine Gliomas
Summary
Intracranial implantation of GL261 cells into C57BL/6 mice produces malignant gliomas that recapitulate many of the hallmarks of human glioblastoma multiforme. We used GL261 cells stably expressing luciferase to allow us to use in vivo imaging to follow tumor progression. The surgery and 3D in vivo imaging are demonstrated.
Abstract
The mouse glioma 261 (GL261) is recognized as an in vivo model system that recapitulates many of the features of human glioblastoma multiforme (GBM). The cell line was originally induced by intracranial injection of 3-methyl-cholantrene into a C57BL/6 syngeneic mouse strain 1; therefore, immunologically competent C57BL/6 mice can be used. While we use GL261, the following protocol can be used for the implantation and monitoring of any intracranial mouse tumor model. GL261 cells were engineered to stably express firefly luciferase (GL261-luc). We also created the brighter GL261-luc2 cell line by stable transfection of the luc2 gene expressed from the CMV promoter. C57BL/6-cBrd/cBrd/Cr mice (albino variant of C57BL/6) from the National Cancer Institute, Frederick, MD were used to eliminate the light attenuation caused by black skin and fur. With the use of albino C57BL/6 mice; in vivo imaging using the IVIS Spectrum in vivo imaging system is possible from the day of implantation (Caliper Life Sciences, Hopkinton, MA). The GL261-luc and GL261-luc2 cell lines showed the same in vivo behavior as the parental GL261 cells. Some of the shared histological features present in human GBMs and this mouse model include: tumor necrosis, pseudopalisades, neovascularization, invasion, hypercellularity, and inflammation 1.
Prior to implantation animals were anesthetized by an intraperitoneal injection of ketamine (50 mg/kg), xylazine (5 mg/kg) and buprenorphine (0.05 mg/kg), placed in a stereotactic apparatus and an incision was made with a scalpel over the cranial midline. A burrhole was made 0.1mm posterior to the bregma and 2.3mm to the right of the midline. A needle was inserted to a depth of 3mm and withdrawn 0.4mm to a depth of 2.6mm. Two μl of GL261-luc or GL261-luc2 cells (107 cells/ml) were infused over the course of 3 minutes. The burrhole was closed with bonewax and the incision was sutured.
Following stereotactic implantation the bioluminescent cells are detectable from the day of implantation and the tumor can be analyzed using the 3D image reconstruction feature of the IVIS Spectrum instrument. Animals receive a subcutaneous injection of 150μg luciferin /kg body weight 20 min prior to imaging. Tumor burden is quantified using mean tumor bioluminescence over time. Tumor-bearing mice were observed daily to assess morbidity and were euthanized when one or more of the following symptoms are present: lethargy, failure to ambulate, hunched posture, failure to groom, anorexia resulting in >10% loss of weight. Tumors were evident in all of the animals on necropsy.
Protocol
1. Cell Culture
- The GL26 cell line was obtained from the Division of Cancer Treatment and Diagnosis (DCTD) National Cancer Institute (NCI), Frederick, MD. To facilitate a quantitative measurement of tumor growth rate GL261 cells were made bioluminescent using the Lentiphos HT System (Clontech Laboratories, Inc., Mountain View, CA) with the Lenti-X HT Packaging Mix (Clontech Laboratories, Inc.) and the FUW-GL plasmid (a generous gift from the laboratory of J.B. Rubin, MD, PhD). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% tetracycline-free Fetal Calf Serum (FCS; Clontech Laboratories, Inc.). GL261 cells were also stably transfected with the gene encoding luc2 using the pGL4.51[luc2/CMV/Neo] vector (Promega Corp, Madison, WI) and FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN) following conditions specified by the manufacturer. The luc2 gene is a codon optimized version of firefly luciferase that provides a significantly higher light output than the standard luc gene. Stable transfectants were selected and maintained in DMEM media containing 10%FCS and 100 μg/ml Geneticin (G418, Invitrogen Corp., Carlsbad, CA).
