Therapeutic Gene Delivery and Transfection in Human Pancreatic Cancer Cells using Epidermal Growth Factor Receptor-targeted Gelatin Nanoparticles
Summary
Type B gelatin-based engineered nanovectors system (GENS) was developed for systemic gene delivery and transfection in the treatment of pancreatic cancer. By modification with epidermal growth factor receptor (EGFR) specific peptide on the surface of nanparticles, they could target on EGFR receptor and release plasmid under reducing environment, such as high intracellular glutathione concentrations.
Abstract
More than 32,000 patients are diagnosed with pancreatic cancer in the United States per year and the disease is associated with very high mortality 1. Urgent need exists to develop novel clinically-translatable therapeutic strategies that can improve on the dismal survival statistics of pancreatic cancer patients. Although gene therapy in cancer has shown a tremendous promise, the major challenge is in the development of safe and effective delivery system, which can lead to sustained transgene expression.
Gelatin is one of the most versatile natural biopolymer, widely used in food and pharmaceutical products. Previous studies from our laboratory have shown that type B gelatin could physical encapsulate DNA, which preserved the supercoiled structure of the plasmid and improved transfection efficiency upon intracellular delivery. By thiolation of gelatin, the sulfhydryl groups could be introduced into the polymer and would form disulfide bond within nanoparticles, which stabilizes the whole complex and once disulfide bond is broken due to the presence of glutathione in cytosol, payload would be released 2-5. Poly(ethylene glycol) (PEG)-modified GENS, when administered into the systemic circulation, provides long-circulation times and preferentially targets to the tumor mass due to the hyper-permeability of the neovasculature by the enhanced permeability and retention effect 6. Studies have shown over-expression of the epidermal growth factor receptor (EGFR) on Panc-1 human pancreatic adenocarcinoma cells 7. In order to actively target pancreatic cancer cell line, EGFR specific peptide was conjugated on the particle surface through a PEG spacer.8
Most anti-tumor gene therapies are focused on administration of the tumor suppressor genes, such as wild-type p53 (wt-p53), to restore the pro-apoptotic function in the cells 9. The p53 mechanism functions as a critical signaling pathway in cell growth, which regulates apoptosis, cell cycle arrest, metabolism and other processes 10. In pancreatic cancer, most cells have mutations in p53 protein, causing the loss of apoptotic activity. With the introduction of wt-p53, the apoptosis could be repaired and further triggers cell death in cancer cells 11.
Based on the above rationale, we have designed EGFR targeting peptide-modified thiolated gelatin nanoparticles for wt-p53 gene delivery and evaluated delivery efficiency and transfection in Panc-1 cells.
Protocol
1. Preparation of Plasmid DNA Encapsulated EGFR-Targeted Gelatin Nanoparticles
- Synthesis of thiolated gelatin
- Thiolated gelatin were synthesized as previous method 2-5 , by covalent conjugation with 2-iminothiolane on primary amino groups of type B gelatin. 1 gram of gelatin was dissolved in 100 ml deionized water and incubated with 20 mg 2-iminothiolane hydrochloride at room temperature for 15 hours.
- Unreacted reagent was removed by dialysis against 5 mM HCl solution, followed by 1 mM HCl solution for 3 hours each. Purified thiolated gelatin was freeze dried and stored at 4°C for further use.
- Preparation of DNA-containing nanoparticles
- 200 mg thiolated gelatin was dissolved in water and pH of solution was adjusted to 7 by addition of 0.2 M NaOH solution. 1mg DNA was added and gently mixed with gelatin solution.
- Chilled ethanol was added slowly into the mixture while stirring solution at high speed. Nanoparticles were formed when solvent composition changed to 75% hydro-alcoholic solution.
- Nanoparticles were further crosslinked by slow addition of 0.1 ml 8% (v/v) glyoxal solution. Unreacted reagents were quenched with 0.5 ml 0.2 M glycine solution.
- Nanoparticles were ultra-centrifuged at 16,000 rpm for 30 minutes. Pellets were washed with deionized water twice and purified nanoparticles were freeze-dried and stored at 4°C.
- Surface modification of nanoparticles
- Nanoparticles were suspended in 0.1 M phosphate buffer (pH 7.4) with concentration of 10 mg/ml and incubated with 2 times weight of methoxy-PEG-succinimidyl carboxy methyl ester (mPEG-SCM, MW 2,000 Da) or maleimide-PEG-succinimidyl carboxy methylester (MAL-PEG-SCM, MW 2,000 Da) for 2 hours at room temperature with slow stirring.
- PEGylated nanoparticles were collected with ultra-centrifugation at 16,000 rpm for 30 minutes. Pellets were washed with deionized water twice and purified nanoparticles were freeze-dried and stored at 4°C.
- MAL-PEG-SCM modified nanoparticles were suspended in 0.1M phosphate buffer (pH 6.5) with concentration of 10mg/ml and incubated with 10% weight of EGFR specific peptide (Y-H-W-Y-G-Y-T-P-Q-N-V-I-G-G-G-G-C) for 6 hours at room temperature with slow stirring.
