Use of Artificial Sputum Medium to Test Antibiotic Efficacy Against Pseudomonas aeruginosa in Conditions More Relevant to the Cystic Fibrosis Lung
Summary
Current diagnostic antimicrobial susceptibility testing relies on the planktonic growth of isolates in nutrient rich, aerobic conditions. Here, we employ an alternative artificial sputum medium to study antimicrobial susceptibility of Pseudomonas aeruginosa biofilms under both aerobic and microaerophilic conditions more representative of the cystic fibrosis lung.
Abstract
There is growing concern about the relevance of in vitro antimicrobial susceptibility tests when applied to isolates of P. aeruginosa from cystic fibrosis (CF) patients. Existing methods rely on single or a few isolates grown aerobically and planktonically. Predetermined cut-offs are used to define whether the bacteria are sensitive or resistant to any given antibiotic1. However, during chronic lung infections in CF, P. aeruginosa populations exist in biofilms and there is evidence that the environment is largely microaerophilic2. The stark difference in conditions between bacteria in the lung and those during diagnostic testing has called into question the reliability and even relevance of these tests3.
Artificial sputum medium (ASM) is a culture medium containing the components of CF patient sputum, including amino acids, mucin and free DNA. P. aeruginosa growth in ASM mimics growth during CF infections, with the formation of self-aggregating biofilm structures and population divergence4,5,6. The aim of this study was to develop a microtitre-plate assay to study antimicrobial susceptibility of P. aeruginosa based on growth in ASM, which is applicable to both microaerophilic and aerobic conditions.
An ASM assay was developed in a microtitre plate format. P. aeruginosa biofilms were allowed to develop for 3 days prior to incubation with antimicrobial agents at different concentrations for 24 hours. After biofilm disruption, cell viability was measured by staining with resazurin. This assay was used to ascertain the sessile cell minimum inhibitory concentration (SMIC) of tobramycin for 15 different P. aeruginosa isolates under aerobic and microaerophilic conditions and SMIC values were compared to those obtained with standard broth growth. Whilst there was some evidence for increased MIC values for isolates grown in ASM when compared to their planktonic counterparts, the biggest differences were found with bacteria tested in microaerophilic conditions, which showed a much increased resistance up to a >128 fold, towards tobramycin in the ASM system when compared to assays carried out in aerobic conditions.
The lack of association between current susceptibility testing methods and clinical outcome has questioned the validity of current methods3. Several in vitro models have been used previously to study P. aeruginosa biofilms7, 8. However, these methods rely on surface attached biofilms, whereas the ASM biofilms resemble those observed in the CF lung9 . In addition, reduced oxygen concentration in the mucus has been shown to alter the behavior of P. aeruginosa2 and affect antibiotic susceptibility10. Therefore using ASM under microaerophilic conditions may provide a more realistic environment in which to study antimicrobial susceptibility.
Protocol
1. Preparation of Artificial Sputum Medium (ASM)
- Add 4 g DNA from fish sperm to 250 ml sterile water very slowly over a period of several hours. The DNA takes several hours to completely dissolve and can be stirred overnight at room temperature.
- Add 5 g mucin from porcine stomach (type II) slowly to 250 ml sterile water until the mucin has dissolved completely. The solution can be stirred overnight at 4 °C.
- Dissolve 0.25 g of each essential and non-essential L-amino acid, with the exception of L-tyrosine and L-cysteine, in 100 ml sterile water. Dissolve 0.25 g of L-cysteine in 25 ml of 0.5 M potassium hydroxide (Mr 56.11 g/mol) and 0.25 g of L-tyrosine in 25 ml sterile water.
- Dissolve 5.9 mg diethylenetriaminepentaacetic acid (DTPA), 5 g NaCl and 2.2 g of KCl in 100 ml of sterile water.
- Combine the DNA, Mucin, L-amino acids, DTPA, NaCl and KCl in a 1 litre bottle.
- Add 5 ml of egg yolk emulsion and fill to approximately 850 ml with sterile water.