- Prior to implantation the cultured cells are harvested by trypsinzation, washed once in DMEM without serum and resuspended in DMEM without serum at a concentration of 1 x 107 cells/ml.
2. Surgery Setup 2
- A sterile environment is maintained throughout the surgery, including all surgical instruments, supplies, gloves, drapes, etc..
- Ten week old C57BL/6-cBrd/cBrd/Cr (albino C57BL/6) mice are purchased from the National Cancer Institute at Frederick Animal Production Program and used at an average weight of 20 grams (NCI, Frederick, MD).
- Animals are anesthetized by intraperitoneal injection of xylazine (5 mg/kg), ketamine (50 mg/kg) and buprenorphine (0.05 mg/kg). Toe pinch is done to ensure that the animal is adequately anesthetized before the surgery is begun. Movement (even if slight) of any part of the animal is an indication of a reduction in the level of anesthesia. Animal is immediately given an additional 3.3 mg/kg xylazine and 26.6 mg/kg ketamine. Body temperature is maintained using a lamp and sterile dressings covering the body.
- Once the animal is properly anesthetized, they are placed on the cushioned bed of the stereotactic headframe (Model 900 Small Animal Stereotaxic, David Kopf Instruments)[Figure 1].
- A small amount (1/4 inch) of AKWA Tears Opthalmic Lubricant Ointment is applied over the corneas.
- The animal is secured in the stereotactic frame by opening the animal’s mouth (by placing your index finger and thumb around the animal’s jaw) and sliding the top front teeth into the indentation in the stereotactic frame. The clamp is tightened to secure the animal’s head into the headset making sure that the head is aligned and the eyes are centered, making sure however not to exert undue force on the animal’s head.
- The O2 hose is taped down near the animal’s nostrils. Oxygen flow rate is 0.5 l/min.
- The lower back and tail is taped down to the stereotactic bed being careful not to compromise respirations.
- The surgical incision site is shaved. Povidine-Iodine Swab Sticks are used to wash the area between the eyes going back to the area between the ears with iodine, making sure the iodine does not drip into the animal’s eyes.
- The cells are prepared for implantation just prior to the surgery and are periodically mixed to ensure they don’t settle.
- A 10 μl, 50 mm World Precision Instrument (WPI), Sarasota, FL, syringe with 26 gauge beveled needle is loaded with the cell inoculum. If the cells seem to be clumped together it may be necessary to reload the syringe.
- The 10 μl syringe is then placed into the UMP3-1 UltraMicroPump micro injector (WPI, Sarasota, FL).
3. Intracranial Implantation 2
- The skin incision is made using a size 15 scalpel blade and toothed forceps. A 10-15 mm incision is made longitudinally from between the animal’s eyes, moving towards the animal’s ears exposing the bregma (junction of the sagittal and coronal sutures at the top of the skull). Be sure to properly identify the bregma for it can be easily confused with the sinus area which is distal to the bregma [Figure 2].
- After the animal is sedated it receives an intra-incisional injection of 0.25% (2.5 mg/ml) bupivacaine.
- A burrhole is made 0.1 mm posterior to bregma and 2.3 mm to right of the midline by slowly twisting a 16 gauge 1½ inch needle by hand, applying little pressure to the needle while twisting until the skull is penetrated and the brain is exposed.
- The syringe needle is moved down into position using the microdrive stereotactic syringe holder until it just touches the brain’s surface. From this position, the needle is advanced into the brain to a depth of 3 mm and kept in place for 3 minutes.
- The needle is withdrawn .4 mm to a total depth of 2.6 mm below the surface of the brain, creating a small pocket where the cells are to be infused. It is optional at this point to take an X-ray image of the needle in the animal to ensure proper placement and depth.
- The cell suspension is infused over 3 minutes using the micro injector set to a volume of 2000 nL (2 μL) with an infusion rate of 667 nL/minute.
- The needle is left in place for 2 minutes to prevent leakage from the site of infusion.
