Imaging InlC Secretion to Investigate Cellular Infection by the Bacterial Pathogen Listeria monocytogenes
Summary
Listeria monocytogenes is a Gram positive bacterial pathogen frequently used as a major model for the study of intracellular parasitism. Imaging late L. monocytogenes infection stages within the context of small-interfering RNA screens allows for the global study of cellular pathways required for bacterial infection of target host cells.
Abstract
Bacterial intracellular pathogens can be conceived as molecular tools to dissect cellular signaling cascades due to their capacity to exquisitely manipulate and subvert cell functions which are required for the infection of host target tissues. Among these bacterial pathogens, Listeria monocytogenes is a Gram positive microorganism that has been used as a paradigm for intracellular parasitism in the characterization of cellular immune responses, and which has played instrumental roles in the discovery of molecular pathways controlling cytoskeletal and membrane trafficking dynamics. In this article, we describe a robust microscopical assay for the detection of late cellular infection stages of L. monocytogenes based on the fluorescent labeling of InlC, a secreted bacterial protein which accumulates in the cytoplasm of infected cells; this assay can be coupled to automated high-throughput small interfering RNA screens in order to characterize cellular signaling pathways involved in the up- or down-regulation of infection.
Introduction
The Gram positive bacterium Listeria monocytogenes is a food-borne pathogen that invades host cells, disrupts its internalization vacuole and replicates in the cytoplasm of host cells1. L. monocytogenes ease of manipulation in the laboratory context (rapid growth, low toxicity for healthy individuals) associated with the persistence of bacterial virulence traits observed in cellular and animal models (hemolytic activity, leukocytosis) allowed its initial use in the 1960s as a major model for the study of intracellular parasitism and for the establishment of the theoretical foundations of cellular immunity against infection2. In the late 1980s and early 1990s, the dissection of the bacterial intracellular cycle3 as well as the molecular characterization of the most important bacterial virulence factors4-7 favored the use of L. monocytogenes as a key molecular tool for the manipulation and study of host cell functions. The presence of avirulent (L. innocua) and virulent (L. monocytogenes) species in the Listeria genus paved the way for comparative genomic studies8 that, together with the recent establishment of the complete L. monocytogenes transcriptome9, have increased our understanding of the evolution of L. monocytogenes as a human pathogen and as a model system for infection studies10.
L. monocytogenes induces its internalization into host cells upon interaction of the bacterial surface proteins InlA and InlB with their host cell receptors E-cadherin and Met, respectively11-12. Initial candidate-based studies led to the identification of the α/β catenins-actin link as a significant component of the InlA-invasion pathway13 and of the phosphoinositide 3-kinase (PI 3-K) as a critical effector of the InlB-dependent invasion cascade14-15. Proteomic and functional-based assays subsequently allowed the identification of novel cytoskeletal elements16 and lipid second messengers17 required for host cell invasion. Transcriptional studies18 and mass spectrometry-based quantitative proteomics19 have recently shed new light concerning the activation of host signaling cascades and the repression of immune responses during L. monocytogenes infection. Systems biology approaches based on the inactivation of large sets of genes (kinomes, complete genomes) by small interfering RNA (siRNA) silencing have recently opened new avenues for the analysis of global host signaling cascades in the context of specific cellular functions, including phagocytosis and pathogen internalization20. Genome-wide siRNA screens have been previously performed to investigate cellular cascades required for infection of L. monocytogenes in phagocytic Drosophila S2 cells21-22, but this type of analysis has not been performed in non-phagocytic cells, which represent critical targets for infection in vivo.
We have optimized a protocol for the microscopical detection of late stages of infection by L. monocytogenes that is suited for high-throughput siRNA studies of bacterial entry within epithelial cells. Our assay takes advantage of a highly invasive L. monocytogenes strain that presents a point mutation in PrfA, the major transcriptional regulator of L. monocytogenes virulence factors6: this mutation (named PrfA*) renders PrfA constitutively active23 and leads to an increased expression of the invasion proteins InlA and InlB, therefore favoring bacterial entry in otherwise poorly infected non-phagocytic cells. Our readout for infection is based on the detection of the cytosolic accumulation of the secreted bacterial protein InlC: this molecule is a pleiotropic effector that is expressed preferentially by intra-cytoplasmic L. monocytogenes9 and which participates not only in the bacterial cell-to-cell spread24 but which also modulates host immune responses25. The fluorescent labeling of InlC secretion by intracellular bacteria not only allows to clearly distinguish infected from non-infected cells, but also represents an end-point readout that can be used to subsequently dissect infection in its different steps: entry, vacuolar escape, cytosolic bacterial proliferation and cell-to-cell spread. This microscopy-based protocol can be coupled to siRNA screens therefore to study cellular pathways involved in the infection of host cells by L. monocytogenes.