- Peptide modified nanoparticles were collected with ultra-centrifugation at 16,000 rpm for 30 minutes. Pellets were washed with deionized water twice and purified nanoparticles were freeze-dried and stored at 4°C.
2. Characterization of EGFR-Targeted Nanoparticles
- Particle size and zeta potential measurement
Nanoparticles were suspended in water with concentration of 1mg/ml. Suspension was analyzed using Zetasizer Nano (Malvern Inc). Particle size analysis was carried out at a scattering angle of 90 degrees at 25°C. Zeta potential was measured at default parameters of dielectric constant, refractive index and viscosity of water at 25°C. - Scanning Electron Microscopy
Lyophilized nanoparticles were mounted on aluminum sample mount and sputter-coated with palladium to enhance conductivity and minimize buildup of charges. Samples were observed for surface morphology under a Hitachi 4800 field emission scanning electron microscope at 3 kV. - Electron spectroscope for chemical analysis (ESCA)
Freeze-dried formulation of control, PEGylated and peptide modified nanoparticles were analyzed by ESCA. It was performed at the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO), University of Washington (Seattle, WA).- Samples were placed in ultrahigh vacuum and exposed to low-energy x-ray beam, which induced an emission of secondary photoelectrons from the surface.
- By plotting number of detected electrons as a function of binding energy, observed spectrum peaks were assigned to each chemical components.
- High-resolution analysis of C1s spectra was performed to determine exact chemical composition from hydrocarbon (C-C or C-H at 285mV), ether (C-O) at 286.4mV), and carbonyl (C=O at 288.1 mV), and relative composition of each functionality was determined by area under curve.
- Stability of encapsulated plasmid
The stability of the encapsulated plasmid DNA was confirmed by running the extracted DNA on pre-cast gels. The nanoparticles were digested with 0.2 mg/ml protease containing PBS (30 min at 37°C) and 0.2 U/ml DNAse (10min at room temperature) separately, simultaneously or sequentially. Samples were then loaded onto 1.2% agarose gel (GP) (E-Gel, Invitrogen, CA) at a concentration of 100ng/well in a 18μL volume per well. Naked plasmid was loaded as control and gel was run at 75 V for 30 minutes. Kodak Digital X-ray Specimen (DXS) System was used to visualize bands with UV transluminescence. - Determination of plasmid loading
Plasmid encapsulated nanoparticles were suspended at 1mg/ml and digested with 0.2mg/ml protease at 37°C for 30 minutes. Solution was centrifuged at 13,000 rpm for 10 minutes and supernatant was collected and tested for plasmid concentration with Picogreen assay (Invitrogen). Encapsulation ratio was calculated by dividing the encapsulated plasmid concentration with the initial loading 0.5% (w/w).
3. In Vitro Transfection Studies in Panc-1 Pancreatic Cancer Cells
- Cell culture conditions
Panc-1 and Capan-1 pancreatic adenocarcinoma cell lines, SKOV3 ovarian adenocarcinoma cell line and NIH-3T3 murine fibroblast cell line were obtained from ATCC. Panc-1 and NIH-3T3 were grown with DMEM supplied with L-glutamine, Pen-strep and 10% fetal bovine serum at 37°C and 5% CO2, while Capan-1 required DMEM supplied 20% fetal bovine serum. SKOV3 was grown in RPMI-1640 supplied with 10% fetal bovine serum. - Western blot analysis for EGFR expression
- Cell lysates were collected from 2 million cells and analyzed for total protein concentration using BCA assay (Pierce). NIH-3T3 was used as negative control and SKOV3 was used as positive control for EGFR expression.
- 10 μg of total protein extract was run on pre-cast sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system at 135V for 90 minutes.
- Subsequently, gel was transferred onto PVDF membrane by iBlot Dry Blotting System (Invitrogen).
- Membrane was blocked with 5% non-fat milk in Tween-containing Tris buffer saline (TBS-t) for 1 hour at room temperature.
- Membrane was cut and incubated with 1:1,000 dilution of primary rabbit beta-actin antibodies and 1:1,000 dilution of primary rabbit EGFR antibodies separately overnight at 4°C.
- Membrane was then washed twice with TBS-t and incubated with 1:2,000 dilutions of secondary anti-rabbit horse-radish peroxidase-conjugated IgG in TBS-t for 1 hour at room temperature.
- After rinsing excess antibodies with TBS-t and water, 4 ml ECL substrate (Pierce, Rockford, IL, USA) was added and incubated with membranes for 5 minutes.
- Chemiluminescent bands were then visualized using Kodak Digital X-ray Specimen (DXS) System.
- Cell viability studies with different formulations
- Panc-1 cells were grown in 96-well plates at 10,000 cells per well in 200μL of supplemented DMEM overnight.