- The needle is slowly withdrawn completely.
- The burrhole is filled with bone wax using a Penfield dissector.
- The incision is sutured using a 4-0 (1.5 Metric) vicryl suture making sure no large gaps are left in the skin. Vicryl is a suture material that dissolves, and therefore sutures do not need to be removed when the incision is healed.
- After surgery the animals are placed in a cage under a heating lamp that is set at the height required to warm the bottom surface of the cage to 30°C. When the animals are fully awake (as judged by normal movement in the cage) they are returned to group housing. Oral ibuprofen is added to their drinking water for 5 days post-operatively. 100 mg of children’s ibuprofen (100mg/5ml) is added to a standard 473 ml rodent water bottle. Animals are observed daily and imaged and weighed every 3 days. Two weeks post-implantation observation is increased to twice per day. Animals are euthanized when they show signs of declining health which includes hunched posture, reduced mobility and visible body weight loss (≥20%). These symptoms are a published response to the tumor and they reproducibly appear approximately 1 day prior to death due to the tumor.
4. In vivo Bioluminescence Imaging 3
- Start the [Living Image] software.
- The IVIS Imaging System is Initialized by clicking the [Initialize IVIS System] button on the bottom right side of the control panel.
- Select the [Luminescent] Imaging Mode on the top left side of the control panel.
- To determine the optimal time of imaging after luciferin injection a kinetic study is necessary [Figure 3]. This description is for firefly luciferase;
- Inject 10μl/g body weight of D-luciferin firefly (15mg/ml in PBS; Caliper Life Sciences Catalog XR-1001 or a similar product from another vendor) into the animal, as described below.
- Wait 3 minutes, and then anesthetize the mouse by placing it into the gas anesthesia chamber (2% Isoflurane gas in O2).
- Turn off the gas anesthesia to the chamber and open the anesthesia valve and the vacuum to the IVIS manifold. Immediately place the sedated animal on the temperature controlled imaging platform, making sure the mouse’s nostril is properly placed in the gas anesthesia manifold. The first image should be taken approximately 5 minutes after the luciferin injection. Up to 5 animals can be imaged at one time in the IVIS Spectrum instrument. If less than 5 animals are to be imaged, it is possible to plug the unused manifold(s) to conserve on isoflurane gas.
- Continue to take images every 3 minutes by creating a sequence for up to an hour to generate a kinetic curve for luciferin expression.
- Click on the [Sequence Setup] button in the control panel.
- The sequence editor appears.
- In the control panel, specify the settings for the first bioluminescent image in the sequence.
- We recommend starting with Medium Binning.
- We also recommend Auto-Exposure to determine the optimal exposure time.
- Select the [Delay] button in the sequence editor and specify a delay time of 3 minutes between each acquisition.
- Click [Add] in the sequence editor. Acquisition parameters are then added to the table.
- Repeat step 4 for each image in the sequence.
- Once the curve is established, the optimal imaging time can be determined by plotting the signal strength (intensity) versus time. Image animals at the time of highest in vivo photon count to get the strongest and most accurate signal.
- Early experiments used intraperitoneal (i.p.) injection of luciferin, however, i.p. injections occasionally resulted in intermittent results showing little to no bioluminescence in the tumor. We hypothesized that the occasional random lack of signal was due to the delivery of the luciferin to the bowel or other internal organs. We therefore began to use subcutaneous (s.c.) luciferin injections and saw greater imaging reproducibility [Figure 3].
- Twenty minutes after s.c. injection, the animals are anesthetized by placing them in a chamber with 2% Isoflurane gas in O2 until they are unresponsive.
- The anesthetized animal(s) are moved to the imaging chamber. Ophthalmic ointment should be used for the kinetic study because of the length of imaging. It is not needed for the other imaging procedures because they are short in duration.
- The image is acquired at medium binning with a 5 minute exposure time. The auto acquisition option may also be used.
- If the signal is saturated and/or faint at medium binning, binning or exposure time can be adjusted.