Protocol
1. Preparation of Cellular and Bacterial Cultures, Transfection Tools and Primary Antibodies
- Prepare a fresh agar plate to isolate individual L. monocytogenes colonies from a bacterial glycerol stock (50% glycerol/50% saturated bacterial liquid overnight culture) kept at -80 °C.
- Using an aluminum rack (kept at -80 °C) to transport a frozen bacterial glycerol stock, streak bacteria on a Brain Heart Infusion (BHI) agar plate.
- Incubate the plate at 37 °C during 48 hr (or until individual bacterial colonies can be isolated).
- Keep this working plate afterwards at 4 °C during a maximum time of 1 month (it will be used to seed liquid cultures).
- In our protocol, we use the L. monocytogenes serotype 1/2a EGDe.PrfA* strain, but as illustrated in Figure 1, the L. monocytogenes serotype 4b P14.PrfA* strain gives similar results.
- HeLa ATCC CCL2 cells are grown in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in the absence of antibiotics.
- Independent pools of scrambled (control) and anti-Met siRNAs are loaded in black 384-well microscopy cell culture plates.
- Dilute 160 pmol of siRNA in 500 μl of RNase-free water and add 5 μl of this solution per well (final concentration: 1.6 pmol).
- Keep this plate at -20 °C for a maximum period of 1 year.
- Polyclonal rabbit antibodies against the bacterial protein InlC have been obtained by immunization using an InlC-GST recombinant protein as described previously25.
2. Reverse siRNA Cell Transfection
- 72 hr before infection bring to room temperature a black 384-well microscopy cell culture plate containing 1.6 pmol siRNA in 5 μl of RNase-free water in each well.
- Centrifuge the plate for 3 min at 300 rcf at room temperature to bring down siRNA which could have been deposited in the walls of the wells.
- Prepare room temperature DMEM supplemented with 0.4% Lipofectamine RNAiMAX.
- Add 25 μl of the DMEM/RNAiMAX transfection medium to each well (do not incubate the transfection medium longer than 20 min before adding it to the siRNA).
- Move the plate back and forth to mix the siRNA with the transfection solution, then keep the plate for 1 hr at room temperature to allow siRNA-Lipofectamine complexes to form.
- Wash HeLa cells from a confluent- or sub-confluent cell culture flask once with 10 ml 37 °C pre-warmed PBS.
- Detach HeLa cells by adding 1 ml 37 °C prewarmed trypsin to the cell culture flask and incubate for 3 to 5 min at 37 °C.
- Re-suspend cells in 10 ml 37 °C prewarmed DMEM supplemented with 16% FBS.
- Count cells and prepare a cell suspension of 12,000 cells per ml in DMEM supplemented with 16% FBS.
- Add 50 μl of the cell suspension to each well in the 384-well plate.
- Move the plate quickly back and forth to distribute cells and let cells settle down for 10 min at room temperature.
- Seal the plate with Parafilm and keep it for 72 hr in a humidified 5% CO2-containing atmosphere at 37 °C.
3. Cellular Infection and Staining
- The day before infection, take a single colony of L. monocytogenes from the BHI agar plate and resuspend it in 5 ml of liquid BHI medium in a 15 ml polystyrene tube.
- Incubate overnight at 37 °C in a shacking device to allow for bacterial growth.
- The day of the infection, wash 1 ml of the overnight L. monocytogenes culture by centrifuging 2 min at 10,600 rcf on a table-top centrifuge.
- Discard the supernatant (which contains the secreted cytotoxin listeriolysin O) and re-suspend the pellet in 1 ml of PBS (repeat the washing step 3 more times).