- Growth medium was replaced with serum free media containing different concentrations of nanoparticles with 0, 0.5, 1, 2, 4, 6 mg/ml. 1mg/ml PEI, a known cytotoxic cationic polymer, was used as positive control.
- Cells were treated with 200μL nanoparticles for 6 hours and then replaced with 20μL MTS reagent and 100μL complete culture media.
- After post incubation for 3 hours at 37°C in 5% CO2, the absorbance of formazan product was measured at 490nm with Biotek SynergyHT plate reader (Winooski, VT).
- The percent viability of cells was expressed as the ratio of absorbance of polymer treated cells relative to negative control (0mg/ml) multiplied by 100 and plotted as function of polymer concentrations.
- Cell trafficking studies
- Rhodamine B isothiocyanate (RBITC) was used to conjugate on thiolated gelatin by reaction with amine group. After dialysis and lyophilization, RBITC labeled thiolated gelatin was used for nanoparticles preparation.
- Before desolvation, 25μL PicoGreen was mixed with 1mg plasmid for 1 minute and labeled plasmids were added to gelatin solution. Different formulations were made following the previous method.
- Panc-1 cells were grown in 6-well plates containing glass cover-slips with 200,000 cells per well. After overnight growth, 2ml of labeled nanoparticles were treated into each well with concentration of 1mg/ml in serum free medium.
- After different time points, from 15 minutes to 6 hours, medium was replaced with culture medium containing 1μg/ml of Hoest 33342 (Invitrogen) for 15 minutes incubation at room temperature. 2ml of 4% paraformaldehyde solution was replaced into each well to fix cells. Cells were then washed with PBS twice.
- Coversilps were mounted on to glass slides. Laser scanning confocal fluorescence microscopy was used to take images of fixed cells.
- Qualitative determination of transfection efficiency by fluorescence microscopy with pEGFP-N1 encapsulated nanoparticles
- pEGFP-N1 plasmids were encapsulated into nanoparticles and suspended in serum free media with concentration equivalent to 10μg plasmids per ml for further treatments.
- Panc-1 cells were grown overnight in 6-well plates containing glass cover-slips with 200,000 cells per well. 2ml of pEGFP-N1 plasmids encapsulated nanoparticles were treated into each well. 20μg of plasmids were mixed with 20μl Lipofectin, cationic lipid transfection reagent, and it was used as positive control, while untreated cells were used as negative control.
- Cells were incubated with different formulations for 6 hours.
- Medium was replaced with culture medium and cells were post-transfected for 24, 48, 72 and 96 hours.
- After post-transfection, medium was replaced with culture medium containing 1μg/ml of Hoest 33342 (Invitrogen) and incubated with cells for 15 minutes at room temperature.
- Coverslips were mounted onto glass slides and expression of GFP in the cells was observed by fluorescence microscope. Differential interference contrast (DIC) and fluorescence images acquired using Olympus BX61 microscope and digital images were processed with Image J software.
- Quantitative determination of transfection efficiency by ELISA with pEGFP-N1 encapsulated nanoparticles
- pEGFP-N1 plasmids were encapsulated into nanoparticles and suspended in serum free media with concentration equivalent to 10μg plasmids per ml for further treatments.
- Panc-1 cells were grown overnight in 6-well plates with 200,000 cells per well. 2ml of pEGFP-N1 plasmids encapsulated nanoparticles were treated into each well. 20μg of plasmids were mixed with 20μl Lipofectin, cationic lipid transfection reagent, which was used as positive control and untreated cells were used as negative control.
- Cells were incubated with different formulations for 6 hours.
- Medium was replaced with culture medium and cells were post-incubated for 24, 48, 72 and 96 hours.
- After post-transfection, cell lysates were collected from each well and analyzed for total protein concentration using BCA assay (Pierce).
- well plate was coated with 100μl of 1:1000 dilutions of monoclonal anti-GFP antibodies in each well. After 2 hours incubation, plate was washed with PBS-0.5%(w/v) Tween-80 for 4 times.
- 300μl TBS blocking buffers were added into each well and incubated for 2 hours. Then plate was then washed with PBS-0.5% (w/v) Tween-80 for 4 times.
- 30μg of proteins of each group were added into the plate and incubated overnight at 4°. Then plate was then washed with PBS-0.5% (w/v) Tween-80 for 4 times.
- 100μl of 1:2400 dilutions of secondary anti-GFP antibodies relative to alkaline phosphatase were added to each well and incubated for 1 hour. Then plate was then washed with PBS-0.5% (w/v) Tween-80 for 4 times.
- 100μl alkaline phosphatase substrates were added to each well and incubated for 30 minutes to 1 hour. Plate was measured with BioTek Synergy HT plate reader for absorbance at 405nm.
- Expressed GFP concentration was reported as nanograms per milligrams of total protein.
- Qualitative determination of transfection efficiency by RT-PCR with wt-p53 plasmid encapsulated nanoparticles
- Wt-p53 plasmids were encapsulated into nanoparticles and suspended in serum free media with concentration equivalent to 10μg plasmid per ml for further treatments.