- Adjust pH to 6.9 with 1 M Tris (pH 8.5; Mr 121.14) and bring the volume to 1 litre with sterile water.
- Sterilize the ASM by filtration using a Vacuubrand ME 2 diaphragm vacuum pump and Millipore Steritop filter units with a pore and neck size of 0.22 μm and 45 mm, respectively. Each Steritop filter unit can be re-used immediately up to three times; however, the filters need to be rinsed twice with sterile water before re-use. The filtration process is slow and can be performed over 2 days. Other versions of ASM have been developed that use the addition of antibiotics instead of filtration11 however, due to possible drug interactions, we do not recommend the method for this particular application.
- Unfiltered and filtered ASM should be stored at 4 °C in the dark. Using fresh ASM is recommended however, it can be kept under these conditions for a maximum of one month.
2. Determination of the Planktonic Sessile Cell Minimum Metabolic Inhibitory Concentration (PSMIC)
- To determine the minimum metabolic inhibitory concentration (PSMIC) values for 15 planktonically grown P. aeruginosa isolates, the microdilution method should be performed, as described in the guidelines of the British Society for Antimicrobial Chemotherapy BSAC12. The antibiotic of choice, in this case tobramycin sulphate, is serially diluted in Luria-Bertani (LB) medium in a 96-well microtitre plate to provide an appropriate range of antibiotic concentrations.
- Dilute overnight cultures of P. aeruginosa in LB to an OD600 of 0.05 (±0.01) and add 100 μl volumes to the wells of the 96-well microtitre plate containing 100 μl of the serially diluted antibiotic. In this case, the final concentrations of tobramycin sulphate ranged between 512 – 0.5 μg/ml. Eight replicates of each antibiotic concentration should be performed.
- Negative control wells for each isolate should be set up, in which no antibiotic is added. Also, eight wells should contain only LB for use as a blank during downstream analysis (Section 2.6).
- Incubate the 96-well microtitre plates for 1 – 2 days at 37 °C without shaking under aerobic or microaerophilic (5 % O2, 10 % CO2, and 85 % N2) conditions. Microaerophilic conditions are obtained using CampyGen gas generation packs in large anaerobic jars.
- Following incubation, bacterial growth is determined by measuring the absorbance of the bacterial culture in each well at a wavelength of 600 nm using a Fluostar Omega microplate reader and the MARS Data Analysis Software.
- Absorption from antibiotic-treated planktonic cultures (Aantibiotic treated planktonic cells) and absorption from the negative controls (Anegative control) should be corrected by subtraction of the background absorbance obtained from the wells containing LB only (Ablank). The percentage inhibition of viability is subsequently calculated as (mean Aantibiotic treated planktonic cells/mean Anegative control)x 100 %. The PSMIC90 is defined as the antibiotic concentration causing 90 % inhibition of planktonic bacterial growth.
- To determine bacterial viability after treatment with the antibiotic of choice, 10 μl of 0.02 % (v/v) resazurin (diluted in distilled water) is added to each well and the plates are incubated under aerobic conditions for 1 – 2 h at 37 °C, while shaking at 150 rpm. Viable cells will reduce the blue resazurin dye to the pink fluorescent resorufin form.
- Following incubation with resazurin, monitor the fluorescence of each well using an excitation wavelength of 540 nm and an emission wavelength of 590 nm in a Fluostar Omega microplate reader. The data should be analysed as described below.
3. Determination of Biofilm Sessile Cell Minimum Inhibitory Concentration (BSMIC)
- Overnight cultures of P. aeruginosa (in this case, 15 isolates of P. aeruginosa are used) should be diluted in LB to an OD600 of 0.05 (±0.01), then further diluted 1:100 in fresh ASM (total volume 1.8 ml).
- The diluted cultures (1.8 ml) should be added to each well of a 24-well tissue culture treated plate. Three wells should contain ASM only for use as a blank during downstream analysis (Section 3.9).
