Rapid Colorimetric Assays to Qualitatively Distinguish RNA and DNA in Biomolecular Samples
Summary
A suite of colorimetric assays is described for rapidly distinguishing protein, RNA, DNA, and reducing sugars in potentially heterogeneous biomolecular samples.
Abstract
Biochemical experimentation generally requires accurate knowledge, at an early stage, of the nucleic acid, protein, and other biomolecular components in potentially heterogeneous specimens. Nucleic acids can be detected via several established approaches, including analytical methods that are spectrophotometric (e.g., A260), fluorometric (e.g., binding of fluorescent dyes), or colorimetric (nucleoside-specific chromogenic chemical reactions).1 Though it cannot readily distinguish RNA from DNA, the A260/A280 ratio is commonly employed, as it offers a simple and rapid2 assessment of the relative content of nucleic acid, which absorbs predominantly near 260 nm and protein, which absorbs primarily near 280 nm. Ratios < 0.8 are taken as indicative of ‘pure’ protein specimens, while pure nucleic acid (NA) is characterized by ratios > 1.53.
However, there are scenarios in which the protein/NA content cannot be as clearly or reliably inferred from simple uv-vis spectrophotometric measurements. For instance, (i) samples may contain one or more proteins which are relatively devoid of the aromatic amino acids responsible for absorption at ≈280 nm (Trp, Tyr, Phe), as is the case with some small RNA-binding proteins, and (ii) samples can exhibit intermediate A260/A280 ratios (~0.8 < ~1.5), where the protein/NA content is far less clear and may even reflect some high-affinity association between the protein and NA components. For such scenarios, we describe herein a suite of colorimetric assays to rapidly distinguish RNA, DNA, and reducing sugars in a potentially mixed sample of biomolecules. The methods rely on the differential sensitivity of pentoses and other carbohydrates to Benedict’s, Bial’s (orcinol), and Dische’s (diphenylamine) reagents; the streamlined protocols can be completed in a matter of minutes, without any additional steps of having to isolate the components. The assays can be performed in parallel to differentiate between RNA and DNA, as well as indicate the presence of free reducing sugars such as glucose, fructose, and ribose (Figure 1).
Introduction
Much of cell biology occurs via molecular interactions involving DNA and RNA.4 These naturally occurring nucleic acids (NAs) interact with one another,5 with proteins,6 and with a host of small-molecule compounds and ligands in vivo (e.g., divalent cations7). The interactions may be short- or long-lived (kinetically), may range from high to moderate to low affinity (thermodynamic strength), and can also exhibit substantial variation in chemical properties and specificity – some associations are quite specific (e.g., DNA···transcription factors, RNA···splicing factors), while other interactions are necessarily far more generic (e.g., DNA···bacterial histone-like HU proteins8). Non-specific interactions with NAs can have practical consequences for in vitro experiments involving mixtures of biomolecules, as it is possible, and even likely, that some NAs will associate with the biomolecules of interest, at least under some subset of the experimental conditions being used (ionic strength, solution pH, etc).
Consider, for instance, production of a protein of interest (POI) via heterologous over-expression of the recombinant protein in Escherichia coli cell culture; such a procedure is routinely performed in virtually any structural biology lab.9 In preparing for further experiments, such as biochemical/biophysical characterization, crystallization, etc., initial efforts generally focus on obtaining a sufficient quantity of the POI in as pure a form as possible, ideally as a chemically homogeneous and biophysically monodisperse specimen. After disruption of host cells, the early stages of a typical purification workflow aim to isolate the POI from E. coli proteins, nucleic acids, cell wall debris, and other components of the cellular lysate. However, host NAs may co-purify with the POI for several physicochemical reasons – a highly basic POI may non-specifically pull-down host DNA/RNA; the POI may have a generic NA-binding activity (e.g., the aforementioned HU); the POI may be a fairly specific NA-binding protein but exhibit cross-reactivity with host RNAs or DNAs; host NAs may interact with a chromatography matrix and thereby simply co-elute with the POI; and so on. Indiscriminate, high-affinity binding of host NAs to a POI can pose a vexing problem because the NA impurities will likely interfere with downstream experiments (e.g., fluorescence anisotropy assays of POI•RNA binding10). Alternatively, unanticipated POI···NA associations also can be viewed fortuitously, as such interactions illuminate the POI’s nucleic acid-binding capacity. Either way, whether NAs are key components or contaminants, one must first quantify and identify the type (DNA, RNA) of co-purifying NAs in preparation for downstream experiments.