- The program files and subsequent image comments are saved in the user’s computer directory.
5. 3D Imaging 3
- Click on the [Imaging Wizard] tab in the [Sequence Setup] window of the control panel.
- Select the [Bioluminescence] imaging mode on the Imaging Wizard start screen and click [Next].
- In the “Bioluminescence – DLIT” window of the imaging wizard, select the [Firefly] reporter probe. The emission / excitation of the selected firefly source spectrum will appear with the six corresponding filter selections to be acquired.
- The last screen will appear with default choices that include Auto exposure acquisition parameters and a of Field of View C. Default settings work very well; however, these settings can be modified if necessary.
- Click [Next] and the sequence editor window will be populated with the sequence of six spectral regions at 20 nm wide filters (560 nm, 580 nm, 600nm, 620 nm, 640 nm, and 660nm). Press [Acquire Sequence]. The first filter (560 nm) will include a structured light pattern using a laser galvanometer to establish surface topography.
- Select the [Surface Topography] tab in the Tool Palette. Surface smoothing can be applied to account for any sharp angles created during the reconstruction process. The default low smoothing is recommended.
- Click [Create] and the tomography analysis box will appear. Draw a crop box that includes the entire animal and then click [Next].
- The threshold tool will appear as a purple mask over the selected region. The mask should automatically be set to match the photograph of the animal. If necessary, adjust the threshold of the mask to more appropriately fit the outline of the animal.
- Click [Finish] and the reconstructed mesh will appear. The reconstruction can then be saved in the results tab.
- Following the creation of the animal’s surface topography, proceed to the [DLIT 3D Reconstruction] drop down on the Tool Palette.
- Under the [Analyze] tab select all six wavelengths to perform the reconstruction. Deselect images that yielded saturated pixels or counts below 600. Leave settings in the [Parameters] tab as default.
- Under the [Properties] tab, “Muscle” should be listed as the default choice for Tissue Properties, and “Firefly” should be listed as the source spectrum.
- Click [Reconstruct] under the Analyze tab, and the 3D reconstruction of the animal’s surface and the corresponding reconstruction of the signal source should appear [Figure 4].
- To determine signal location and intensity, select the Voxels button on the 3D Tools tab of the Tool Palette.
- Draw a square around all displayed voxels and the total flux measurements is shown in the bottom of the volume tab.
- Click on [Center of Mass] to identify the signal location of the selected voxels. Coronal, sagittal and transaxial slices of the animal will appear and using the [Measurement Cursor Display] the distance from the animal’s surface to the voxel center can be measured.
- Please refer to the Living Image Software User’s Manual for further information on co-registration of organ atlases and other advanced features.
6. Data Analysis 3
- After the image is acquired and saved, access the program file by clicking the [Browse] button and selecting the file.
- The image information can be found under [View] → [Image Information].
- The intensity of the signal can be quantified by selecting the Region Of Interest (ROI) [ROI Tools] button. Make sure the image is analyzed under the [Photon] mode by selecting “Photon” on the drop-down list on the top left corner of the image control panel.
- Select the [Measurement ROI] button from the “Type” drop-down list. Select the ROI shape of interest; options include Circle, Square, and or Grid. Cover all areas of intensity on the acquired image.
- The ROI position is set by dragging the ROI shape selection to the region containing the bioluminescent signal.
- The signal intensity of the ROI is computed by clicking the [Measure] button. The ROI label displays the intensity. ROI’s can be managed and saved using the Living Image software.
7. Representative Results:
Successful cell implantation is evident when implanted cells are detectable using the IVIS spectrum on the day of surgery. Both GL261-luc and GL261-luc2 cells are detectable, however, the luc2 gene will provide a higher level of bioluminescence [Figure 5]. Images taken soon after implantation may have non-specific signaling on the animal’s paws and nose which should be disregarded as background. Signal located at the site of implantation is real and the signal will increase with time [Figure 6]. The decline in signal intensity on day 6 is reproducible and most likely due to loss of tumor take by some of the implanted cells. Quantitative measurements of tumor burden are reproducible in that they should rise steadily until the animal ultimately succumbs to the disease. However, growth curves will largely depend on the state of implanted cells, and or unavoidable minor variability in the implantation procedure. Our laboratory has elected to image implanted animals every three days.
