Analysis of Apoptosis in Zebrafish Embryos by Whole-mount Immunofluorescence to Detect Activated Caspase 3
Summary
Certain genetic perturbations or exposure to toxins can disrupt normal developmental processes leading to death of specific cell types. The analysis of activated Caspase 3 by whole-mount immunofluorescence in zebrafish embryos reveals stage- and tissue-specific localization of cells specifically undergoing apoptosis.
Abstract
Whole-mount immunofluorescence to detect activated Caspase 3 (Casp3 assay) is useful to identify cells undergoing either intrinsic or extrinsic apoptosis in zebrafish embryos. The whole-mount analysis provides spatial information in regard to tissue specificity of apoptosing cells, although sectioning and/or colabeling is ultimately required to pinpoint the exact cell types undergoing apoptosis. The whole-mount Casp3 assay is optimized for analysis of fixed embryos between the 4-cell stage and 32 hr-post-fertilization and is useful for a number of applications, including analysis of zebrafish mutants and morphants, overexpression of mutant and wild-type mRNAs, and exposure to chemicals. Compared to acridine orange staining, which can identify apoptotic cells in live embryos in a matter of hours, Casp3 and TUNEL assays take considerably longer to complete (2-4 days). However, because of the dynamic nature of apoptotic cell formation and clearance, analysis of fixed embryos ensures accurate comparison of apoptotic cells across multiple samples at specific time points. We have also found the Casp3 assay to be superior to analysis of apoptotic cells by the whole-mount TUNEL assay in regard to cost and reliability. Overall, the Casp3 assay represents a robust, highly reproducible assay in which to analyze apoptotic cells in early zebrafish embryos.
Introduction
Here we describe how to perform whole-mount immunofluorescence in early zebrafish embryos to detect cells with activated Caspase 3 (referred to hereafter as the Casp3 assay). The overall goal of this method is to identify the relative abundance and location of apoptotic cells in zebrafish from the 4-cell stage to 32 hr-post-fertilization (hpf).
Both intrinsic (mitochondria-mediated) and extrinsic (death-receptor-mediated) apoptotic pathways mediate cell death through activation of the caspase cascade1-2. Initiator caspases normally exist as monomers within the cell, and once recruited to pathway-specific platforms in response to a pro-apoptotic signal, they become activated dimers. Activated initiator caspases (Caspase 9; intrinsic pathway, and Caspase 8; extrinsic pathway) then target the predimerized, inactive effector caspases (Caspase 3, 6, 7) for cleavage to form active heterotetramers. Active effector caspases then cleave cellular targets with specific tetrapeptide residues to initiate the specific proteolytic destruction that culminates in the ultrastructural hallmarks of apoptosis, such as chromatin condensation and membrane blebbing, and cell surface signals that mediate packaging of the cell for engulfment by phagocytes in an immune silent manner. In mammalian cells, active effector caspases are thought to perform the majority of cellular cleavage events that mediate apoptosis through both the intrinsic and extrinsic pathways1-2. The protocol described herein uses an antibody that specifically binds the cleaved, heterotetrameric form of Caspase 3, but not the inactive Caspase 3 dimer.
One of the advantages of the Casp3 assay is the ability to visualize the spatial distribution of apoptotic cells within the context of the whole embryo. Therefore, tissues of interest can be examined for the presence of apoptotic cells, and if antibodies or fluorescent transgenic strains exist that label specific cell types, then it is possible to pinpoint the exact identity of cells undergoing apoptosis through colabeling experiments. Several groups have successfully used activated Caspase 3 as a marker for apoptosis in zebrafish embryos (e.g. 3-7). Additional whole-mount techniques to identify apoptotic cells in zebrafish embryos include the TUNEL assay8 and acridine orange (AO) staining9. We have used all three methods to identify apoptotic cells in zebrafish embryos4,6,10.
