Analyzing Murine Schwann Cell Development Along Growing Axons
Summary
Here we describe a Schwann cell (SC) migration assay in which SCs are able to develop along extending axons.
Abstract
The development of peripheral nerves is an intriguing process. Neurons send out axons to innervate specific targets, which in humans are often more than 100 cm away from the soma of the neuron. Neuronal survival during development depends on target-derived growth factors but also on the support of Schwann cells (SCs). To this end SC ensheath axons from the region of the neuronal soma (or the transition from central to peripheral nervous system) to the synapse or neuromuscular junction. Schwann cells are derivatives of the neural crest and migrate as precursors along emerging axons until the entire axon is covered with SCs. This shows the importance of SC migration for the development of the peripheral nervous system and underlines the necessity to investigate this process. In order to analyze SC development, a setup is needed which next to the SCs also includes their physiological substrate for migration, the axon. Due to intrauterine development in vivo time-lapse imaging, however, is not feasible in placental vertebrates like mouse (mus musculus). To circumvent this, we adapted the superior cervical ganglion (SCG) explant technique. Upon treatment with nerve growth factor (NGF) SCG explants extend axons, followed by SC precursors migrating along the axons from the ganglion to the periphery. The beauty of this system is that the SC are derived from a pool of endogenous SC and that they migrate along their own physiological axons which are growing at the same time. This system is especially intriguing, because the SC development along axons can be analyzed by time-lapse imaging, opening further possibilities to gain insights into SC migration.
Protocol
1. Preparation of Collagen Gels
- Prepare a stock medium, containing 455 μl 10x MEM, 112 μl 7.5% NaHCO3, 50 μl glutamine and NaOH. The concentration and the amount of the NaOH depends on the rat- tail collagen preparation (see 1.2), the final volume of the stock medium being 1,000 μl.
- Prepare rat-tail collagen according to Ebendal (1). Store the Collagen solution at 4 °C. A degradation of the collagen solution could not be observed. After preparation of a new batch, estimate the NaOH concentration needed for the stock medium (see 1.1). Use the pH-indicator of the medium for titration of the amount and concentration of NaOH. 800 μl acidic rat-tail collagen have to turn a shade of red when mixed with 210 μl of the stock medium. If the concentration of NaOH is too low the collagen solution will turn yellow, due to the pH indicator of the medium. Add the collagen to the medium and mix without producing bubbles. Perform these steps on ice. The polymerization occurs within 2 hr after the collagen solution and the stock medium have been mixed and the new solution is incubated at 37 °C. Test this before starting an experiment. 1.3) Mix 800 μl of collagen solution with 210 μl of stock medium (see 1.2). Place 100 μl of this solution in wells of an 8 well chamber slide. Incubate the slides at 37 °C, 5% CO2 and humid conditions in a cell culture incubator. Prepare the collagen gels under a cell culture bench and sterile conditions. The collagen gelatinizes within 2 hr.
2. Dissection of Embryonic SCGs
- Harvest embryos of time pregnant mice (E16-18) from the uterus and keep them in PBS until further needed.
- Dissect one embryo from the amnion. Decapitate the embryo caudal of the clavicular level. Fix the head on a silicon bottom dish with “insect-needles”. The ventral side of the head has to face to you and you have to see the submandibular region. The fixation enables easier preparation.
- Remove the skin of the submandibular region left and right and with it the sub-cutaneous connective tissue.
- Remove the submandibular salivary glands from their location to clear the view onto the hyoid, the infrahyoidal muscles and the sternocleidomastoid muscle.
- Remove these muscles carefully to clear the view onto the larynx and the trachea. Remove the trachea and the larynx. The carotid arteries are then visible on both sides on the prevertebral muscles. The SCG is located in the bifurcation of the carotid arteries and has an oval shape. Carefully remove the ganglia and place them in PBS. Remove the ganglia gently from the dissection tools if they are sticking to them. It is beneficial to use forceps as well as a needle holder mounted with an “insect needle” for the SCG removal.