Several analytical methods exist for detecting and quantitating NAs in a sample. Most of the available methods are fundamentally either spectrophotometric (e.g., A260 absorbance values and A260/A280 ratios), fluorometric (e.g., binding of thiazole orange or other fluorescent dyes to NA), or colorimetric (susceptibility of nucleosides to chemical reactions that yield chromophores absorbing in the uv-vis region of the electromagnetic spectrum), as recently described by De Mey et al.1 However, the crucial step of identifying the type of polynucleotide as RNA or DNA is beyond the scope of many of these quantitation approaches. Here we provide a set of colorimetric assays to rapidly identify the types of NA components in a proteinaceous sample.
The protocols described here can be efficiently executed without additional steps of isolating the potential NA impurities, and rely on Benedict’s assay for reducing sugars11, the orcinol assay for pentoses12,13, and diphenylamine reactions14,15 of 2′-deoxypentoses (Figures 1 and 2). The Benedict’s test (Figure 2a) utilizes the ability of the linear, open-chain (aldehyde) form of an aldose sugar to reduce Cu2+, with concomitant oxidation of the sugar’s carbonyl to a carboxylate moiety and production of Cu2O as an insoluble red precipitate. This reaction will test positive with free reducing sugars such as aldoses and ketoses (which convert to the corresponding aldoses via enediol intermediates), but not with pentose sugars that are locked into cyclic form as part of the covalent backbone of a DNA or RNA polynucleotide. Because of the minimalistic requirement of a free hemiacetal functionality, other compounds that could test positive in this assay – and therefore act as potential interferents – include α-hydroxy-ketones and short oligosaccharides (e.g. the disaccharide maltose). Both the Bial’s orcinol (Figure 2b) and Dische’s diphenylamine (Figure 2c) reactions are based on initial destruction of the polynucleotide backbone, via depurination of the nucleoside and further acid- or base-catalyzed hydrolysis of the parent nucleotides, to yield furan-2-carbaldehyde (furfural) derivatives; these derivatives then react with either a polyhydroxy phenol such as orcinol (Bial’s) or diphenylamine (Dische’s) reagents to form colored condensation products of largely unknown chemical structure. The DNA versus RNA specificity of the Dische’s assay stems from the fact that the pentose sugar must be 2′-deoxygenated in order to be susceptible to oxidation to ω-hydroxylevulinyl aldehyde, which further reacts with diphenylamine under acidic conditions to yield a bright blue condensate (Figure 2c). Using the streamlined protocols described here, we have found that these sugar-specific colorimetric reactions can differentiate between RNA and DNA, and will also indicate the presence of free reducing sugars such as glucose, fructose, or ribose in a biomolecular sample.
Protocol
1. Benedict’s Assay for Reducing Sugars
- Prepare a suitable quantity of Benedict’s reagent – 940 mM anhydrous sodium carbonate, 588 mM sodium citrate dehydrate, 68 mM copper (II) sulfate pentahydrate. This reagent can be stored at room temperature (RT) for at least six months with no noticeable change in reactivity.
- The above reagent is 6x. Thus, for 600 μl reactions, add 100 μl of Benedict’s reagent to a clean 1.5 ml microcentrifuge tube (e.g., Eppendorf brand), per sample to be assayed.
- Add anywhere from 10 μl to 500 μl of sample to this tube; the optimal volume can be determined based on the intensity of color formation in an initial trial run. If sufficient sample is available then begin such trials at the maximal possible sample volume (i.e., five-sixths of the overall reaction volume, 500 μl of sample in this case), and then dilute in subsequent assays.
- Add ddH2O to the tube to bring the final volume to 600 μl; mix the solution by vortexing or pipetting.
- Incubate the samples for 20 min in a boiling water bath.
- Remove the heated sample from the bath and allow it to cool at RT for 10 min.
- Centrifuge the sample tube at > 9,300 x g (~10,000 rpm in an FA45-24-11 Eppendorf fixed-angle rotor) for 5 min in order to sediment any particulate material; this step is more important for quantitative rather than qualitative studies.
- Aliquot the supernatant from this tube into a clean cuvette.
- Blank the uv-vis spectrophotometer with water.