- Panc-1 cells were grown overnight in 6-well plates with 200,000 cells per well. 2ml of wt-p53 plasmids encapsulated nanoparticles were treated into each well. 20μg of plasmids were mixed with 20μl Lipofectin, cationic lipid transfection reagent, which was used as positive control and untreated cells were used as negative control.
- Cells were incubated with different formulations for 6 hours.
- Medium was replaced with culture medium and cells were post-incubated for 48 hours.
- mRNA were extracted from each well by using High Pure RNA isolation kit (Roche, Indianapolis, IN) and measured with Nanodrop 2000 (Thermo Scientific, Wilminton, DE).
- RT-PCR was done by using QIAGEN One-step RT-PCR kit (QIAGEN, Valencia, CA). Primers for p53, Bax, Bcl-2, Beta-actin, DR5, Apaf-1, PUMA, Survivin were synthesized by Eurofins MWG Operon (Huntsville, AL).
- PCR products were evaluated with gel electrophoresis and the pixels of cDNA bands were analyzed with ImageJ software.
- Quantitative determination of therapeutic efficiency with wt-p53 palsmid encapsulated nanoparticles
- Wt-p53 plasmid was encapsulated into nanoparticles and suspended in serum free media with concentration equivalent to 10μg plasmids per ml for further treatment.
- Panc-1 cells were grown overnight in 6-well plates with 200,000 cells per well. 2ml of wt-p53 plasmids encapsulated nanoparticles were treated into each well. 20μg of plasmids were mixed with 20μl Lipofectin, cationic lipid transfection reagent, which was used as positive control and untreated cells were used as negative control.
- Cells were incubated with different formulations for 6 hours.
- Medium was replaced with culture medium and cells were post-incubated for 24, 48, 72 and 96 hours.
- Chromatin Condensation/Membrane Permeability/Dead Cell Apoptosis Kit (Invitrogen, Carlsbad, CA) was used to label apoptotic cells, necrotic cells and live cells with different dyes.
- iCys Research Imaging Cytometer from CompuCyte (Westwood, MA) was used to analyze and compare apoptosis levels after treatment. Based on the fluorescence microscopic images, the intensities of all colors was recorded and plotted versus counts and the percentages for different populations were calculated.
- Compared to the negative control, which means there was no treatment for the cells, the apoptotic cells fold changes were calculated out and listed in the graph.
- 3.8.8 Apo-ONE Homogeneous Caspase-3/7 Assay kit (Promega, Madison, WI) was used to examine the pro-apoptotic activity after transfection of wt-p53 plasmid. 1mg/ml PEI was used as a negative control to eliminate all the apoptotic activity. After post-transfection, cells were treated with rhodamine 110, bis-(N-CBZL-aspartyl-L-glutamyl-L-valyl-L-aspartic acid amide; Z-DEVD-R110), which is a substrate of caspase 3/7, for up to 18 hours.
- Plate was measured with BioTek Synergy HT plate reader for fluorescence at 490/520 nm. Based on the intensity of green fluorescence, pro-apoptotic activity could be evaluated.
4. Representative Results
1. Synthesis and Chatacterization of EGFR Targeted Nanoparticles
EGFR targeting peptide-modified nanoparticles were synthesized as the scheme showed in figure 1. The nanoparticles prepared by desolvation were characterized for particle size and zeta potential. The average size and surface charge of the particles prepared from thiolated gelatins with different degrees of thiolation are listed in Table 1. The mean particle diameters of different nanoparticles were between 150-250 nm. Thiolated nanoparticles have smaller size compared to gelatin nanoparticles, might due to the disulfide bridge formation inside particles. With different surface modifications, sizes of nanoparticles have increased. The zeta potentials of different formulations were around -20 mV. With SEM analysis, the sizes, surface morphology and spherical shape of nanoparticles were observed and corresponding to Zetasizer result. DNA loading efficiencies in gelatin nanoparticles and thiolated gelatin nanoparticles were higher than 95% (Table 1).
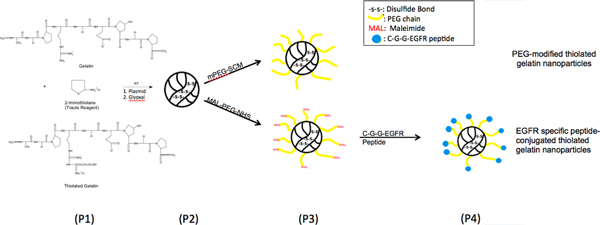
Figure 1. Chemical Reaction Scheme, illustrating surface modification of thiolated gelatin nanoparticles with epidermal growth factor receptor (EGFR) binding peptide through a poly(ethylene glycol) (PEG) spacer. Please click here to see a larger version of this figure.