- Once in PBS, store the ganglia at room temperature in PBS until you dissected enough ganglia. Then clean the ganglia from blood vessels and connective tissue. Perform the SCG dissection and cleaning under a dissecting microscope placed under a laminar flow bench.
- Finally, place the explanted ganglia onto the gelatinized collagen gels with the help of syringe needles mounted on a syringe for easier handling.
- Incubate the SCG explants on the gelatinized collagen in a cell culture incubator at 37 °C, with 5 % CO2 and humid conditions.
3. Treatment of SCG Explants
- After 1-2 hr incubation, cover the SCG explant containing collagen with 100 μl Neurobasal cell culture medium containing B27 supplement, glutamine and antibiotics. Now the wells of the chamber slides contain a volume of 200 μl (100 μl gel and 100 μl medium). Add another 200 μl of the medium, however now containing NGF (60 ng/ml) to facilitate axonal growth. Accordingly the final NGF concentration is 30 ng/ml.
-
- To investigate the onset of SC migration, add additional factors directly at day in vitro 0 (DIV0). Dissolve the factors in the NGF containing medium in the double concentration needed (2).
- In order to analyze the impact on SCs, which already started migrating along axons, add additional factors at DIV3 until DIV4 (potential stop of the experiment). To this end, carefully remove 180 μl of the solution, at DIV3, and add 200 μl new medium containing NGF and the additional factors of interest at the double concentration (2).
4. Time-lapse Imaging
Use an inverted microscope for analyses. Various objectives can be used, defining the field of view and magnification. One important aspect, however, is the working distance of the used objective, as imaging of the SCG explants is performed through the glass slide and the collagen gel. The recording frame rate of 1/10-30 minutes showed good results (2). However, this aspect has to be adjusted to the scientific question. A normal CCD camera can be used for image acquisition. For time lapse imaging a cell culture incubation chamber has to be attached to the stage of the microscope. Incubate the tissue during imaging at 37 °C, with 5% CO2 and humid conditions. Start the incubation system one hour before the start of imaging. This allows the microscope parts (e.g. objective) including the chamber slide to adjust to the temperature and prevents temperature induced drifts. Define specific areas of analyses (within on explant and between different explants) with the help of the microscope software and a software-controlled motorized stage (multiposition setup) for simultaneous analyses of different applied conditions to the SCG explants. For time-lapse imaging wildtype tissue as well as tissue from transgenic animals can be used marking the SCs (s100b:GFP) (3). For imaging fluorophores a fluorescent light source has to be implemented in the micropscope setup. Use standard filters.
5. Quantification of SC migration distances
- At the end point (DIV4 for instance), fix the collagen gels with 4% PFA in the chamber slide for 3-4 hr. Consecutively wash the collagen gels 5 times for 10 min with PBS. Apply standard protocols for immunocytochemistry (2). If you are new to the preparation of SCGs, verify the identity of the ganglion as a sympathetic ganglion by immunohistochemistry against tyrosine hydroxylase (TH) a common marker for catecholaminergic cells. During immunohistochemistry (after the primary antibody), transfer the explant containing collagen gels to 24 well plates, which have a greater volume, which is beneficial for washing the gels.
- After immunohistochemistry, carefully place the collagen gels on glass slides. Carefully remove the remaining solution and let the collagen dry/shrink to two dimensions. This allows easy measurements of distances (2). After this, mount the samples.
- Finally, measure migration distances with the help of Fiji (NIH) software. No special plugins are needed to this end. To allow correct measurements in μm, check the correct imaging metadata and the settings under the Fiji, image and scale. Use Fiji also for quantification of cells.