- Measure the absorbance of this sample at 475 nm.
2. Bial’s Orcinol Assay for Pentose Sugars
- Prepare a suitable quantity of fresh Bial’s reagent – 24.2 mM orcinol monohydrate (see Figure 2b for the structure of this compound), 6 M HCl, 0.025% w/v ferric chloride hexahydrate. Note: For extended storage, the Bial’s reagent can be prepared as two separate stock solutions: (i) Reagent A [0.05% w/v FeCl3•6H2O in concentrated HCl] and (ii) Reagent B [422 mM orcinol monohydrate prepared in 95% ethanol]. Reagent A can be stored at RT for six months; Reagent B can be stored at 4 °C for one month, covered with foil to limit light exposure. These stock solutions are mixed in a 15 (A) : 1 (B) v/v ratio prior to use.
- The above reagent is 2x. Thus, for 1.0 ml reactions, add 500 μl of Bial’s reagent to a clean 1.5 ml microcentrifuge tube, per sample to be assayed.
- Add anywhere from 10 μl to 500 μl of sample to this tube. As per note 1.3 (above), if sufficient sample is available then begin trial reactions using the maximal possible sample volume (i.e., one-half the overall reaction volume, 500 μl of sample in this case) and dilute from there.
- Add ddH2O to the tube to bring the final volume to 1.0 ml; mix the solution by vortexing or pipetting.
- Incubate the samples for 20 min in a boiling water bath.
- Remove the heated sample from the bath and allow it to cool at RT for 10 min.
- Centrifuge the sample tube at > 9,300 x g (~10,000 rpm in an FA45-24-11 Eppendorf fixed-angle rotor) for 5 min in order to sediment any particulate material; this step is more important for quantitative rather than qualitative studies.
- Aliquot the supernatant from this tube into a clean cuvette for visual inspection.
- For semi-quantitative analysis, blank the uv-vis spectrophotometer with water and measure the absorbance of the cuvette sample at 660 nm.
3. Dische’s Diphenylamine Assay for 2′-deoxypentose Sugars
- Prepare a suitable quantity of Dische’s diphenylamine reagent – 60 mM diphenylamine, 11 M glacial acetic acid, 179 mM sulfuric acid, 0.62% v/v ethanol. This reagent can be prepared in advance and stored at RT in a dark container, or covered with foil, to limit light exposure. Due to light sensitivity the reagent should not be stored indefinitely, though in practice it can be prepared every two to three months with no apparent change in reactivity.
- The above reagent is 2x. Thus, for 1.0 ml reactions, add 500 μl of Dische’s reagent to a clean 1.5 ml microcentrifuge tube, per sample to be assayed.
- Add anywhere from 10 μl to 500 μl of sample to this tube. As per note 1.3 (above), if sufficient sample is available then begin trial reactions using the maximal possible sample volume (i.e., one-half the overall reaction volume, 500 μl of sample in this case) and dilute from there.
- Add ddH2O to the tube to bring the final volume to 1.0 ml; mix the solution by vortexing or pipetting.
- Incubate the samples for 20 min in a boiling water bath.
- Remove the heated sample from the bath and allow it to cool at RT for 10 min.
- Centrifuge the sample tube at >9,300 x g (~10,000 rpm in an FA45-24-11 Eppendorf fixed-angle rotor) for 5 min in order to sediment any particulate material; this step is more important for quantitative rather than qualitative studies.
- Aliquot the supernatant from this tube into a clean cuvette for visual inspection.
- For semi-quantitative analysis, blank the uv-vis spectrophotometer with water and measure the absorbance of the cuvette sample at 600 nm.
4. Further Usage Notes
- The following classes of molecules are suitable reference compounds for positive and negative control reactions for each assay:
- Benedict’s – Positive = free ribose, fructose, glucose; Negative = RNA, DNA, ATP, etc. (any saccharide lacking a free reducing sugar functionality)
- Bial’s – Positive = RNA (e.g. baker’s yeast extract), ribose, ATP, UMP; Negative = bovine serum albumin (BSA) or any other protein
- Dische’s – Positive = DNA (e.g. calf thymus); Negative = RNA, ATP, etc. (any non-2′-deoxygenated nucleotide)
- These assays have been found to be fairly resilient to compounds often used in protein purification; for instance, common salts such as NaCl, (NH4)2SO4, and K2SO4 did not interfere, and the reactions appear generally unaffected by the contents of the diluent. Detergents or chaotropic agents such as urea may affect the reactivity of the assays if the pH is highly basic; a neutral to acidic pH range is generally most optimal for the Bial’s and Dische’s assays because of the underlying reaction chemistry (see the text and the protons in Figure 2b, c). Potentially suboptimal reaction conditions, suspected interferents, etc. should be tested on a case-by-case basis, using positive and negative control experiments with reference compounds.