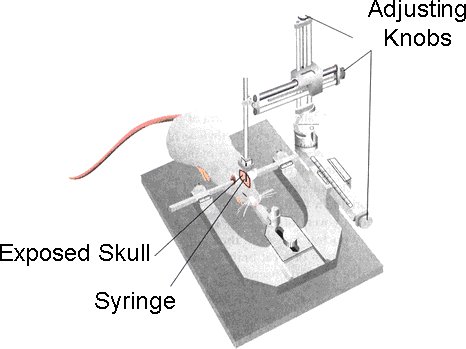
Figure 1. After the mouse is properly anesthetized, it is placed in the stereotactic frame. The mouse head is secured using the mouth clamp.
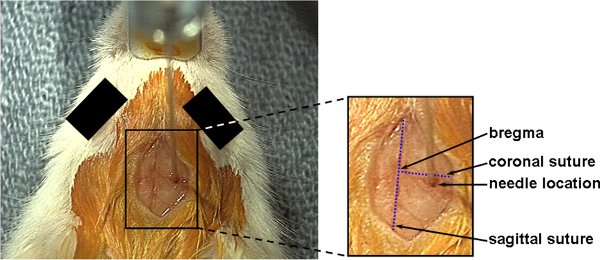
Figure 2. After the skin is opened the main anatomical landmarks are identified including include the bregma, the coronal and sagittal sutures. The burrhole is made 2.3mm to the right of the bregma by slowly twisting a 16 gauge 1½ inch needle with a small amount of pressure until the skull is penetrated and the brain is exposed.
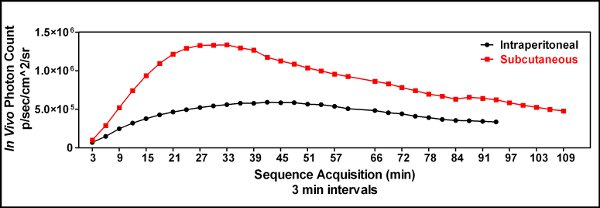
Figure 3. A kinetic comparison of subcutaneous ( ) versus intraperitoneal (
) versus intraperitoneal ( ) luciferin injection was performed to demonstrate the utility of a subcutaneous luciferin injection and to identify the optimal time to image following luciferin administration. Three minutes after the luciferin was injected the mouse was sedated, placed in the IVIS Spectrum instrument and imaged every 3 minutes for up to an hour and every 6 minutes after that to generate a kinetic curve of bioluminescence. This demonstrated that a subcutaneous route of luciferin administration was superior to an intraperitoneal injection in our hands, and that the optimal time to image the animals was approximately 25 minutes following the luciferin injection when GL261-luc cells were used.
) luciferin injection was performed to demonstrate the utility of a subcutaneous luciferin injection and to identify the optimal time to image following luciferin administration. Three minutes after the luciferin was injected the mouse was sedated, placed in the IVIS Spectrum instrument and imaged every 3 minutes for up to an hour and every 6 minutes after that to generate a kinetic curve of bioluminescence. This demonstrated that a subcutaneous route of luciferin administration was superior to an intraperitoneal injection in our hands, and that the optimal time to image the animals was approximately 25 minutes following the luciferin injection when GL261-luc cells were used.
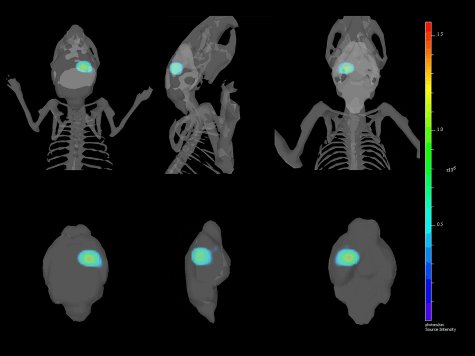
Figure 4. Multiple views of a 3-Dimensional reconstruction of the intracranial implantation of GL261-luc2 cells co-registered with the mouse skeleton and brain.