Representative Results
Axonal growth is facilitated from SCG explants upon treatment with NGF (4) (movie scheme S1 Figure 1 scheme). This process is easily visible by any inverted microscope and can be followed by time-lapse imaging (movie S2). If a scientist is new to dissecting SCG from mouse embryos we strongly recommend a validation of the technique by a simple anti- tyrosine hydroxylase (TH) immunohistochemistry. TH is a common marker for catecholaminergic neurons (in this case sympathetic neurons) and does also label axons (Figure 2B). By this means axonal lengths can be analyzed in this system (2).
Importantly, after axonal growth has been induced, a wave of migrating cells can be observed (movie S2 and S3). Migrating cells were validated as S100 immuno positive SC (2). In addition a transgenic mouse line, in which GFP is under the control of the human s100b promoter fragment (3) can be used to directly analyze the labeled SC population (2) (movie S4 and S5). With the help of this transgenic line these experiments can also be performed with confocal- or light sheet microscopy (not performed here).
For analyses of SC during migration, we recommend an area of analysis with a low axonal density (Figure 1 close-up DIV3 and DIV4) and thereby a small population of SC per area of analysis. This facilitates best possibilities to see cell morphology and cellular behavior (movie S3).
SC migration distances can be measured at the end of an experiment (e.g. DIV4). To this end DAPI nuclear labeling can be performed on fixed tissue and distances from the leading SC nuclei to the border of the explant can be measured with the help of Fiji (NIH software) (Figure 2 scheme and DAPI labeling) (2). In addition also SC proliferation or SC death can be analyzed. To this end immunohistochemistry for pHH3 or for activated Caspase 3 can be performed for example, identifying mitotic or apoptotic cells respectively (2).
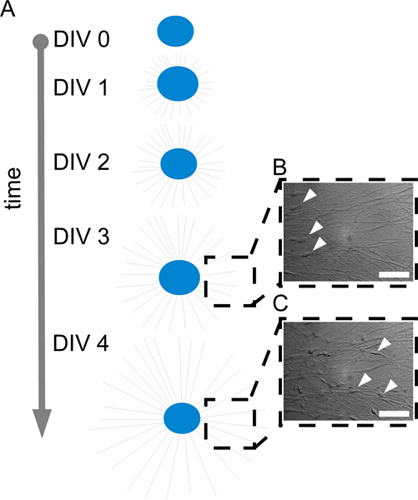
Figure 1. A: Scheme showing axonal growth from an explanted SCG over time (DIV0 -DIV4). B/C: brightfield images, recorded during time-lapse imaging showing close ups of one region of an SCG explant at DIV3 (B) and DIV4 (C) (scale-bar = 100 μm).
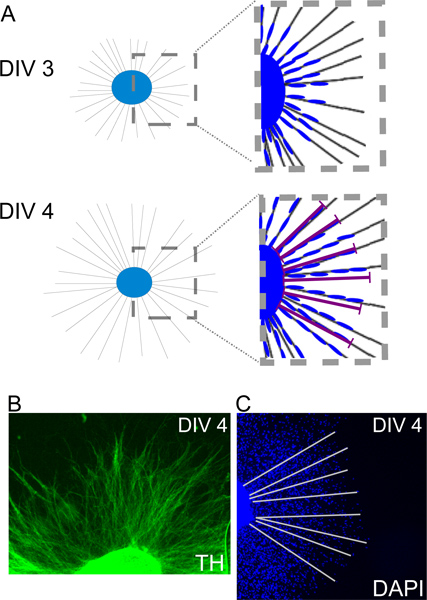
Figure 2. A: Scheme showing SC (blue) along extended axons (grey), grown from an explanted SCG (light blue) at DIV3 and DIV4. Migration distances can be measured on DAPI nuclear-labeled samples by measuring the distance from the nucleus of the leading SC to the border of the explanted SCG at multiple locations (C). TH immunohistochemistry (DIV4) should be performed if a scientist is new to SCG dissection. A positive labeling clearly identifies the explanted ganglion as a sympathetic ganglion (B). TH immunohistochemistry also enables analysis of axons grown from explanted SCGs.