- Read the bacterial optical density at 600 nm and estimate the number of bacteria (OD=1 is equivalent to 1E9 bacteria/ml).
- Prepare the adequate L. monocytogenes dilution in DMEM supplemented with 1% FBS: using highly invasive strains like EGDe.PrfA*, we suggest the use of 5E4 bacteria in 30 μl of medium per well (the multiplicity of infection is estimated as 25 for 2,000 cells).
- Remove the cell culture medium in each well (80 μl) and replace it by adding 30 μl of the L. monocytogenes-containing medium.
- Centrifuge the plate at 200 rcf for 5 min at room temperature to synchronize the infection process.
- Incubate the plate for 1 hr in a humidified 5% CO2-containing atmosphere at 37 °C on a prewarmed aluminum block.
- Remove the L. monocytogenes-containing medium from each well and add 30 μl of prewarmed DMEM supplemented with 10% FBS and 40 μg/ml gentamicin to kill extracellular L. monocytogenes.
- Incubate for 4 hr in a 5% CO2-containing atmosphere at 37 °C on a prewarmed metal block.
- Prepare a solution of PBS supplemented with 8% formaldehyde (this solution should be prepared fresh so that the formaldehyde monomers, rather than the paraformaldehyde polymers, are used).
- Without discarding the cell culture medium, add 30 μl of PBS supplemented with 8% formaldehyde (final concentration: 4%) and incubate for 15 min at room temperature.
- Remove the fixative and wash the cells three times with 80 μl of PBS per well (keep the cells in a final volume of 80 μl of PBS per well).
- Prepare a 1:250 dilution of the rabbit anti-InlC serum in PBS supplemented with 0.2% saponin.
- Add 10 μl of the primary antibody solution to each well after removing the PBS and incubate for 30 min at room temperature.
- Discard the primary antibody solution and wash four times with 40 μl of PBS per well.
- Dilute the secondary Alexa Fluor 546-coupled anti-rabbit antibody (1:250), the DAPI solution (1:1,500) and phalloidin-Dy647 (1:150) in PBS supplemented with 0.2% saponin.
- Add 10 μl of this secondary staining solution to each well and incubate for 30 min at room temperature.
- Discard the secondary staining solution and wash four times with 40 μl of PBS per well (leave a final volume of 40 μl in each well and seal the plate).
- The plate can be imaged immediately or can be stored at 4 °C protected from light (cover with aluminum foil) for subsequent analysis.
4. Image Acquisition and Analysis
- Acquire images in three different channels (350 nm, 546 nm and 647 nm) using a 10X objective mounted on an automated microscope (acquire preferentially 9 images per well).
- The InlC signal can be measured using image analysis software like CellProfiler that allows automated segmentation of nuclei and cell bodies using the DAPI and the phalloidin staining, respectively.
Representative Results
Fluorescent labeling of cytoplasmic InlC provides a robust readout for cell infection by L. monocytogenes, as illustrated in Figure 1: the central cell in the micrograph is highly infected by the strain P14.PrfA*23 as it can be observed in the phase contrast image (arrowheads, Figure 1A) and it is confirmed by the DAPI signal where individual bacteria can be clearly distinguished (Figure 1B). The InlC staining (superposed to the DAPI staining in Figure 1C) depicts how this secreted protein densely accumulates in the cytosol of infected cells and allows an unambiguous detection of the infected host cell morphology. Staining of the actin cytoskeleton with fluorescent phalloidin (Figure 1D) provides information concerning the morphology of the complete inoculated cellular monolayer and further illustrates how neighboring non-infected cells (cell nuclei marked with an asterisk) do not display any InlC labeling. It is worth to mention that, since InlC cytosolic levels are dependent on the number of cytoplasmic bacteria, we have noted a correlation between the InlC signal detected by immunofluorescence and the number of L. monocytogenes per cell: as observed in Figure 1, neighboring cells infected with low bacterial numbers display a reduced InlC staining which can be quantified. Interestingly, it is also possible to observe that InlC is easily detected in the protrusions formed by bacteria caught in the process of cell-to-cell spread (Figure 1C) as observed also with the actin staining (Figure 1D). Of note, bacteria are not directly stained in our protocol but since the 488 nm channel is not used, L. monocytogenes can be labeled using a specific anti-bacterial serum; otherwise, GFP-expressing bacteria can alternatively be used.