- Secure the 24-well plates with laboratory parafilm and incubate for 3 days under aerobic or microaerophilic conditions at 37 °C, while shaking at 75 rpm. Microaerophilic conditions should be obtained using CampyGen gas generation packs in large anaerobic jars.
- Dilute the antibiotic of choice, in this case tobramycin sulphate, to provide an appropriate concentration range in fresh ASM. In this instance, final concentrations ranged between 512 – 1 μg/ml. Add each concentration of the antibiotic, in volumes of 200 μl, to the appropriate wells of the 24-well plates. Four replicates of each antibiotic concentration should be performed. Biofilms not exposed to the antibiotic of choice were used as a negative control.
- Secure the 24-well plates with laboratory parafilm and incubate under aerobic or microaerophilic conditions for a further 24 h at 37 °C, while shaking at 75 rpm.
- After incubation in the presence of the antibiotic of choice, disrupt the bacterial biofilms using 100 μl of 100 mg/ml cellulase (diluted in 0.05 M citrate buffer [9.6 g/l Citrate.H20 in water and pH to 4.6 with NaOH]) and incubate the 24-well plates under aerobic conditions at 37 °C, while shaking at 150 rpm for 1 h. If required, biofilms could be further disrupted by manual pipetting at this stage.
- To determine the metabolic activities of the bacterial cells released from the disrupted biofilms, 100 μl of 0.02 % (v/v) resazurin (diluted in distilled water) should be added to each well of the 24-well plates and incubated for 1 – 2 h at 37 °C, while shaking at 150 rpm.
- Following incubation with resazurin, measure the fluorescence of each well using an excitation wavelength of 540 nm and an emission wavelength of 590 nm in a Fluostar Omega microplate reader and the MARS Data Analysis Software.
- Fluorescence from the antibiotic-treated biofilms (Fantibiotic-treated biofilms) and fluorescence from the negative controls (Fnegative control) should be corrected by subtraction of the background fluorescence obtained from the wells containing ASM only (Fblank). The percentage inhibition of viability is subsequently calculated as (mean of Fantibiotic treated biofilms/mean Fnegative control) x 100 %. The BSMIC90 is defined as the antibiotic concentration causing 90 % inhibition of metabolic activity.
4. Representative Results
ASM biofilm formation is possible in small (2 ml) volumes and the biofilms are fully formed within 3 days (Figure 1A). This can be demonstrated by rigorously pipetting the biofilm, which should be difficult to disrupt. The microcolonies are comparable to those grown in larger volumes4 (Figure 1B). Figure 2 shows major differences between cells grown planktonically and in a biofilm as detected by electron microscopic image analysis. Biofilm cultures clearly show considerable levels of extracellular matrix surrounding the cells and individual structures within the biofilm are difficult to identify.
Several studies suggest that the biofilm lifestyle can affect antimicrobial susceptibility13, 14. Our small scale ASM assay can be used to determine the BSMIC of multiple antibiotics for multiple isolates at the same time. The workflow of the assay is shown in Figure 3. The effect of antibiotics on bacterial cell viability can be measured using the resazurin assay. Antibiotics, in this case tobramycin, can be added to the established biofilm and incubated for 24 h. After this the biofilm is disrupted and resazurin is added.
Metabolically active cells can reduce the resazurin dye resulting in a colour change from blue (resazurin) to pink (resorufin) 15. Figure 4A shows an example assay in which P. aeruginosa was incubated with different concentrations of tobramycin before biofilm disruption and addition of resazurin in a microtitre plate. The blue non-fluorescent colour indicates non-viable cells, whereas viable cells reduce the dye to the pink fluorescent form, resorufin. The SMIC can then be calculated by converting fluorescence into percentage remaining bacterial viability. Figure 4B shows the change in % viability with increasing tobramycin concentration. 10% viability was chosen as a cut-off in order to calculate the SMIC90.