TUNEL (Terminal deoxynucleotidyl transferase dUTP nick end labeling) staining takes advantage of the Terminal deoxynucleotidyl Transferase (TdT) enzyme that is used to add nucleotides to 3’ hydroxyl ends of DNA during VDJ recombination in lymphocytes11. TdT adds dUTP-biotin to free 3’ hydroxyl groups in the DNA, which are abundant primarily in apoptotic cells. Tissues are then exposed to avidin-peroxidase, and apoptotic cells are stained upon addition of the substrate 3-amino-9-ethylcarbazole. The TUNEL assay was used to perform an extensive analysis of apoptotic cells during embryonic development in wild-type zebrafish tissues12. Similar to the Casp3 assay, the TUNEL assay is only performed on fixed embryos. We have found the Casp3 assay to be more consistent and less expensive than the TUNEL assay.
AO staining was first described as a mechanism to specifically identify apoptotic cells in Drosophila13. AO stains both the cytoplasm and nucleus of apoptotic, but not necrotic, cells. AO is well known to bind nucleic acids14, and this is thought to account for the nuclear staining. However, AO does not stain the chromatin of live cells; rather, it appears that specific changes occurring during apoptosis permit intercalation and staining of condensed apoptotic nuclei15. AO staining has the advantage of rapid apoptotic analysis of live embryos. However, documentation of samples must be performed quickly due to the dynamic nature of apoptotic cell formation and clearance. On the other hand, simultaneous fixation of all samples provides a snapshot of apoptosis at a given time across all samples and is not subject to this temporal variable. Thus, since analysis of apoptotic cells by AO staining is not compatible with fixed embryos, the Casp3 assay is preferable for experiments in which photographic documentation and/or quantitation is required across multiple samples.
Protocol
1. Preparation of Solutions
- Prepare 500 ml of 1x PBST (1x PBS, 0.1% Tween-20), and filter sterilize. 1x PBST can be kept at room temperature (RT) indefinitely as long as there are no signs of bacterial growth.
- CAUTION: Prepare 50 ml of 4% paraformaldehyde (PFA) in a chemical hood by dissolving powdered PFA in 1x PBS at 60 °C for 1 hr and covering in aluminum foil to protect from light. Store at 4 °C for 1 month, or at -20 °C for 2 years.
- Prepare 500 ml of fresh 1x PDT (1x PBST, 0.3% Triton-X, 1% DMSO). 1x PDT can be stored at 4 °C for up to one month.
- Prepare 50 ml of blocking buffer (1x PBST, 10% heat-inactivated fetal bovine serum, 2% bovine serum albumin). Create 10 ml aliquots, and store at -20 °C for up to 1 year. Once thawed, blocking buffer can be stored at 4 °C for up to 48 hr.
- Prepare 100 ml of 4% methylcellulose by heating 30 ml of water to 80 °C. Add 4 g of methylcellulose to the hot water, and mix using a stir bar on a room-temperature stir plate. Thorough mixing usually takes between 4 hr to overnight. Add the remainder of (unheated) water, and mix at 4 °C for 1 hr. Store at RT.
- If using embryos between tailbud stage and 32 hpf, prepare 50 ml of 10 mg/ml pronase, and store for up to 1 week at 4 °C. Otherwise, store in 10 ml aliquots at -20 °C for up to 1 year.
- Place 50 ml of 100% methanol at -20 °C.
- Prepare 60x egg water by dissolving 280 g Instant Ocean Sea Salt in 2 L of distilled water (as in Westerfield16). Prepare 1x egg water by diluting the 60x stock in deionized water.
2. Preparation of Embryos
- Collect zebrafish embryos in a Petri dish in 1x egg water between the 4-cell stage and 32 hpf.
- For embryos between the 4-cell and tailbud stages, proceed to step 2.2.
- For embryos between tailbud stage and 32 hpf, remove chorions by adding 100 μl of 10 mg/ml pronase to embryos in 10 ml of 1x egg water.
- Note: Pronase activity can be variable between batches. Thus, final pronase concentration should be optimized for dechorionation when new batches are obtained.
- Let incubate at room temperature for 30 min.