Using imaging analysis tools such as the public software CellProfiler (Broad Institute, www.cellprofiler.org), it is possible to segment cells and to measure the InlC signal in individual infected cells, which can be used to estimate an infection index for a specific experimental condition. As shown in Figure 2, the labeling of the nuclei with DAPI (Figure 2A) and the actin cytoskeleton with phalloidin (Figure 2C) provide the information required for cellular segmentation: the nuclei are used as reference objects for the identification of individual cells (Figure 2B) and the cytoplasm is subsequently identified using a spread function from the nuclei that takes into account the cytoskeletal signal (Figure 2D). Finally, the intensity of the InlC signal can be quantified for each identified cellular object (Figures 2E and 2F) to estimate the number of infected cells by setting a threshold for the InlC intensity that we consider as negative. The infection index is calculated as the number of infected cells divided by the total number of cells in the population.
Our protocol can be coupled to siRNA screens to investigate the function of large panels of target molecules in the infection of host cells by L. monocytogenes. Controls are required to validate the efficiency of transfection: for example, siRNA targeting Kif11 is a commonly used control for transfection that leads to cell death, and therefore the absence of cells at the end of the assay is an indication that transfection proceeded efficiently. To specifically address the L. monocytogenes invasion process, siRNA inactivation of the cellular receptor Met in HeLa cells leads to a major inhibition of bacterial entry into host cells and leads to very low levels of InlC-positive cells in an inoculated cellular monolayer (Figure 3A) as compared to cells treated with a scrambled siRNA control (Figure 3B). The function of candidate unknown molecules can be investigated with our assay using this known cell molecule as standard.
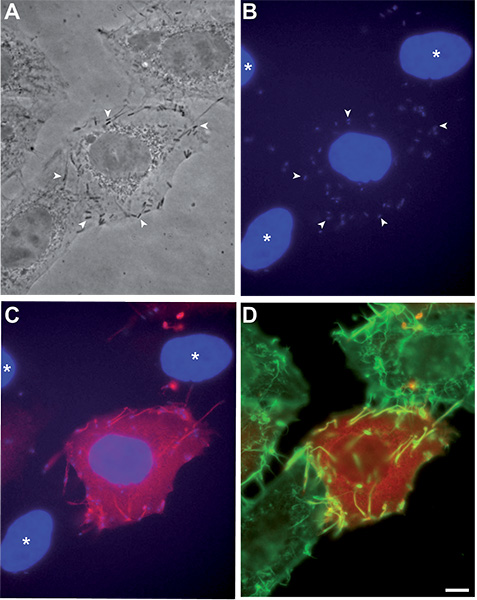
Figure 1. Detection of InlC labeling in HeLa cells infected with L. monocytogenes strain P14.PrfA*. HeLa CCL2 cells have been infected as described in our protocol, processed for immunofluorescence and imaged using a 63X objective. (A) Phase contrast image, several P14.PrfA* bacteria are indicated by arrowheads. (B) DAPI staining (blue), the same individual bacteria as in (A) are labeled by arrowheads. (C) Superposition of the DAPI staining (blue) and the InlC staining (red), the nuclei of uninfected cells are labeled by an asterisk. (D) Superposition of the InlC (red) and actin (green) signals. Bar: 5 μm.
Click here to view larger figure.

Figure 2. Segmentation analysis of HeLa cells infected with L. monocytogenes strain EGDe.PrfA*. HeLa CCL2 cells were infected as described in our protocol, processed for immunofluorescence and imaged using a 10X objective. (A) DAPI signal. (B) Superposition of the DAPI signal (blue) displayed in (A) and actin signal (red) displayed in (C) showing the segmentation of the nuclei (view enlarged inset). (C) Actin signal. (D) Same image as (B) showing the segmentation of the cells. (E) InlC signal. (F) Same image as (D) with superposition of the InlC staining (yellow). Bar: 50 μm.
Click here to view larger figure.