Under aerobic conditions, the tobramycin SMIC90 values are higher for cells grown as a biofilm than those of planktonic cultures. Table 1 shows the variation in PSMIC90 and BSMIC90 for all isolates tested. Table 2 shows that under aerobic conditions, a dramatic increase in resistance to tobramycin (2 to >32 fold increase in SMIC) was observed for most isolates when grown in ASM (biofilm mode) compared to LB (planktonic mode). In addition, biofilms grown under microaerophilic conditions exhibited an increased SMIC of between 2 and >128-fold when compared to biofilms grown under aerobic conditions.
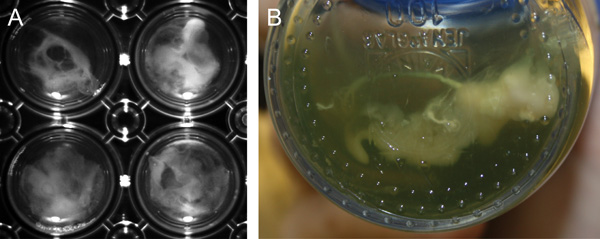
Figure 1. Biofilm formation of P. aeruginosa in ASM P. aeruginosa strain PAO1 forms macroscopically visible clumps (microcolonies) when grown in ASM. A, Biofilm formation in 30 ml ASM cultures (large-scale) after 7 days growth in screw cap glass Duran flasks . B, Biofilm formation in 2 ml ASM cultures (small-scale) after 3 days growth in 24-well polystyrene plates.
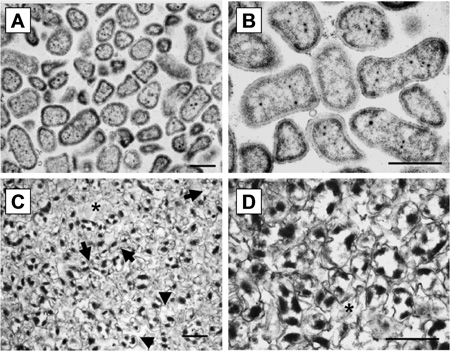
Figure 2. TEM micrographs of ASM biofilms A/C TEM micrograph (x;27,000) of PAO1 grown planktonically and in ASM, respectively, B/D TEM micrograph (x57,000) of PAO1grown planktonic and in ASM, respectively. Planktonically grown bacteria were cultivated overnight in LB broth. Biofilms were cultivated for 7 days in 30 ml ASM cultures. Black arrows refer to cells within the biofilm and stars refer to extracellular spaces. Scale bars = 1 μm.
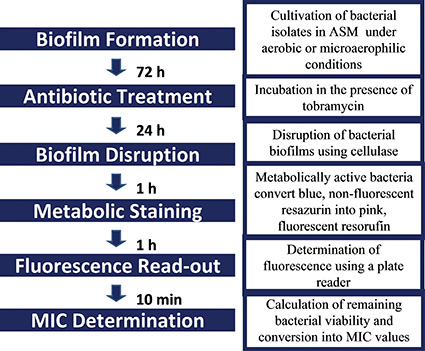
Figure 3. Workflow of the ASM biofilm antimicrobial susceptibility assay.
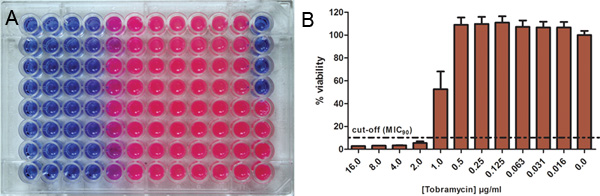
Figure 4. Use of resazurin for the determination of antibiotic susceptibilities Bacterial cells were incubated with different concentrations of the antibiotic and the remaining metabolic activity was determined using resazurin. A, The blue non-fluorescent oxidized form of resazurin indicates non-viable cells and is reduced by metabolic active cells to pink fluorescent resorufin. B, Fluorescence intensity is converted into percentage of remaining bacterial viability. 10% viability was chosen as cut-off in order to calculate the MSMIC90. Click here to view larger figure.