- Pour off egg water / pronase solution into waste, and slowly add back fresh 1x egg water.
- Rinse 2x by pouring off existing egg water (and free-floating chorions), and gently add new egg water.
- Note: Most chorions should be removed at this point. With a transfer pipet, gently suck embryos up and down until remaining chorions are removed.
- Transfer up to 40 embryos to a 1.5 ml tube. Rinse 2 times with 1 ml of 1x PBST.
- Remove PBST, leaving liquid just above embryos (approximately 100 μl volume should remain in the tube). Add 1 ml of 4% PFA. Lay tubes on their side (so embryos spread out along length of tube) in an appropriate-size container for ease of transport (e.g. a Petri dish or a pipet box top), and place on a gentle rocker at 4 °C overnight or up to a week.
- For embryos between the 4-cell and tailbud stages, manually dechorionate the fixed embryos with forceps.
3. Immunofluorescence Procedure
- Place tubes in a tube rack so that all embryos sink to the bottom of the tube. Remove PFA (to appropriate waste container) leaving liquid just above level of embryos. Following fixation (step 2.3), embryos should never be exposed to air in order to prevent background antibody staining.
- Add 1 ml of ice-cold 100% methanol (to permeabilize) drop-wise. Embryos are now fragile after the fixation, and extra care must be taken to prevent them from tumbling within the tube during all subsequent removal/addition of liquids. Lay tubes on their sides, and place at -20 °C for 2 hr. Alternatively, embryos can be left at -20 °C for up to 24 hr.
- Place tubes in a rack to collect embryos at the bottom of the tube. Remove methanol, leaving liquid just above level of embryos.
- Carefully add 1 ml 1x PDT. Remove PDT, and add 1 ml of fresh 1x PDT. Lay tubes on their sides and gently rock at RT for 30 mins. Remove PDT, and repeat 30 min wash with fresh PDT.
- Remove PDT until liquid is just above the level of the embryos, and add 500 μl blocking buffer.
- Lay embryos on their side in a transport container, and ensure that embryos do not travel more than 2/3 up the length of the tube. Otherwise, embryos get caught in the lid of the tube and outside of the liquid.
- Carefully place transport container with tubes on a very-gentle rocker at RT for 1 hr.
- Stand tubes upright in a tube rack, and open all tubes to check for embryos stuck on the lid. For any tubes with embryos on the lid, close the top, and gently rock the tube by hand to bring all embryos back into the liquid.
- Add 1 μl rabbit anti-activated Caspase 3 antibody (1:500 dilution).
- If performing a colabeling experiment, simply add the appropriate dilution of the second nonrabbit-derived antibody in addition to the anti-activated Caspase 3 antibody.
- Carefully lay embryos on their side, and gently rock at room temperature for 2 hr. Alternatively, embryos can be incubated at 4 °C overnight.
- Stand tubes in a tube rack. Remove block/antibody solution. Add 1 ml of 1x PDT. Be sure to invert tube by hand to collect any embryos stuck on the lid of the tube. Then, open all tubes to ensure all embryos have been removed from the lid.
- Repeat step 3.4.
- Repeat steps 3.5-3.10, but substitute the anti-activated Caspase 3 antibody with the appropriate dilution of an anti-rabbit fluorophore-conjugated antibody that absorbs blue light and emits green light. However, any fluorophore-conjugated antibody with a user-preferable absorption/emission spectra can be used here. After addition of the secondary antibody, it is important to keep embryos in the dark for the remainder of the assay. Cover the transport container with aluminum foil during the incubation steps, but do not worry about light exposure when changing buffers.
- If performing a colabeling experiment, simply add the appropriate dilution of a complementary fluorophore-conjugated secondary antibody at this step.
- Note: If the user wishes to colabel active-Caspase 3 with Gfp-expressing cells, then the rabbit anti-active Caspase 3 antibody should be followed here by a nongreen-fluorescing anti-rabbit secondary antibody, and the anti-GFP antibody should be followed by a green-fluorescing nonrabbit secondary antibody. This will eliminate confusion if some Gfp fluorescence remains visible after fixation of embryos.