Characterization of Nanoparticles
| Formulation | Nanoparticle Diameter (nm) | Zeta Potential (mV) | Plasmid DNA Loading Efficiency (%) |
| Gel NP | 151.4 ± 23.5 | -17.1 ± 5.23 | 95.6 ± 2.2 |
| SH-Gel NP | 132.6 ± 17.9 | -24.6 ± 5.16 | 97.0 ± 3.8 |
| SH-Gel-PEG | 179.0 ± 30.9 | -22.3 ± 9.50 | 95.8 ± 6.5 |
| SH-Gel PEG Peptide | 230.8 ± 41.5 | -18.1 ± 4.02 | 94.8 ± 5.1 |
Table 1. Particle size, surface charge, and plasmid DNA encapsulation efficiencies of control and EGFR-targeted gelatin and thiolated gelatin nanoparticles.
High-resolution C1S scans of electron spectroscopy for chemical analysis (ESCA) was used to analyze surface component of thiolated gelatin (SH-Gel NP), PEG-modified thiolated gelatin (SH-Gel PEG) and EGFR targeting peptide-modified thiolated gelatin nanoparticles (SH-Gel PEG Peptide). The results in Table 2 showed peak intensities of the C-H (hydrocarbon), C-O (ether), and C=O (carbonyl) groups at 285.0, 286.3, and 288.1 eV, respectively. The ether C-O signal has increased after PEG modification and decreased after peptide conjugation. While nitrogen composition has decreased after PEG modification and increased after peptide modification, which confirmed the presence of EGFR-targeting peptide on the nanoparticles. ESCA analysis has further confirmed PEG and peptide surface modification.
Electron Spectroscopy for Chemical Analysis of Nanoparticles Surface composition
| Formulation | C 1s (%) | O 1s (%) | N 1s (%) |
| SH-Gel NP | 59.3±0.8 | 22.9±0.5 | 12.9±0.1 |
| SH-Gel-PEG | 58.2±0.6 | 28.0±1.2 | 9.5±0.7 |
| SH-Gel PEG Peptide | 56.7±0.8 | 25.9±0.7 | 12.3±0.6 |
| Formulation | C-C (%) | C-O, N (%) | C=O (%) |
| SH-Gel NP | 51.5 | 26.6 | 21.9 |
| SH-Gel-PEG | 17.1 | 63.1 | 19.8 |
| SH-Gel PEG Peptide | 33.1 | 42.8 | 24.1 |
Table 2. C1S high-resolution scans of electron spectroscopy for chemical analysis (ESCA)
In order to examine the stability of encapsulated plasmid, nanoparticles were treated with protease or DNAse separately, simuntaneously or sequentially. After electrophoresis, the results in Figure 2 have shown that plasmid DNA encapsulated in all the nanoparticles are protected by nanoparticles and stable, comparable to naked plasmid DNA. These studied have shown that all these nanoparticles could encapsulate and preserve the plasmid structure after encapsulation.
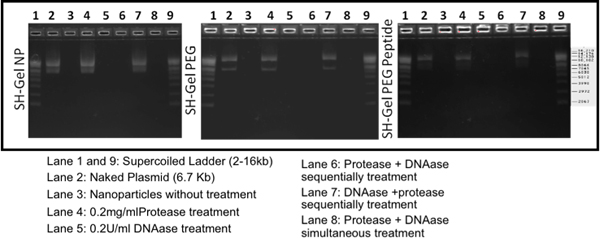
Figure 2. Stability of plasmid DNA encapsulated in thiolated gelatin, PEG-modified thiolated gelatin, and EGFR peptide-modified thiolated gelatin nanoparticles by agarose gel electrophoresis. The nanoparticles were treated with 0.2 mg/ml of protease to prove plasmid DNA encapsulation within the nanoparticle matrix
2. Baseline EGFR Expression in Pancreatic Cancer Cells
Two human pancreatic adenocarcinoma cell lines (Panc-1 and Capan-1) were analyzed by western blot for EGFR expression. Human ovarian adenocarcinoma (SKOV3) and murine fibroblast ((NIH-3T3) cells were chosen as positive and negative controls, respectively. Beta-actin was analyzed as protein loading control. Panc-1 cells have shown higher EGFR expression compared to Capan-1 and this cell line was then used for the following in vitro studies
3. Cytotoxicity of Control and Surface-Modified Thiolated Gelatin Nanoparticles
In order to evaluate the cellular interaction of nanoparticles, cytotoxicity assays were carried out after treatment with nanoparticles. Based on the results in Figure 3, both the control and the surface-modified nanoparticles were relatively safe and biocompatible in Panc-1 cells even at high concentrations, with comparison to PEI. The following studies were carried out with 1mg/ml nanoparticles.