| Strains | PSMIC 90 (μg/ml) 1 | BSMIC 90 (μg/ml) 1 | ||
| Aerobic | Microaerophilic2 | Aerobic | Microaerophilic 2 | |
| PAO1 | 4 | 4 | 8 | >512 |
| Liverpool Epidemic Strain (LES) isolates | ||||
| LESB5821 | 8 | 64 | 64 | 128 |
| LES40022 | 32 | 128 | 8 | 256 |
| LESB25 | 16 | 32 | 256 | 512 |
| LESB55 | 16 | 64 | 64 | >512 |
| LESB64 | 16 | 64 | >512 | >512 |
| LES43122 | 4 | 8 | 32 | >512 |
| LESB49 | 16 | 64 | 64 | 256 |
| LES109 | 32 | 128 | 32 | >512 |
| Non-LES isolates | ||||
| 49461 | 16 | 32 | 16 | >512 |
| 59032 | 0.5 | 2 | 4 | >512 |
| 59073 | >512 | >512 | >512 | >512 |
| 59076 | 16 | 32 | 32 | >512 |
| 27 | 8 | 16 | 4 | >512 |
| 45 | 16 | 32 | 4 | >512 |
Table 1. Susceptibility of P. aeruginosa to tobramycin.
1For determination of PSMICs and BSMICs tobramycin was used in 2-fold serial dilutions
ranging from 512 – 0.5 μg/ml (n = 8 for each concentration) and 512 – 1 μg/ml (n = 4 for each
concentration), respectively;; PSMICs were determined using the standard
microdilution method1.
2Microaerophilic conditions were 5 % O2, 10 % CO2, and 85 % N2.
| Strain | PSMIC90/BSMIC90fold change1 | |||
| PSMICaerobic → PSMICmicroaerophilic |
BSMICaerobic → BSMICmicroaerophilic |
PSMICaerobic → BSMICaerobic |
PSMICmicroaerophilic → BSMICmicroaerophilic |
|
| PAO1 | 0 | >64 | 2 | 128 |
| LES isolates | ||||
| LESB58 | 8 | 2 | 8 | 2 |
| LES400 | 4 | 32 | 0.25 | 2 |
| LESB25 | 2 | 2 | 16 | 16 |
| LESB55 | 4 | >8 | 4 | >8 |
| LESB64 | 4 | ND | >32 | >8 |
| LES431 | 2 | >16 | 8 | >64 |
| LESB49 | 4 | 4 | 4 | 4 |
| LES109 | 4 | 16 | 0 | >4 |
| Non-LES isolates | ||||
| 49461 | 2 | >32 | 0 | >16 |
| 59032 | 4 | >128 | 8 | >256 |
| 59073 | ND | ND | ND | ND |
| 59076 | 2 | >16 | 2 | >16 |
| 27 | 2 | >128 | 0.5 | >32 |
| 45 | 2 | >128 | 0.25 | >16 |
Table 2. Fold change of PSMICs and BSMICs to tobramycin.
1ND, not determined; values in bold indicate SMIC fold changes >10.
Discussion
In this study we used a novel in vitro model based on ASM to replicate P. aeruginosa biofilm conditions within the CF lung4. The model was modified successfully for small-scale, high-throughput testing of antimicrobial agents.
The critical steps of this assay are:
- Consistent preparation of the ASM media and maintaining sterility. We have devoted long hours to optimising the way in which each component is added to achieve reproducible results every time. Filtration of the ASM is slow but is preferable to autoclaving, which may damage the mucin component. We do not advise the addition of antibiotics, as suggested by others 11 because this may impose considerable selective pressures, drive mutations, induce prophage lysis 16 and significantly alter the expression of multiple bacterial genes.
- The assay should be optimised according to the volume of ASM used. Shaking speeds are increased for smaller volumes and a shorter biofilm life-cycle is observed.