4. Analysis of Apoptotic Cells by Fluorescence Microscopy
- Analyze embryos in 1x PDT in a small Petri dish or glass spot plate under a fluorescent microscope using the appropriate light filter. If positive controls have bright fluorescence with minimal background, then proceed to step 4.2. (If not, consult the troubleshooting section in the Discussion, and repeat the protocol with new embryos.) Embryos can be stored at 4 °C in 1x PDT for up to 48 hr.
- Pour 4% methylcellulose in a 3.5 cm Petri dish, just enough to cover the bottom of the dish. Alternatively, a glass spot plate or depression slide can be used with a large drop of methylcellulose in the depression. Carefully suck embryos from the bottom of the tube into a clean transfer pipet and transfer to the Petri plate for visualization.
- Orient an embryo of interest for documentation using an angled probe. There will likely be some variability between embryos from the same sample, so it is important to take images that represent fluorescence intensity and pattern observed in the majority of the embryos from that sample. Documentation should be performed within 48 hr of completing the assay to prevent accumulation of non-specific background fluorescence. Residual 4% methylcellulose will not interfere with subsequent PCR reactions required for genotyping of embryos.
- Note: To aid in immobilization of an embryo at a certain angle, it is helpful to gently push the embryo to the bottom of the dish so that the yolk or some other part of the embryo (depending on the desired view) is resting on the bottom of the dish. Then, use the probe to move either the embryo or the immediately surrounding methylcellulose (while simultaneously visualizing under the microscope) to obtain the desired angle for documentation.
Representative Results
The Casp3 assay gives clear evidence of apoptotic cells from the 4-cell stage to 32 hpf. Figure 2A shows that injection of embryos with high concentrations of mRNA encoding potent pro-apoptotic bh3-only proteins, such as Bim or Noxa, quickly gives rise to active-Caspase 3 by the 4-cell stage of development. Embryonic death subsequently occurs within 1 hr4. Note that intensity of fluorescence is moderate, but results are conclusive. At older stages, fluorescence intensity is more robust. For instance, Figure 2B shows bright active-Caspase 3-positive cells in sphere-stage embryos that have been injected with moderately toxic levels4 of mRNA encoding the pro-apoptotic bh3-only gene puma. Messenger RNA encoding the anti-apoptotic bcl-2 gene was coinjected to prove that injection of puma mRNA induces apoptosis through the intrinsic, mitochondria-mediated pathway.
IR-induced apoptosis in zebrafish requires p53-mediated transcription of bh3-only genes such as puma5-6. The delay in IR-induced apoptosis in Figure 3 is consistent with the requirement for p53-mediated transcription.
The Casp3 protocol should give rise to a robust signal-to-noise ratio. Figure 4 compares the optimal Casp3 assay result to two common suboptimal results that arise when critical steps are compromised (see Discussion): A weak signal-to-noise ratio due to background fluorescence in animal tissue and yolk fluorescence that distracts from the signal-of-interest.
Colabeling experiments using transgenic GFP-expressing lines approximate tissues undergoing apoptosis (Figure 5).
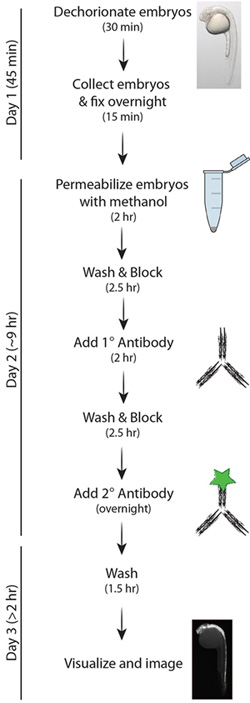
Figure 1. Simplified schematic of the Casp3 protocol.