Representative Results
Results are shown in Figure 3 for application of these colorimetric assays to known reference compounds. Representative qualitative data are shown for the Benedict’s (a), Bial’s orcinol (b), and Dische’s diphenylamine (c) assays, and standard curves for these three assays are shown in Figure 4. In panels 3(a-c), the left panels show positive/negative control experiments using suitably reactive/unreactive analytes; these visual results are shown in situ, in the cuvettes as described in Protocols 1-3 (above). The right sub-panels show a titration series for each respective analyte. In the Benedict’s assay (a), the positive control is ribose (0.42 mg/ml), while negative controls are water, a generic protein (0.75 mg/ml BSA), and two non-reducing sugars (DNA at 0.75 mg/ml and RNA at 12.5 mg/ml). In the orcinol assay (b), the positive controls are ribose (0.15 mg/ml), RNA (7.5 mg/ml), and DNA (0.45 mg/ml), while water and BSA protein (0.45 mg/ml) are negative controls. In the diphenylamine assay (c), calf thymus DNA (0.45 mg/ml) is shown as a positive control, and water, yeast extract RNA (7.5 mg/ml), ribose (0.15 mg/ml), and BSA (0.45 mg/ml) serve as negative controls. Finally, panel (d) illustrates the robustness of the assays by showing the Dische’s reaction with samples of varying heterogeneity: two concentrations of DNA are shown as a positive control in the left two samples (cuvettes 1-2), DNA + RNA mixtures (at various ratios) are shown in the next three samples (3-5), and the final three samples show DNA in the absence (sample 6) or presence (samples 7-8) of the nucleic acid-binding protein ‘Hfq’.16 Note that the positive result of the Dische’s assay is preserved for DNA-containing samples even in the presence of ‘contaminating’ RNA or protein.
Quantitative Analysis: The dilution ranges in the Figure 3(a-c) panels vary because each reaction has a distinct visual detection limit, depending on the type of sugar being assayed. Spectrophotometric, rather than visual, detection can be used to improve the measurement ranges, e.g. as shown in Figure 4 for the (a) Benedict’s, (b) Bial’s, and (c) Dische’s assays. Though the protocols described here are intended as primarily qualitative assays, the Beer-Lambert relation between absorbance and concentration enables at least semi-quantitative estimation of the sugar or nucleic acid content. As an example of a standard curve for calibration of the Benedict’s test, a linear regression fit of absorbance (475 nm) against ribose concentration is shown in Figure 4(a); error bars indicate the standard deviation of n = 3 replicates, and the squared correlation coefficient is provided. The deviation from linearity at very low ribose concentrations (e.g. the 0.28 mM datum) reflects the limited sensitivity of this assay. Though the assays are primarily intended for qualitative studies, the following guidelines are suggested for semi-quantitative analysis:
- For the Benedict’s assay (Figure 4a): The reaction readout (A475nm) was found to be linear at least over the range 0.04 → 0.5 mg/ml analyte, as performed in triplicate.
- For the Bial’s assay (Figure 4b): The absorbance data (A660nm) were linear at least over the range 0 → 0.5 mg/ml analyte (baker’s yeast RNA), the assay having been performed in triplicate (R2 = 0.99 linear regression coefficient). Though the samples were centrifuged for clarification before absorbance measurements, no precipitant was found at these low concentrations of analyte and therefore the centrifugation step was not strictly necessary. The assay deviates from linearity at analyte concentrations beyond this range.
- For Dische’s assay (Figure 4c): The A600nm response was found to be linear at least over the range 0.15 → 0.75 mg/ml of analyte (calf thymus DNA), the assay having been performed in triplicate (R2 = 0.99). The plot of absorbance versus [analyte] began to plateau at 0.90 mg/ml DNA, indicating the boundary of the linear response region.