An obvious application of the small-scale ASM biofilm model is the more realistic determination of biofilm antimicrobial susceptibilities (BSMIC90). Anaerobic and microaerophilic niches are present in the CF lung and there is evidence that oxygen is limited deep within mature biofilms2, 17. Here we demonstrate that 10/14 clinical P. aeruginosa isolates from CF patient sputa exhibit a considerable (4 – ≥ 128 fold) decrease in sensitivity to tobramycin under microaerophilic conditions in ASM. The results of this study suggest that antibiotics, such as tobramycin, might be less effective against P. aeruginosa infections in the CF lung than indicated by conventional susceptibility testing methods. These results reflect previous studies on the antimicrobial susceptibility of biofilms 10. Small-scale ASM assays thus provide a simple high throughput platform for generating meaningful antibiotic susceptibility data to better inform therapeutic decisions. The assay is limited in the same way as conventional antibiotic susceptibility testing in that single colonies are picked for screening that may not be representative of the whole population. However, we believe that an approach (i) using non-surface attached biofilm growth and (ii) applicable to microaerophilic conditions, represents a clear alternative and a potential improvement to existing methods. We conclude that this assay is an appropriate model to study P. aeruginosa biofilm populations. Further testing in clinical settings would ascertain whether antibiotic susceptibilities based on biofilm-grown P. aeruginosa could lead to different antibiotic choices with potentially improved microbiological and clinical outcomes. Similar investigations using classic biofilm models have shown that BSMIC values lead to different recommendations for antibiotic treatment5,17.
In addition to testing for the effectiveness of anti-infective agents, the ASM system represents a cheap, simple and reproducible alternative to animal models for studies such as those aimed at understanding the diversification of P. aeruginosa populations. We have observed extensive heterogeneity in natural populations of P. aeruginosa recovered from CF patient sputa 18, 19. Similar phenotypic and genotypic diversification can be observed during growth in ASM4 (and our unpublished data), making it an attractive in vitro model of the CF lung conditions. The relative simplicity of the ASM model makes it easy to design long-term adaptation experiments aimed, for example, at monitoring the effects of antibiotics or other stresses on P. aeruginosa population divergence. In addition, other bacterial pathogens can be grown in ASM. For example, Fouhy et al. 2007 have used ASM to study biofilm formation by S. maltophillia20.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We acknowledge the support of the United Kingdom National Institute for Health Research, the Dr Hadwen Trust for Humane Research, the UK’s leading medical research charity funding exclusively non-animal research techniques to replace animal experiments, and the Wellcome Trust (Grant 089215). We also acknowledge Novartis Pharmaceuticals UK Ltd (unrestricted educational grant).
Materials
| Name of reagent | Company | Catalogue number |
| DNA from fish sperm | Sigma-Aldrich | 74782 |
| Mucin from porcine stomach, type II | Sigma-Aldrich | M2378 |
| L-Alanine | Acros Organics | 102830250 |
| L-Arginine | Sigma-Aldrich | A5006 |
| L(+)-Asparagine monohydrate | Acros Organics | 175271000 |
| L(+)-Aspartic acid | Acros Organics | 105041000 |
| L-Cysteine | Sigma-Adrich | 168149 |
| L(+)-Glutamic acid | Acros Organics | 156211000 |
| L-Glutamine | Sigma-Aldrich | G3126 |
| Glycine | Acros Organics | 220911000 |
| L-Histidine | Sigma-Adrich | H8000 |
| L-Isoleucine | Sigma-Aldrich | I2752 |
| L-Leucine | Sigma-Aldrich | L8000 |
| L(+)-Lysine monohydrochloride | Acros Organics | 125222500 |
| L-Methionine | Sigma-Aldrich | M9625 |
| L-Phenylalanine | Acros Organics | 130310250 |
| L-Proline | Sigma-Aldrich | P0380 |
| L-Serine | Acros Organics | 132660250 |