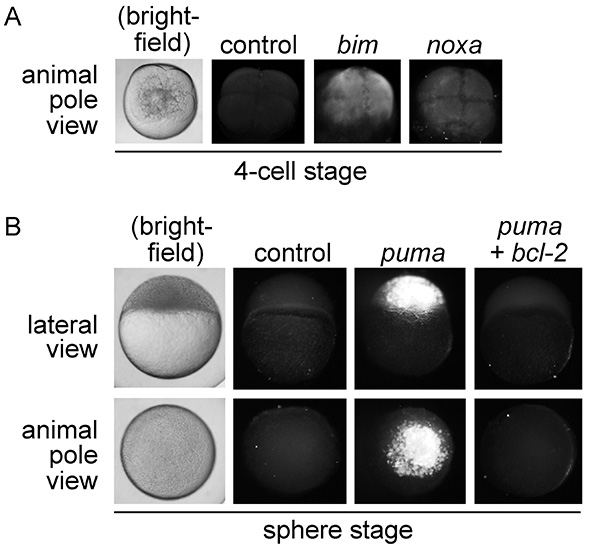
Figure 2. (A) Analysis of apoptosis in 4-cell stage embryos. Embryos were injected at the one-cell stage of development with 50 pg of mRNA encoding egfp (control) or the pro-apoptotic bh3-only genes bim and noxa. At the 4-cell stage (approximately one hr-post-injection, hpi), embryos were analyzed by immunofluorescence to detect activated Caspase 3. Bim and noxa mRNA induce rapid apoptosis in zebrafish embryos immediately following injection which, if embryos are left to develop, leads to death by 2 hpi4. A brightfield image of a wild-type 4-cell stage embryo is shown for comparison. (B) Analysis of apoptosis in sphere-stage zebrafish embryos. Embryos were injected at the one-cell stage of development with mRNA encoding either 50 pg egfp (control), 5 pg puma plus 45 pg egfp (puma), or 5 pg puma plus 45 pg bcl-2 (bcl-2). At sphere stage (approximately 4 hpi), embryos were analyzed by the Casp3 assay. Both lateral and animal pole views are shown to visualize apoptotic cells. Rescue of puma-induced apoptosis by bcl-2 indicates that injection of puma mRNA induces apoptosis specifically through the intrinsic, mitochondria-mediated pathway. A brightfield image of a wild-type sphere-stage embryo is shown for comparison.
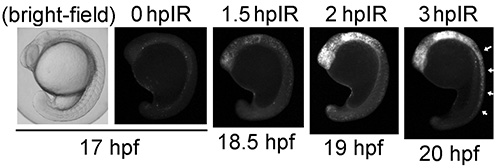
Figure 3. Time course of ionizing radiation (IR)-induced apoptosis between 17-20 hpf. Zebrafish embryos were irradiated with 8 Gy IR at 17 hpf and analyzed at 0, 1.5, 2, and 3 hr-post-IR (hpIR) by the Casp3 assay. Arrows at 20 hpf show accumulation of apoptotic cells in the spinal cord at 3 hpIR. A brightfield image of a 17-hpf embryo is shown for comparison.