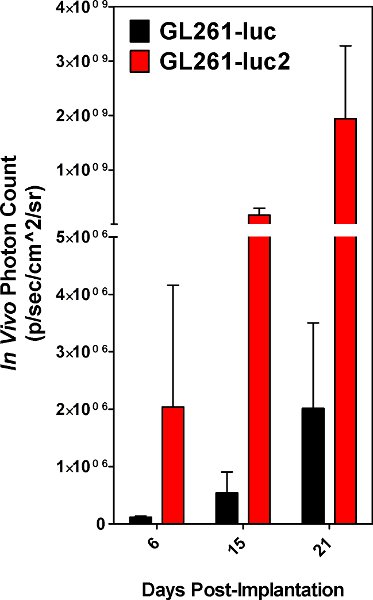
Figure 5. Photon counts obtained from tumors resulting from GL261-luc cells vs GL261-luc2 cells. Results are an average of 5 animals.
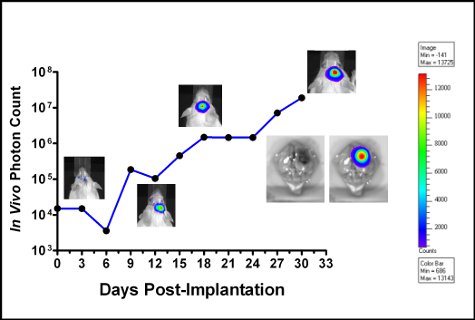
Figure 6. Graph of GL261-luc tumor cell growth in an albino C57BL/6 mouse. Bioluminescence was measured every 3 days and plotted as in vivo photon count versus days post-implantation. Photographs show bioluminescence at various time points. Coloration is an indication of bioluminescence (pixel intensity) which is relative to tumor cell number (color bar is shown to the right). After the animal succumbed to the disease the brain was dissected and luciferin was added topically to obtain the ex vivo image shown in the figure inset.
Discussion
The cell inoculum is infused at a depth of 2.6 mm from the surface of the brain after creating a 0.4 mm pocket. To ensure proper placement and depth of needle an X-ray can be taken using a C-Arm or similar X-ray image intensifying device; however, this is optional. Complications of the surgery may arise if the animal is not properly sedated, at which point the animal may move during cell infusion. This can cause leakage of cell mix or bleeding from needle track. Leakage of cells causes ectopic growth of tumor cells. It is also important not to puncture the ventricle which can be done if the burrhole is made medial to the 2.3 mm described in the protocol 4. Proper placement of the needle and cell spread was tested by infusing a mouse with 2 µl methylene blue dye and dissecting the brain tissue to check the location of the infused dye.
In this protocol we have used the IVIS Spectrum in vivo imaging system and the Living Image software (v 4.0) designed for use with this instrument. (Caliper Life Sciences). Any comparable in vivo imaging system and image analysis tools can be used to obtain similar results. These systems provide some advantages over traditional magnetic resonance imaging (MRI) to follow the growth of an experimental intracranial tumor. The most obvious is the relative costs of the 2 instruments – animal MRI machines are far more expensive and typically require the services of a skilled MRI technician. In vivo imaging such as what was described here can be done by the end user. The bioluminescence data is quantitative, while quantitation of MRI data is time-consuming and somewhat inexact. Furthermore, MRI images show edema and inflammation in addition to tumor cells and it can be difficult to separate tumor from treatment effect. For these reasons obtaining accurate volumetric measurements of growing tumor can be a challenge. Bioluminescence requires ATP, therefore only living tumor cells contribute to the tumor size data. Despite this, there are some advantages to MRI if a machine is available. Cells do not have to be labeled with a bioluminescent marker to be visualized by MRI. The ability to visualize peri-tumoral edema may be an advantage for some experimental protocols. A strength of both technologies is that using one does not preclude the use of the other, so data can be obtained on the growth of the tumor as well as the presence of peri-tumoral edema and inflammation from the same animal when both technologies are available to the researcher.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We would like to acknowledge Dr. Joshua B. Rubin for the generous gift of the plasmids for the lentivirus system as well as Mahil Rao for helpful suggestion on the preparation of the GL261-luc cells.