- Practical considerations for quantitative or semi-quantitative analysis: To remove precipitated material that would otherwise interfere with absorbance readings, all three assays require centrifugation at maximal speeds (25,000 x g, or whatever limit can be withstood by the microfuge tubes) for reasonable lengths of time (e.g., 20 min); this is especially important at higher analyte concentrations. After centrifugation, clarified samples can be transferred to a cuvette or microplate (e.g., 96-well microtiter plates) for convenient absorbance measurements.
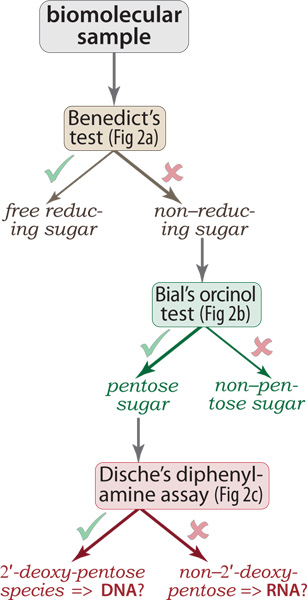
Figure 1. Decision tree for application of the assays. Starting with a potentially heterogeneous sample of biomolecules (e.g., from whole-cell lysates), the colorimetric assays described here can be used to determine if the mixture contains non-reducing sugars (Benedict’s test). If so, then Bial’s orcinol test further reveals whether the population of non-reducing sugars contains pentose rings (as in DNA or RNA), versus hexoses (e.g., glucose, other pyranoses) and possibly yet other aldoses. Finally, if the sample contains at least a moderate fraction of DNA then the 2′-deoxyribose ring will react (upon acidification and heating) with Dische’s diphenylamine reagent, yielding a visibly blue condensation product. Note that this diagram is a decision tree, not a flowchart: the logic of the assay results is shown serially, but the assays can be executed in parallel.
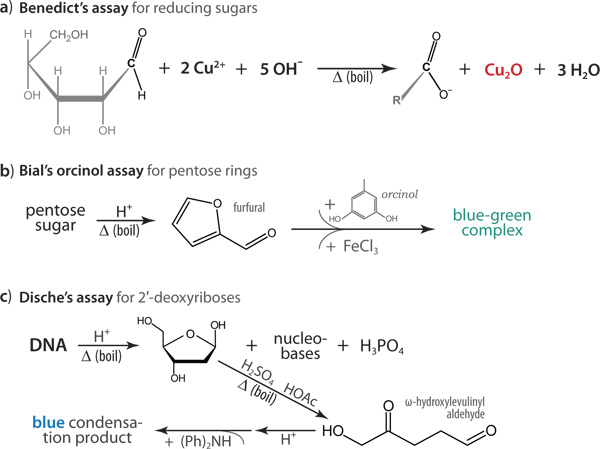
Figure 2. Underlying chemical reactions are shown for the colorimetric assays, with the detectable colored product indicated alongside the corresponding reactions. (a) An insoluble, red precipitate of Cu2O is the positive result of Benedict’s assay for reducing sugars. (b) Upon heating and acidification, sugars containing pentose rings will decompose to furfural, which then reacts with orcinol (Bial’s reagent) to yield a soluble blue-green adduct. In Dische’s assay, lack of a hydroxyl substituent at the 2′ position enables a ring-opening oxidation reaction, the aldehyde product of which further reacts with diphenylamine [(Ph)2NH] to yield a bright blue product. Chemical structures remain unknown for the large, multi-ring condensation products of the orcinol (b) and diphenylamine (c) reactions.

Figure 3. Application of the colorimetric assays to reference compounds and heterogeneous samples. Sample results are shown for the Benedict’s (a), Bial’s (b), and Dische’s (c) colorimetric assays. In (a) → (c), the left sub-panels show positive/negative controls using suitably reactive/unreactive analytes and the right sub-panels show a titration series for each analyte. Also shown is an illustrative panel of Dische reactions (d) wherein the analyte varies in heterogeneity – either DNA alone (left), DNA/RNA mixtures of differing ratios (middle), or DNA in the presence of a nucleic acid-associated protein (‘Hfq’). These panels are further described in the Representative Results section of the text.