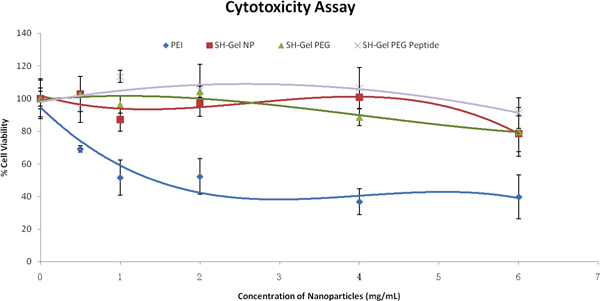
Figure 3. Percent cell viability as a function of nanoparticle formulation concentrations in Panc-1 cells as evaluated by tetrazolium dye (MTS) assay
4. Receptor Mediated Cell Uptake in Panc-1 Cells
To confirm surface accessibility of EGFR-targeting peptide and receptor-mediated endocytotic uptake of nanoparticles, a system was designed by labeling each component with different fluorescence for visualization of nanoparticles uptake and trafficking in cells. With this labeling system, plasmid DNA, nanoparticles and cell nucleus could be identified. Laser scanning confocal fluorescence microscopy was used to take images at different time points, from 15 minutes to 6 hours. By comparing the images of different formulations, peptide conjugated gelatin nanoparticles showed the fast uptake and plasmid release within 30 minutes. This result further proved that EGFR peptide-conjugated nanoparticles underwent facilitated endocytosis with quick interaction between the EGFR specific peptide and EGFR receptors on cell surface, which was much faster, compared to other nanoparticles, which underwent non-specific endocytosis.
Cell Trafficking Study
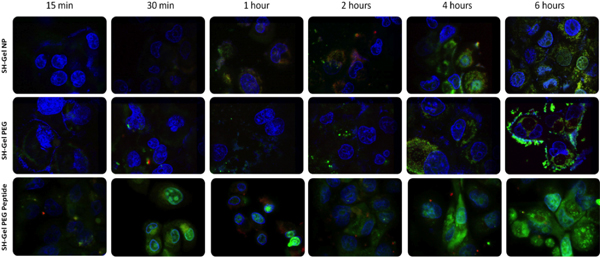
Figure 4. Confocal fluorescence microscopy analysis of DNA-encapsulated nanoparticle uptake and trafficking in Panc-1 cells. (red=rhodamine-labeled nanoparticles, green=PicoGreen-labeled plasmid DNA, and blue=DAPI-labeled nucleus). The laser power was 7 times less in last four figures of lower panel.
5. Qualitative and Quantitative In Vitro Transfection with Enhanced Green Fluorescent Protein
ELISA in Figure 5 and fluorescence microscopic analysis in Figure 6 were used to measure qualitative and quantitative GFP tranfection efficiency in Panc-1 cells upon administration of unmodified, PEG-modified and EGFR peptide-modified thiolated gelatin nanoparticles. Plasmids delivered by EGFR-targeted nanoparticles resulted in the highest level of GFP expression after 48 hours relative to other controls, including Lipofectin-complexed DNA.
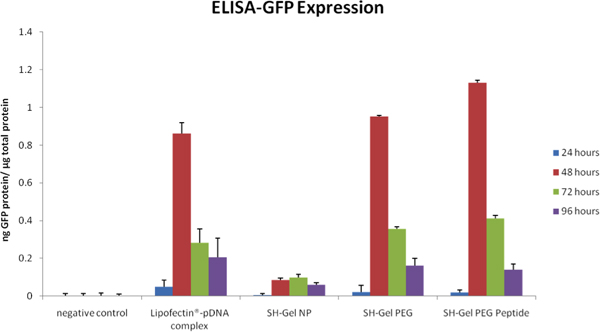
Figure 5. GFP expression analyzed by ELISA plotted as a function of time post-administration of plasmid DNA in control and EGFR-targeted nanoparticles.
Fluoresence Microscopic Analysis for GFP transfection
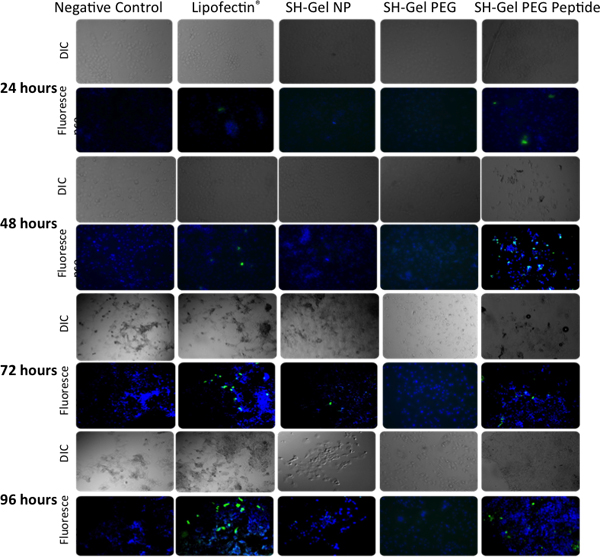
Figure 6. Qualitative analysis of green fluorescent protein expression in Panc-1 cells by epifluoresence microscopy after 24, 48, 72 and 96 hours post-transfection with EGFP-N1. Lipofectin-DNA complex was used as a positive control.