Movie S1. scheme showing axonal growth from an explanted SCG over several days in vitro. Click here to view movie.
Movie S2. Axonal growth and SC migration from an NGF treated SCG explant. Imaging started at DIV2. Recording frame rate was 1/30 min. Scale-bar = 100 μm. Click here to view movie.
Movie S3. Close up of a peripheral area of an SCG explant. Due to a low axonal density single SC can easily be analyzed. Imaging start was DIV3. Recording frame rate was 1/10 min. Scale- bar = 100 μm. Click here to view movie.
Movie S4. The combination of brightfield and fluorescence enables visualization of S100 GFP positive SCs and the axons. Recording frame rate was 1/10 min. Scale- bar = 100 μm. Click here to view movie.
Movie S5. SC migration at the border of an SCG explant. Here only the fluorescence channel is shown enabling easier interpretation of S100 GFP positive SCs. Recording frame rate was 1/10 min. Scale- bar = 100 μm. Click here to view movie.
Discussion
The development of the peripheral nervous system is an exciting process. When the development is completed, axons are ensheathed by SCs along the entire length, which can, in humans, often be over 100 cm. To this end the correct number of the required SCs has to be established during development and the SCs also have to move along extending axons to the periphery to ensure the complete axonal coverage. This holds true for myelinated but also for unmeylinated axons. In both cases all axons are in contact with SCs and depend on their support. To study SC development, assays are needed which take the axonal compartment into account and therefore mimic the in vivo development to a better extent than scratch assays (5), Boyden assays (6) or chamber assays can do. In some assays the axonal compartment was taken into account already. For example sections of sciatic nerves were used as routes for the SCs to migrate along (7, 8), or SCs were co-cultured with neurons along which axons they were observed to migrate (9).
The SCG explant SC migration assay, however, has even more advantages. It is especially interesting because SCs are developing along their own physiological axons, and these are themselves still growing. Furthermore the technique is easy to learn and requires only a bit of technical dissection skills and does not require a complex co-culture system of neurons and SCs. A quite similar setup, however not using SCGs but rather DRGs, was proposed by Gumy and colleagues (10). However, to exploit the full possibilities of such assay, an inverse microscope setup is needed enabling time-lapse imaging. It is important to have the possibility to do “multi- position experiments”, in order to be able to analyze differentially treated SCG explants, situated in a chamber slide, at the same time, enabling the work with optimal controls side by side with the factors of interest (2). For analyzes we only used conventional light- and conventional fluorescence (for transgenic s100b:GFP mice) microscopy. The technique, however, can also easily be used with confocal- or even light sheet microscopy (not performed here). Time-lapse recordings can be used to identify specific cellular properties/behaviors during migration. So far only wildtype and transgenic mice have been used (2). However, transfection of SCs and thereby alteration of pathways with RNAi for example should be feasible. With this technique it could even be possible to analyze the SC-axon interplay of normal and altered SC on the same or neighboring axon. Regarding migration distances, SC proliferation and SC survival, end point analyses can be easily performed by immunohistochemistry with DAPI nuclear labeling and Fiji (NIH) software. However, eventually even life-reporter for proliferation (11) and cell death (12,13) could be used, thereby enabling a direct readout during imaging.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We want to thank Urmas Arumae for sharing a collagen protocol and Jutta Fey and Ursula Hinz for excellent technical assistance. Furthermore we want to thank Christian F. Ackermann, Ulrike Engel and the Nikon Imaging Center at the University of Heidelberg and also Joachim Kirsch for kindly help for the video shoting. The work was partially funded through the Deutsche Forschungsgemeinschaft (SFB 592).