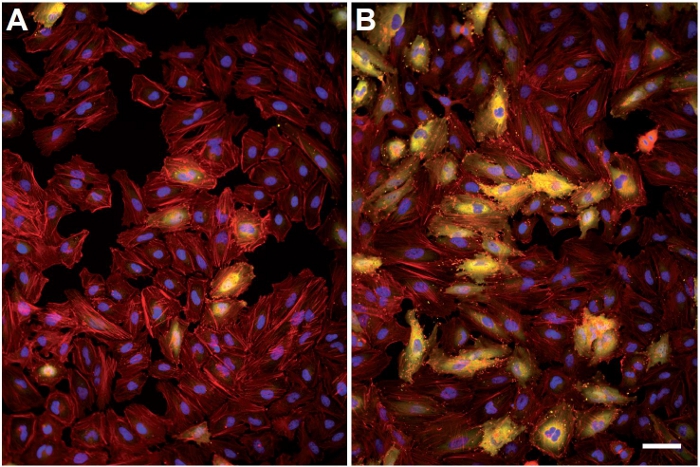
Figure 3. Variation of the InlC labeling between cells inactivated for Met and control cells. HeLa CCL2 cells were initially reverse-transfected with a pool of four siRNAs targeting the host cell receptor Met; 72 hr after transfection, cells were infected as described, processed for immunofluorescence and imaged using a 10x objective. (A) Superposition of the DAPI (blue), actin (red) and InlC (yellow) in cells inactivated for Met. (B) Same channels as in (A) concerning cells treated with a scrambled siRNA control. Bar: 50 μm. Click here to view larger figure.
Discussion
Several parameters are critical for the success of our InlC-detection protocol, including the use of healthy cell lines displaying a sufficiently large cytoplasm to allow an unambiguous detection of the InlC signal. In the assay we present in this article we propose the use of HeLa CCL2 cells, which are particularly well suited for our assay due to the extension of their cytosolic space; other HeLa clones such as HeLa Kyoto cells display a smaller cytoplasm but can be used with our infection protocol (HeLa Kyoto cells are particularly well adapted for segmentation analysis since cells do not overlap with neighboring cells, which is often the case for HeLa CCL2 cells). Cells presenting a very small cytoplasm such as polymorphonuclear cells or macrophages are not recommended to use in combination with our protocol.
A limitation of an imaging protocol as the one we present in here is the fact that a minimal amount of cells should be infected, in a given monolayer in control conditions, in order to provide the system with enough resolutive power: otherwise, if few cells are infected, it becomes difficult to determine changes in infection rates, especially when investigating the function of potential molecular candidates that are expected to reduce infection. This limitation represents a real problem when using HeLa cells, which express Met as the only surface receptor for the L. monocytogenes protein InlB and are therefore poorly invaded by most L. monocytogenes strains. One alternative is to use a very high multiplicity of infection (MOI>100) which may increase the number of invasive bacteria but also increases the risk of cell damage due to higher levels in the cell culture medium of the bacterial pore forming toxin listeriolysin O (LLO), which displays a very potent cytotoxic activity. Other alternative is the use of super-invasive strains such as the EGDe.PrfA* strain presented in our protocol, which allows the use of a low MOI (<25) and reduces the amount of LLO-dependent cytotoxic effects; results obtained with a L. monocytogenes super-invasive strain can be subsequently validated using other less virulent bacterial strains in other types of assays (see below). A third alternative is to use a different cell line which expresses both E-cadherin and Met: it is the case for cell lines such as the trophoblast-like Jeg3 or BeWo cells, or the colon carcinoma LoVo cells, which can be invaded by both the InlA- and the InlB-dependent entry pathways; in these cell lines, higher rates of cell infection can be reached using L. monocytogenes strains such as EGDe, the parental strain of EGDe.PrfA*. It should be taken into account, however, that redundancy of functions in both pathways can lead to compensation effects in one pathway when an effector is inactivated in the other pathway, rendering the analysis of results difficult. It is also important to mention that cells should not be over-infected in order to provide the system with a good dynamic range allowing the detection of events that increase infection: in our particular protocol, our MOI leads to an optimal infection rate of 30%.