6. In Vitro Tranfection with Wild-Type p53 Plasmid in Panc-1 Cells
Wild-type p53 plasmids pORF-hp53, with EF-1α / HTLV hybrid promoter were extracted from E. coli and encapsulated into nanoparticles to study the apoptotic therapeutic effect. Panc-1 cells were treated with particles for 6 hours and post-transfected for additional 24, 48, 72, and 96 hours.
Since p53 could induce apoptosis in cells and in order to accomplish this function, many downstream transcription factors would be involved and directly regulated by expression of wt-p53. Among them, Bax, caspase-3, caspase-9, DR5, PUMA and Apaf-1 would be up-regulated by expression of p53 and while Bcl-2, survivin would be down-regulated. In order to examine the levels of these transcription factors, mRNA was extracted from Panc-1 cells after 48 hours post-transfection and used for RT-PCR. The products were evaluated with gel electrophoresis and bands were analyzed with ImageJ. Based on the results showed in Figure 7, survivin decreased significantly with the treatment of EGFR targeted thiolated gelatin nanoparticles compared to other treatments, no obvious change was seen in Bcl-2, Bax and expression of caspase-3, caspase-9, DR5, PUMA and Apaf-1increased with targeted nanoparticles treatment.
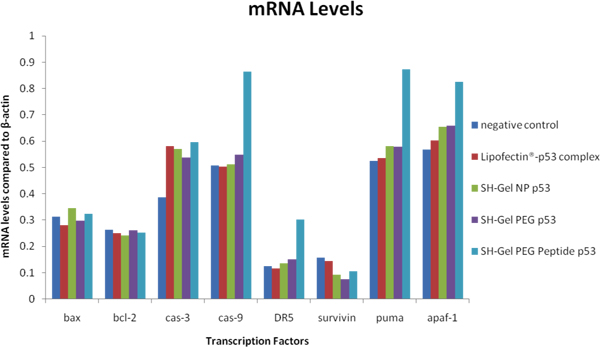
Figure 7. The mRNA levels of downstream factors of wt-p53 expression were compared by RT-PCR after 48 hours post-transfection.
After wt- p53 transfection, Chromatin Condensation/ Membrane Permeability/ Dead Cell Apoptosis kit was used to differentiate apoptotic cells, necrotic cells and live cells with different dyes. iCys Research Imaging Cytometer from CompuCyte (Westwood, MA) was used to analyze and compare apoptosis levels after treatment. Compared to the negative control, apoptotic cells fold changes were calculated out and listed in Figure 8. EGFR targeted thiolated gelatin nanopaticles have showed the highest apoptotic cell population after post-transfection. Analysis of caspase 3/7 activity also showed that EGFR-targeted nanoparticles had rapid internalization and highest level of apoptotic activity in Panc-1 cells.
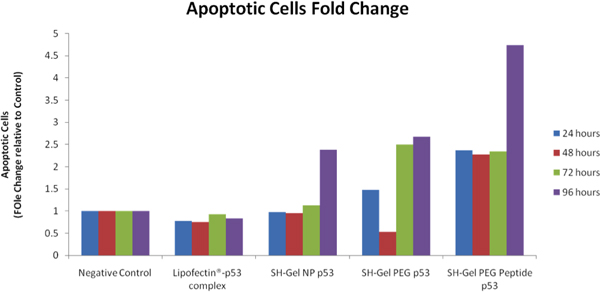
Figure 8. Cytometric analysis of pro-apoptotic activity in control wt-p53 transfected Panc-1 cells using iCys° Imaging Cytometer
Discussion
Control and EGFR targeted thiolated gelatin nanoparticles were prepared with efficient DNA encapsulation and stability. The particle sizes of all of these systems were in the range of 150-250 nm in diameter. Zeta potential has proved that this system is a slightly negative system. With SEM analysis, the sizes of nanoparticles were the same with Zetasizer result. ESCA analysis could confirm PEG and peptide surface modification.
Western blot analysis showed that Panc-1 cells had a high EGFR expression levels and this cell line was used for the in vitro studies. Both the control and the surface-modified nanoparticles were relatively less cytotoxic in Panc-1 cells as compared to PEI.
Cell trafficking studies showed rapid uptake and plasmid release of EGFR-targeted nanoparticles in Panc-1 cells. Delivery of reporter plasmids DNA expressing with EGFR-targeted nanoparticles resulted in highest levels of GFP expression relative to other controls, including Lipofectin-complexed DNA. With the same system, transfection with wt-p53 plasmid triggered the downstream apoptotic pathway and induced rapid apoptosis in Panc-1 cells.
These preliminary results suggest that the EGFR-targeted thiolated gelatin nanoparticles can serve as a safe and efficient DNA delivery system for gene therapy as a treatment for pancreatic cancer.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This study was supported by the National Cancer Institute’s Alliance in Nanotechnology for Cancer’s Center for Cancer Nanotechnology Excellence (CCNE) grant U54-CA151881.