We thank Students Supporting Brain Tumor Research (SSBTR), The Barrow Neurological Foundation and The Wallace Foundation for their generous support.
Experiments on animals were performed in accordance with the guidelines and regulations set forth by the Institutional Animal Care and Use Committee of St. Joseph’s Hospital and Medical Center.
Materials
| Name of the reagent or supply | Company | Catalogue number | Comments |
| GL261-luc2 Bioware Ultra | Caliper Life Sciences | GL261-luc2 | |
| Dulbecco’s Modified Eagle Medium (DMEM) | Invitrogen | 10313039 | |
| Geneticin (G418) | Gibco (Invitrogen) | 11811-023 | |
| Fetal Calf Serum (FCS) | Invitrogen | 26140079 | |
| Phospate Buffered Saline (PBS) | Invitrogen | 70011044 | |
| C57BL/6-cBrd/cBrd/Cr Mice | NCI-Frederick | ||
| AKWA Tears Lubricant Opthalmic Ointment | Akorn Inc | 17478-062-35 | |
| Ketaset (ketamine hydrochloride) | Wyeth | 11570775 | |
| Sedazine (xylazine hydrochloride) | Wyeth | 10031894 | |
| Small Animal Stereotaxic Instrument | Kopf Instruments | 900 | |
| UltraMicroPump with SYS-Micro4 Controller | World Precision Instruments | UMP3-1 | If not available, it is possible to infuse manually |
| 10μl syringe with 26 gauge beveled needle | World Precision Instruments | SGE010RNS | |
| Adison Forceps | World Precision Instruments | 500092 | |
| Penfield Dissector | Codman | 65-1015 | |
| 16g 1½ Precision Glide Needle | Beckton, Dickinson and Company (BD) | 305198 | |
| Surgical Blade Handle | BD | 371030 | |
| Size 15 Blade | BD | 371315 | |
| 4-0 Vicryl Suture | Ethicon | VCP496G | |
| Bone Wax | Medline | DYNJBW25 | |
| Povidine-Iodine Swab Sticks | Medline | MD93901 | |
| D-Luciferin Potassium Salt | Caliper Life Sciences | 122796 | |
| Forane (Isoflurane) | Baxter | 1001936060 | |
| OPMI Pentero Microscope | Carl Zeiss, Inc. | Any surgical microscope will suffice | |
| Xenogen IVIS Spectrum with optional anesthesia system | Caliper Life Sciences |
Riferimenti
- Candolfi, M. Intracranial glioblastoma models in preclinical neuro-oncology: neuropathological characterization and tumor progression. J Neurooncol. 85, 133-148 (2007).
- Stafford, P., Abdelwahab, M. G., do, K. i. m., Preul, Y., Rho, M. C., M, J., Scheck, A. C. The ketogenic diet reverses gene expression patterns and redudes reactive oxygen species levels when used as an adjuvant therapy for glioma. Nutr Metab. 7, (2010).
- Caliper Life Sciences, Inc. . Living Image Software Version 4.0. VivoVision Systems. , (2010).
- Paxinos, G. . The Mouse Brain in Stereotaxic Coordinates. , (2001).
- Jouanneau, E., Poujol, D., Gulia, S., Le, M. I., Blay, J. Y., Belin, M. F. Dendritic cells are essential for priming but inefficient for boosting antitumour immune response in an orthotopic murine glioma model. Cancer Immunol Immunother. 55, 254-267 (2006).