| L-Threonine | Acros Organics | 138930250 |
| L(-)-Tryptophan | Acros Organics | 140590250 |
| L-Tyrosine | Acros Organics | 140641000 |
| L-Valine | Sigma-Aldrich | V0500 |
| Diethylenetriaminepentaacetic acid | Sigma-Aldrich | 32318 |
| NaCl | Fisher Scientific | S/3160/60 |
| KCl | BDH | BDH0258 |
| KOH | BDH | BDH0262 |
| Egg yolk emulsion | Sigma-Aldrich | 17148 |
| ME 2 diaphragm vacuum pump | Vacuubrand | 696126 |
| Steritop filters (Pore size: 0.22 μm, Neck size: 45 mm) | Millipore | SCGPT10RE |
| Luria-Bertani medium | Sigma | L2897 |
| 96-well microtitre plates | Sarstedt | 82.1581 |
| 24-well tissue culture-treated plates | Iwaki | 3820-024 |
| CampyGen gas generation packs | Oxoid | CN0025 |
| Microaerophilic chamber | Oxoid | HP0011 |
| Tobramycin sulphate | Sigma-Aldrich | T1783 |
| Cellulase, from Aspergillus niger | Sigma-Aldrich | 22178 |
| Resazurin | Sigma-Aldrich | 199303 |
| Citrate.H20 | BDH | BDH0288 |
| Fluostar omega microplate reader | BMG-Labtech | SPECTROstar Omega |
Riferimenti
- Andrews, J. M. BSAC standardized disc susceptibility testing method (version 8). J. Antimicrob. Chemother. 64, 454-489 (2009).
- Worlitzsch, D. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317-325 (2002).
- Smith, A. L., Fiel, S. B., Mayer-Hamblett, N., Ramsey, B., Burns, J. L. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest. 123, 1495-1502 (2003).
- Sriramulu, D. D., Lunsdorf, H., Lam, J. S., Romling, U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667-676 (2005).
- Garbe, J. Characterization of JG024, a pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 10, 301 (2010).
- Naughton, S. Pseudomonas aeruginosa AES-1 exhibits increased virulence gene expression during chronic infection of cystic fibrosis lung. PLoS One. 6, e24526 (2011).
- Moskowitz, S. M., Foster, J. M., Emerson, J. C., Gibson, R. L., Burns, J. L. Use of Pseudomonas biofilm susceptibilities to assign simulated antibiotic regimens for cystic fibrosis airway infection. J. Antimicrob. Chemother. 56, 879-886 (2005).
- Field, T. R., White, A., Elborn, J. S., Tunney, M. M. Effect of oxygen limitation on the in vitro antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa grown planktonically and as biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 24, 677-687 (2005).
- Bjarnsholt, T. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547-558 (2009).
- Hill, D. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J. Clin. Microbiol. 43, 5085-5090 (2005).
- Fung, C. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 59, 1089-1100 (2010).
- Andrews, J. M. BSAC standardized disc susceptibility testing method (version 8). The Journal of antimicrobial chemotherapy. 64, 454-489 (2009).
- Rybtke, M. T. The implication of Pseudomonas aeruginosa biofilms in infections. Inflamm Allergy Drug Targets. 10, 141-157 (2011).
- Moreau-Marquis, S., Stanton, B. A., O’Toole, G. A. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21, 595-599 (2008).
- Ahmed, S. A., Gogal, R. M., Walsh, J. E. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J. Immunol. Methods. 170, 211-224 (1994).
- Fothergill, J. L. Effect of antibiotic treatment on bacteriophage production by a cystic fibrosis epidemic strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 426-428 (2011).
- Singh, P. K. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 407, 762-764 (2000).
- Mowat, E. Pseudomonas Aeruginosa Population Diversity and Turnover in Cystic Fibrosis Chronic Infections. Am. J. Respir. Crit. Care. Med. , (2011).
- Fothergill, J. L., Mowat, E., Ledson, M. J., Walshaw, M. J., Winstanley, C. Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J. Med. Microbiol. 59, 472-481 (2010).
- Fouhy, Y. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 189, 4964-4968 (2007).
- Winstanley, C. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 19, 12-23 (2009).
- Carter, M. E. A subtype of a Pseudomonas aeruginosa cystic fibrosis epidemic strain exhibits enhanced virulence in a murine model of acute respiratory infection. J. Infect. Dis. 202, 935-942 (2010).