Materials
| Name of the reagent | Company | Catalogue number | Comments |
| Type B gelatin, Bloom 225 | Sigma-Aldrich | G9391 | |
| 2-iminothiolane hydrochloride | Sigma-Aldrich | I6256 | |
| pEGFP-N1 plasmid | Elim Biopharm | N/A | |
| pORF-hp53 E. coli | Invivogen | porf-hp53 | |
| Glyoxal solution (40wt. % in H2O) | Sigma-Aldrich | 128465 | |
| Glycine | Sigma-Aldrich | 410225-250G | |
| QIA filter Plasmid Mega kit | Qiagen | 12281 | |
| Beckman LE 80K Ultracentrifuge | Beckman | N/A | |
| FreeZone 6 Liter Console Freeze Dry Systems | Labconco | 7753020 | |
| mPEG-SCM, MW 2,000 Da | Laysan | mPEG-SCM-2K-1g | |
| MAL-PEG-SCM, MW 2,000 Da | Jenkem Technology | A5001-1 | |
| Zetasizer Nano | Malvern | Zetasizer Nano ZS | |
| Hitachi 4800 field emission scanning electron microscope | Hitachi | S-4800 UHR FE-SEM | |
| Quant-iT PicoGreen dsDNA Reagent and Kits |
Invitrogen | P7589 | |
| Lipofectin Transfection Reagent | Invitrogen | 18292011 | |
| DMEM | Mediatech cellgro | 10 013 CM | |
| RPMI | Mediatech cellgro | 50 020 PB | |
| Pierce BCA Protein Assay Kit | Thermo Scientific | 23225 | |
| iBlot Dry Blotting System | Invitrogen | IB1001 | |
| XCell SureLock Mini-Cell and XCell II Blot Module Kit CE Mark | Invitrogen | EI0002 | |
| Pierce ECL Western Blotting Substrate | Thermo Scientific | 32109 | |
| Kodak Digital X-ray Specimen (DXS) System | Kodak | N/A | |
| CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS) | Promega | G3580 | |
| BioTek SynergyHT plate reader | BioTek | N/A | |
| Nanodrop 2000 | Thermo Scientific | N/A | |
| One-step RT-PCR kit | Qiagen | 210212 | |
| Chromatin Condensation/Membrane Permeability/Dead Cell Apoptosis Kit | Invitrogen | V23201 | |
| Apo-ONE Homogeneous Caspase-3/7 Assay kit | Promega | G7790 | |
| Hybaid PCR Sprint Thermal Cycler | Thermo Scientific | N/A | |
| EGF Receptor Antibody | Cell signaling | 2232 | |
| β-Actin Antibody | Cell signaling | 4967 | |
| Anti-rabbit IgG, HRP-linked Antibody | Cell signaling | 7074 | |
| Mouse Monoclonal GFP Antibody | Novus Biologicals | NB600-597 | |
| Goat Polyclonal GFP antibody (Alkaline Phosphatase) | Novus Biologicals | NB600-1502 | |
| Phosphatase Substrate Kit | Thermo Scientific | 37620 |
Riferimenti
- Vimalachandran, D. Genetics and prevention of pancreatic cancer. Cancer. Control. 11, 6-14 (2004).
- Kommareddy, S., Amiji, M. Preparation and evaluation of thiol-modified gelatin nanoparticles for intracellular DNA delivery in response to glutathione. Bioconjug. Chem. 16, 1423-1432 (2005).
- Kommareddy, S., Amiji, M. Poly(ethylene glycol)-modified thiolated gelatin nanoparticles for glutathione-responsive intracellular DNA delivery. Nanomedicine. 3, 32-42 (2007).
- Kommareddy, S., Amiji, M. Antiangiogenic gene therapy with systemically administered sFlt-1 plasmid DNA in engineered gelatin-based nanovectors. Cancer. Gene. Ther. 14, 488-498 (2007).
- Kommareddy, S., Amiji, M. Biodistribution and pharmacokinetic analysis of long-circulating thiolated gelatin nanoparticles following systemic administration in breast cancer-bearing mice. J. Pharm. Sci. 96, 397-407 (2007).
- Kaul, G., Amiji, M. Tumor-targeted gene delivery using poly(ethylene glycol)-modified gelatin nanoparticles: in vitro and in vivo studies. Pharm. Res. 22, 951-961 (2005).
- Bardeesy, N., DePinho, R. A. Pancreatic cancer biology and genetics. Nat. Rev. Cancer. 2, 897-909 (2002).
- Li, Z. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB. J. 19, 1978-1985 (2005).
- McCormick, F. Cancer gene therapy: fringe or cutting edge. Nat. Rev. Cancer. 1, 130-141 (2001).
- Green, D. R., Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature. 458, 1127-1130 (2009).
- Barton, C. M. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br. J. Cancer. 64, 1076-1082 (1991).