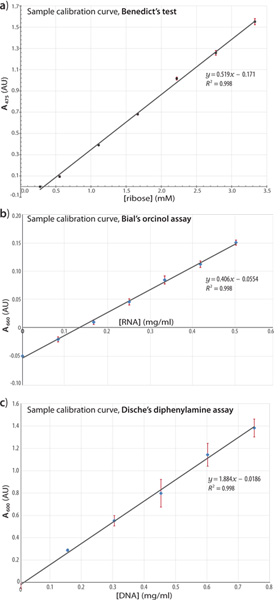
Figure 4. Standard curves from assays with reference compounds. Calibration curves are shown for the Benedict’s (a), Bial’s (b), and Dische’s (c) assays, depicting a representative portion of the linear response region for each assay. The linear regression fits and corresponding correlation coefficients are indicated for each assay. Standard baker’s yeast RNA (b) and calf thymus DNA (c) were the analytes in these titration series. Click here to view larger figure.
Discussion
The colorimetric assays presented here offer a simple approach to rapidly assess the chemical nature of biomolecular mixtures, such as are encountered when purifying proteins, RNAs or complexes from whole-cell lysate in preparation for further studies. As structural biology pursues more native-like assemblies, progressively greater challenges, such as sample heterogeneity, will be posed by the intricate and multi-component complexes. Supramolecular assemblies are often only marginally stable, and their successful isolation may demand less stringent purification conditions (e.g., as found for spliceosomal snRNP complexes17,18). Under such milder conditions both authentic and spurious POI···NA interactions are more likely to persist and thereby interfere in downstream assays with a target protein or nucleic acid. A necessary first step is identification of the types of chemical components in these samples.
Methods to detect RNA, DNA, and protein vary depending on quantity and concentration of analytes, available resources and time constraints and, perhaps most importantly, one’s prior knowledge about the likely chemical composition of the analytes. Proteinaceous material can be detected by many well-established methods, including those that are spectroscopic (A280), hydrodynamic (gel electrophoresis), chemical/colorimetric19 (e.g., biuret test for peptide bonds) and, with great sensitivity and accuracy, via mass spectrometry. Similarly, NAs can be detected by spectroscopic (A260), fluorometric, or chemical assays (described here). Each type of method features characteristic strengths and weaknesses. For instance, PicoGreen, SYBR-Gold, and other cyanine-based fluorescent dyes are quite sensitive to nucleic acid (many orders of magnitude beyond the colorimetric assays), exhibit various degrees of selectivity (e.g., PicoGreen for double-stranded DNA, SYBR-Gold binds most NAs), and also feature broad dynamic ranges for quantitation purposes. However, these approaches are not without limits: the dyes require more advanced equipment for detection (transilluminators for gels, spectrofluorometers for batch samples), versus simple visual readout of colorimetric assays; the dyes are treated as mutagens, akin to ethidium bromide; and there are restrictions on assay conditions (e.g., a recommended pH range of 7-8.5 for SYBR-Gold; a 30% reduction of PicoGreen signal intensity at >200 mM NaCl). Thus, colorimetric and fluorescence-based assays are complementary: nucleic acid dyes could be particularly useful in case of negative results with the rapid colorimetric assays, or as a way to more carefully (quantitatively) expand on initial results from colorimetric assays. A fundamental benefit of the colorimetric NA protocols, in addition to simplicity of use, is that they rely solely on the intrinsic covalent structure of NAs rather than fold/3D structures, reactivities, or any other properties of the biopolymer that might vary with sequence.
The protocols described here are robust against most difficulties, particularly when performed with simple visual inspection of the reaction products; special care must be taken for more quantitative (spectrophotometric) analyses. For instance, in the Benedict’s assay the red precipitant (ppt) that forms in the presence of high concentrations of ribose must be removed prior to spectrophotometric analysis; this is readily achieved via centrifugation. For the Dische’s assay, the addition of diphenylamine reagent is accompanied by formation of a white ppt that can be solubilized by heating; a green ppt that forms in the presence of DNA may interfere with spectrophotometric measurements and must be removed prior to quantitative analysis. In addition, each assay is based on chemical reactions that are susceptible to potential interferents, as mentioned in the Introduction and further detailed below. Finally, we note that a high relative concentration of RNA in a DNA/RNA mixture can mask the expected positive result of Dische’s assay; this false negative for DNA stems from the fact that a high mole fraction of RNA also reacts, via furfural intermediates, with the Dische’s reagents, but yields a colorless product rather than the blue adduct produced by reaction with the 2′-deoxypentose sugar of DNA.