Materials
| Name of Reagent/Material | Company | Catalogue Number | Comments |
| 10x MEM | Gibco | 21430 | |
| Sodium Bicarbonate (7.5%) | Gibco | 25080 | |
| Glutamine | Gibco | 25030 | |
| NaOH | Merck | 109137 | |
| NGF | Roche | 1058231 | R&D#556-NG-100 |
| Neurobasal Medium | Gibco | 21103 | |
| B27 Supplement | Gibco | 17504 | |
| antibiotics | Gibco | 15640 | |
| d-PBS | Gibco | 14040 | |
| insect needles | FST | 26002-20 | |
| syringe needle | Braun | BD # 300013 | |
| 8 well chamber slide | Lab tek | 177402 |
Riferimenti
- Ebendal, T., Rush, R. A. . Use of collagen gels to bioassay nerve growth factor activity. IBRO Handh, (1989).
- Heermann, S., SchmÃcker, J., Hinz, U., Rickmann, M., Unterbarnscheidt, T., Schwab, M. H., Krieglstein, K. Neuregulin 1 type III/ErbB signaling is crucial for Schwann cell colonization of sympathetic axons. PloS One. 6 (12), e28692 (2011).
- Zuo, Y., Lubischer, J. L., Kang, H., Tian, L., Mikesh, M., Marks, A., Scofield, V. L. Fluorescent proteins expressed in mouse transgenic lines mark subsets of glia, neurons, macrophages, and dendritic cells for vital examination. The Journal of Neuroscience. The Official Journal of the Society for Neuroscience. 24 (49), 10999-11009 (2004).
- Levi-Montalcini, R., Booker, B. EXCESSIVE GROWTH OF THE SYMPATHETIC GANGLIA EVOKED BY A PROTEIN ISOLATED FROM MOUSE SALIVARY GLANDS. Proceedings of the National Academy of Sciences of the United States of America. 46 (3), 373-384 (1960).
- Meintanis, S., Thomaidou, D., Jessen, K. R., Mirsky, R., Matsas, R. The neuron-glia signal beta-neuregulin promotes Schwann cell motility via the MAPK pathway. Glia. 34 (1), 39-51 (2001).
- Yamauchi, J., Miyamoto, Y., Chan, J. R., Tanoue, A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. The Journal of cell biology. 181 (2), 351-365 (2008).
- Anton, E. S., Weskamp, G., Reichardt, L. F., Matthew, W. D. Nerve growth factor and its low-affinity receptor promote Schwann cell migration. Proceedings of the National Academy of Sciences of the United States of America. 91 (7), 2795-2799 (1994).
- Mahanthappa, N. K., Anton, E. S., Matthew, W. D. Glial growth factor 2, a soluble neuregulin, directly increases Schwann cell motility and indirectly promotes neurite outgrowth. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 16 (15), 4673-4683 (1996).
- Yamauchi, J., Chan, J. R., Shooter, E. M. Neurotrophins regulate Schwann cell migration by activating divergent signaling pathways dependent on Rho GTPases. Proceedings of the National Academy of Sciences of the United States of America. 101 (23), 8774-8779 (2004).
- Gumy, L. F., Bampton, E. T. W., Tolkovsky, A. M. Hyperglycaemia inhibits Schwann cell proliferation and migration and restricts regeneration of axons and Schwann cells from adult murine DRG. Molecular and Cellular Neurosciences. 37 (2), 298-311 (2008).
- Sakaue-Sawano, A., Kurokawa, H., Morimura, T., Hanyu, A., Hama, H., Osawa, H., Kashiwagi, S. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 132 (3), 487-498 (2008).
- Park, D., Don, A. S., Massamiri, T., Karwa, A., Warner, B., MacDonald, J., Hemenway, C. Noninvasive imaging of cell death using an Hsp90 ligand. Journal of the American Chemical Society. 133 (9), 2832-2835 (2011).
- Shcherbo, D., Souslova, E. A., Goedhart, J., Chepurnykh, T. V., Gaintzeva, A., Shemiakina, I. I., Gadella, T. W. J., Lukyanov, S., Chudakov, D. M. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnology. 9, 24 (2009).