Alternative methods to study the invasion of host cells by L. monocytogenes include the classical gentamicin invasion assay26 which is based on the killing of extracellular bacteria by the addition of gentamicin to the cell culture medium after an initial period of bacterial infection (similar to the procedure presented in the first part of our infection protocol) and invasion is scored by plating the surviving intracellular bacteria on agar plates and counting the number of colony forming units (CFUs) that grow on these plates. This method presents the advantage of using the most direct readout for infection, which is the precise number of invasive bacteria that are actually protected from the gentamicin treatment in the cytoplasmic space of host cells; however, this method presents a single readout for infection and once the host cells are lysed to release intracellular CFUs, there is no longer a record to obtain information concerning the actual status of cells at the end point of the experiment. Our protocol is based on an indirect method, the detection of the secreted InlC protein, but as mentioned previously, the levels of cytoplasmic InlC correlate with the number of cytosolic L. monocytogenes (Figure 1). Moreover, the intrinsic nature of our microscopy assay allows to clearly establish the precise status of the cells at the end of the infection: we can therefore extract information concerning global cellular morphology, actin cytoskeleton distribution, cell cycle phase, etc. and perform high content analysis of the infection. One important feature that can be extracted from this type of analysis is the population context and its influence on infection: indeed, recent work from Pelkmans and coworkers 27 demonstrated that the particular population context of a given cell (for example, its position from the periphery within a group of cells in an islet) affects several cellular functions including endocytosis and susceptibility to virus infection. Our assay presents therefore the potential for the extraction of this information.
Our method presents an additional advantage: since InlC is a late readout of infection due to its secretion by cytoplasmic bacteria, host signaling pathways affecting the levels of InlC accumulation in host cells could potentially affect not only bacterial entry but also vacuolar escape, cytosolic proliferation and eventually cell-to cell spread. Hence, our method can be used as a primary readout for global infection and can be coupled to secondary assays to dissect the specific infection step that has been perturbed by the primary hit identified through siRNA screens.
Finally, it is important to mention that our method can be upscaled and fully automated using plate washer devices, increasing therefore the number of samples analyzed in a single experiment while decreasing the level of results variation when compared to experiments carried out manually.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Research in P. Cossart laboratory is supported by the Pasteur Institute, the Institut National de la Santé et de la Recherche Médicale, the Institut National de la Recherche Agronomique, ERC Advanced Grant (233348), the Agence Nationale de la Recherche (Grant MIE-SignRupVac), the Louis-Jeantet Foundation and the Fondation Le Roch Les Mousquetaires. A.K. is a recipient of a scholarship from the Pasteur-Paris University International Doctoral Program/Institut Carnot Maladies Infectieuses. We acknowledge support by grant 51RT 0_126008 for the Research and Technology Development (RTD) project InfectX in the frame of SystemsX.ch, the Swiss Initiative for Systems Biology (to C.D.).
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Bacto Brain Heart Infusion | BD | 237500 | For liquid BHI preparation |
| Bacto Agar | BD | 214010 | Supplement to liquid BHI for BHI agar plates |
| Heat-Inactivated Fetal Bovine Serum | Biowest | 51830-500 | |
| DMEM | Invitrogen | 61965-026 | |
| Lipofectamine RNAiMax | Invitrogen | 13778-100 | |
| Gentamicin | Sigma | G1397-10ML | |
| Formaldehyde (16%) | EMS | 15710 | Prepare fresh before each experiment |
| Anti-Rabbit Alexa Fluor 546 | Invitrogen | A-11035 | |
| DAPI | Invitrogen | D-1306 | |
| Phalloidin Dy647 | Dyomics | 647-33 | |
| siRNA Scramble | Dharmacon | D-001810-10 | |
| siRNA Met | Dharmacon | L-003156-00-0005 | |
| Black 384-well microscopy cell culture plate | Corning | 3985 | |
| AxioObserver Z1 microscope | Zeiss | 431007 9901 | |
| sCMOS camera | Andor | Neo | |
| Metamorph analysis software | Molecular Devices | 4000 | |
| CellProfiler analysis software | Broad Institute | Public software available at http://www.cellprofiler.org/ |
Riferimenti
- Pizarro-Cerdá, J., Kühbacher, A., Cossart, P. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harbor Perspectives in Medicine. 2, 1-17 (2012).