Potential Interferents: Beyond the obvious case of sugars, lipids and proteins are two other biomolecules that could hypothetically cause trouble with these colorimetric assays due to cross-reactivity (false positives) or masked reactivity (false negatives). In principle, several types of cellular lipids could conceivably interfere, including glycosylglycerols and other glycerolipids conjugated to sugars, glycosphingolipids (e.g., cerebrosides), saccharolipids (e.g., glucosamine derivatives), lipopolysaccharides, and so on. In practice, these classes of lipids are unlikely to pose a problem in working with lipophilic proteins because they are relatively rare, versus the much more abundant phospholipids, and therefore below the detection limits of our assays. Because most of the aforementioned cellular lipids are built upon hexoses (galactose, glucose) rather than pentoses, they should not interfere as false-positives in the Bial’s or Dische’s assays. Free monosaccharides that may give false-positives include fructose, galactofuranose, or other furanoses. On a related note, interference from unwanted sugars could become an issue for recombinant proteins expressed with maltose-binding protein (MBP) tags. In the affinity chromatography steps used to purify such fusion constructs, maltose is used to elute the protein, and it is conceivable that residual maltose in downstream preparations could interfere with the Benedict’s assay (it is the most general/nonspecific of the assays; the glucose units of maltose should not react under the Bial’s or Dische’s). Thus, one would have to exercise caution in the post-chromatographic steps to insure that residual maltose did not yield spurious results. We have not tested glucosylated proteins or other glycoproteins, but we suspect that it would be difficult to obtain a clear positive result for the presence of sugars on such proteins (or glycolipid, proteoglycan, or other glycoconjugates) because we expect that the glycan moieties would lie below the sensitivity limit of our assays. In principle, a solution containing a free aldohexose such as glucose, liberated from a glucosylated protein via acid hydrolysis, would test positive by the Benedict’s assay; similarly, a glycoprotein-derived pentose, such as xylose, should yield a positive result in the Bial’s assay. However, in practice a positive result for glycoproteins would require extremely high protein concentrations, even for multiply glycosylated polypeptides, because of the low sugar/protein molar ratio.
The assay protocols described here can be extended and adapted to handle the limits of small sample size – i.e., small volumes or low concentrations of analytes. For limited sample quantities we have found that the reactions can be scaled down to the 100-μl range (e.g., using PCR tubes and a thermocycler for the incubation steps). For small-volume samples at low concentrations (near the detection limit), a spectrophotometer equipped with a plate-reader can be used; in such scenarios, the only anticipated difficulty would be removal of any unwanted precipitant prior to absorbance measurements. Despite the limited detection range of simple visual inspection, an advantage of the approach described here is its efficiency and rapidity, with samples capable of being analyzed instantly upon heating.
Applications of the protocols presented here include the identification of co-purifying compounds, assessment of RNA or DNA purity, and the detection of residual NA or sugar contamination in protein samples. For example, unwanted NA detected by our assays can be removed from a protein prep via chemical means (e.g., alkaline hydrolysis of RNA) or enzymatic digestion (e.g., nuclease treatment). Although there are alternative approaches to such applications, the methods described here are highly efficient in terms of both time and cost, and can therefore be easily integrated into an experimental workflow. The early identification of co-purifying compounds as free reducing sugar, RNA, DNA, or protein can guide the design of downstream purification steps, versus laborious trial and error studies. A common step following our protocols might be the isolation of RNA from DNA and protein, via thiocyanate-phenol-chloroform extraction, or recovery of DNA alone (by excluding thiocyanate).20 To assess the purity of the NAs resulting from phenol-chloroform extractions, these colorimetric assays can be used as a simple alternative to electrophoresis or DNase treatment. In this and other ways, the colorimetric assays described here can be combined with well-established methods for protein and NA determination to achieve a rapid and robust system for elucidating the RNA, DNA, and protein components in heterogeneous biomolecular samples.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was funded by the University of Virginia and the Jeffress Memorial Trust (J-971). We thank L. Columbus, K. Jain, and P. Randolph for helpful discussions and critical reading of the manuscript.
Materials
| Reagent or equipment | Supplier/company | Catalog number | Comments, notes |
| Anhydrous sodium carbonate | Fisher Scientific | S263 | |
| Sodium citrate dihydrate | Sigma | S-4641 | |
| Copper (II) sulfate pentahydrate | VWR | VW3312-2 | |
| Orcinol monohydrate | Sigma-Aldrich | O1875 | |
| Concentrated HCl | VWR | BDH3030 | |
| Ferric chloride hexahydrate | Sigma | F-2877 | |
| Diphenylamine | Aldrich | 112763 | |
| Glacial acetic acid | Fisher Scientific | A28 | |
| Sulfuric acid | Sigma-Aldrich | 258105 | |
| Ethanol | Koptec | V1101 | |
| Ribose | Sigma | R-7500 | prep at 1% w/v in H2O |
| Ribonucleic acid from baker’s yeast (S. cerevisiae) | Sigma | R6750 | prep at 10 mg/ml in H2O; store at -20 °C |
| Deoxyribonucleic acid (sodium salt), from calf thymus | Sigma | D1501 | prep at 10 mg/ml in H2O; store at 4 °C |
|
Reagents, Equipment & Safety Materials are listed in the following table in the order in which they appear in the Protocol section. Unless otherwise noted (above), all reagents can be stored at ambient room temperature and lighting. For any items not listed below (e.g., microcentrifuge tubes), the usual make / model / variety found in a standard biochemical laboratory can be used (e.g., Eppendorf brand 1.5 ml microfuge tubes). Standard plastic microfuge tubes should be used for steps involving centrifugation (e.g., to sediment particulate material near the end of each protocol). No particular material is preferable, as long as the tubes can be sealed; the typical polypropylene tubes found in biochemistry laboratories work well. In terms of safety concerns and waste disposal, standard laboratory precautions (safety glasses, fume hoods) should be exercised in pre-paring, working with, and disposing of solutions containing concentrated acetic, hydrochloric, or sulfuric acids. For organic reagents such as orcinol or diphenylamine, nitrile gloves are preferable to the common latex (natural rubber) variety. |
Riferimenti
- De Mey, M., et al. Comparison of DNA and RNA quantification methods suitable for parameter estimation in metabolic modeling of microorganisms. Anal. Biochem. 353, 198-203 (2006).
- Desjardins, P., Conklin, D. NanoDrop Microvolume Quantitation of Nucleic Acids. J. Vis. Exp. (45), e2565 (2010).
- Ausubel, F. M. . Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. , (1999).
- Voet, D., Voet, J. G. . Biochimica. , (2011).
- Adams, R. L. P., Knowler, J. T., Leader, D. P. . The Biochemistry of the Nucleic Acids. , (1986).
- Rice, P. A., Correll, C. C. . Protein-nucleic acid interactions: Structural biology. , (2008).
- Bowman, J. C., Lenz, T. K., Hud, N. V., Williams, L. D. Cations in charge: Magnesium ions in RNA folding and catalysis. Curr. Opin. Struct. Biol. , (2012).
- Balandina, A., Kamashev, D., Rouviere-Yaniv, J. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 277, 27622-27628 (1074).
- Graslund, S., et al. Protein production and purification. Nat. Methods. 5, 135-146 (2008).
- Pagano, J. M., Clingman, C. C., Ryder, S. P. Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA. 17, 14-20 (2011).
- Benedict, S. R. A reagent for the detection of reducing sugars. J. Biol. Chem. 277, e5 (1908).
- Endo, Y. A simultaneous estimation method of DNA and RNA by the orcinol reaction and a study on the reaction mechanism. J. Biochem. 67, 629-633 (1970).
- Almog, R., Shirey, T. L. A modified orcinol test for the specific determination of RNA. Anal. Biochem. 91, 130-137 (1978).
- Dische, Z. New color reactions for determination of sugars in polysaccharides. Methods Biochem. Anal. 2, 313-358 (1955).
- Burton, K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochemical Journal. 62, 315-323 (1956).
- Vogel, J., Luisi, B. F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578-589 (2011).
- Deckert, J., et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell Biol. 26, 5528-5543 (2006).
- Stevens, S. W., et al. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 9, 31-44 (2002).
- Sapan, C. V., Lundblad, R. L., Price, N. C. Colorimetric protein assay techniques. Biotechnol. Appl. Biochem. 29 (Pt. 2), 99-108 (1999).
- Chomczynski, P., Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 1, 581-585 (2006).