- Mackaness, G. B. Cellular resistance to infection. Journal of Experimental Medicine. 116, 381-406 (1962).
- Tilney, L. G., Portnoy, D. A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. Journal of Cell Biology. 109, 1597-1608 (1989).
- Mengaud, J., Chenevert, J., Geoffroy, C., Gaillard, J. L., Cossart, P. Identification of the structural gene encoding the SH-activated hemolysin of Listeria monocytogenes: listeriolysin O is homologous to streptolysin O and pneumolysin. Infection and Immunity. 55, 3225-3227 (1987).
- Gaillard, J. L., Berche, P., Frehel, C., Gouin, E., Cossart, P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 65, 1127-1141 (1991).
- Mengaud, J., Dramsi, S., Gouin, E., Vazquez-Boland, J. A., Milon, G., Cossart, P. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Molecular Microbiology. 5, 2273-2283 (1991).
- Kocks, C., Gouin, E., Tabouret, M., Berche, P., Ohayon, H., Cossart, P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 68, 521-52 (1992).
- Glaser, P., et al. Comparative genomics of Listeria species. Science. 294, 849-852 (2001).
- Toledo-Arana, A., et al. The Listeria transcriptional landscape: from saprophytism to virulence. Nature. 459, 950-956 (2009).
- Cossart, P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proceedings of the National Academy of Sciences USA. 108, 19484-19491 (1073).
- Mengaud, J., Ohayon, H., Gounon, P., Mege, R. -. M., Cossart, P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 84, 923-932 (1996).
- Shen, Y., Naujokas, M., Park, M., Ireton, K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 103, 501-510 (2000).
- Lecuit, M., Hurme, R., Pizarro-Cerda, J., Ohayon, H., Geiger, B., Cossart, P. A role for alpha-and beta-catenins in bacterial uptake. Proceedings of the National Academy of Sciences USA. 97, 10008-10013 (2000).
- Ireton, K., et al. A role for phosphoinositide 3-kinase in bacterial invasion. Science. 274, 780-782 (1996).
- Ireton, K., Payrastre, B., Cossart, P. The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositide 3-kinase. Journal of Biological Chemistry. 274, 17025-17032 (1999).
- Pizarro-Cerdá, J., Jonquières, R., Gouin, E., Vandekerckhove, J., Garin, J., Cossart, P. Distinct protein patterns associated with Listeria monocytogenes InlA- or InlB phagosomes. Cellular Microbiology. 4, 101-115 (2002).
- Pizarro-Cerdá, J., Payrastre, B., Wang, Y. -. J., Veiga, E., Yin, H. L., Cossart, P. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cellular Microbiology. 9, 2381-2390 (2007).
- Hamon, M. A., et al. Histone modifications induced by a family of bacterial toxins. Proceedings of the National Academy of Sciences USA. 104, 17555-17559 (2007).
- Ribet, D., et al. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature. 464, 1192-1195 (2010).
- Rämet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B., Ezekowitz, R. A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 416, 644-648 (2002).
- Agaisse, H., Burrack, L. S., Philips, J. A., Rubin, E. J., Perrimon, N., Higgins, D. E. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 309, 1248-1251 (2005).
- Cheng, L. W., Viala, J. P., Stuurman, N., Wiedemann, U., Vale, R. D., Portnoy, D. A. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proceedings of the National Academy of Sciences USA. 102, 13646-13651 (2005).
- Ripio, M. T., Domínguez-Bernal, G., Lara, M., Suárez, M., Vazquez-Boland, J. A. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. Journal of Bacteriology. 179, 1533-1540 (1997).
- Rajabian, T., et al. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nature Cell Biology. 11, 1212-1218 (2009).
- Gouin, E., et al. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IκB kinase subinit IKKα. Proceedings of the National Academy of Sciences USA. 107, 17333-17338 (2010).
- Pizarro-Cerdá, J., Lecuit, M., Cossart, P. Measuring and analysing invasion of mammalian cells by bacterial pathogens: the Listeria monocytogenes system. Methods in Molecular Microbiology. 31, 161-177 (2002).
- Snijder, B., Sacher, R., Rämö, P., Damm, E. M., Liberali, P., Pelkmans, L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 461, 520-523 (2009).