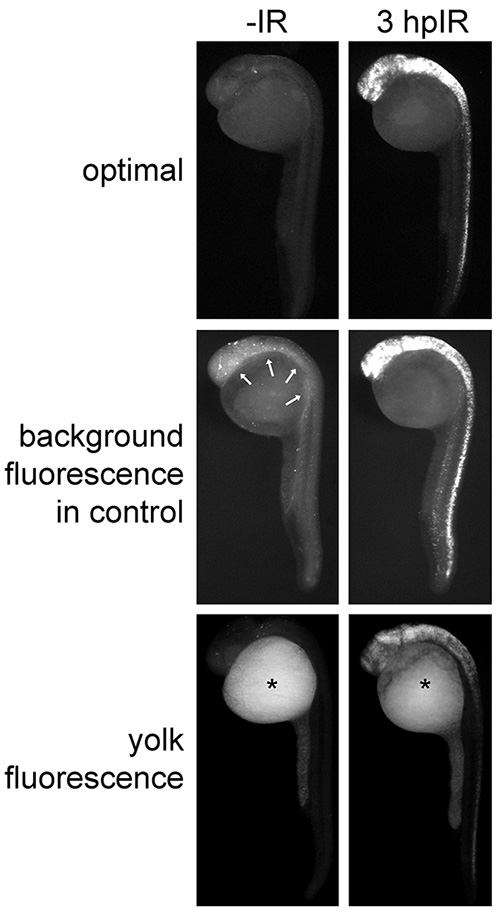
Figure 4. Optimal and sub-optimal Casp3 immunofluorescence results on 27-hpf embryos. Zebrafish embryos were irradiated (or not) at 24 hpf with 15 Gy and analyzed by the Casp3 assay. The “optimal” group shows ideal results in which apoptotic cells brightly fluoresce, and nonapoptotic cells and the yolk show minimal background fluorescence. Arrows in the “background fluorescence in control” group denote excessive background fluorescence in the animal tissue of control embryos. Asterisks in the “yolk fluorescence” group demonstrate excessive nonspecific fluorescence in the yolk.
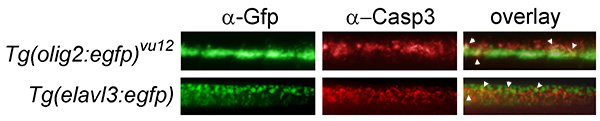
Figure 5. Colabeling experiments approximate tissue specificity of IR-induced apoptosis. The Tg(olig2:egfp)vu12 transgenic line labels motor neurons and oligodendrocytes17, and the Tg(elavl3:egfp) line labels all differentiating neurons18. Each transgenic line was irradiated with 15 Gy at 24 hpf and analyzed 3 hr later using both mouse anti-GFP and rabbit anti-active-Caspase 3 primary antibodies, followed by green- and red-fluorescing secondary antibodies, respectively. A magnification of the spinal cord (located dorsal to the end of the yolk extension of 27-hpf, 3-hpIR embryos) is shown. Apoptotic cells appear to be partially localized to both Tg(elavl3:egfp) and Tg(olig2:egfp)vu12 populations (see arrowheads in overlay), suggesting that neurons and oligodendrocytes are sensitive to IR-induced apoptosis.
Discussion
The Casp3 assay as presented in this protocol is meant to be straightforward, and if performed as instructed herein, should give rise to a robust signal-to-noise ratio. The critical steps of this protocol include the use of fresh 1x PDT buffer for all washes, sufficient length of wash steps in PDT buffer following incubation of embryos in the primary antibody, gentle handling of embryos, and meticulous accounting for all embryos within tubes or transfer pipets during all steps of the protocol.
In our experience, the following are the most common reasons for lack of a signal on positive controls: 1) 1x PBST is substituted for 1x PDT during washing steps, 2) 1x PDT buffer is older than one month, 3) the wrong secondary antibody was used, 4) embryos are older than 32 hpf and/or 5) more than 40 embryos were analyzed in one tube. When inconsistent results or variability arises within samples, this is usually because 1) care was not taken to ensure that all embryos remained within liquid at all times during the procedure, and/or 2) embryos were not incubated under sufficient rocking. High levels of background noise are often visualized as a general fluorescent haze arising throughout the animal tissue of the embryo (see Figure 4, “background fluorescence”). A poor signal-to-noise ratio can arise when 1) embryos were not washed for sufficient time following the primary antibody, and/or 2) embryos were left in PFA or methanol for longer than the recommended time. If embryos fail to remain intact by the end of the assay: 1) solutions were not added drop-wise following fixation with PFA, and/or 2) rocker speed during incubation steps was set too high.
The whole-mount visualization inherent in the Casp3 assay is helpful for spatial localization of apoptotic cells. However, tissue-specificity is approximate and will likely rely on colabeling techniques (see Figure 5) and/or embryo sectioning to pinpoint the exact identity of cells undergoing apoptosis. In the protocol described here, colabeling is limited by the requirement for the specific rabbit-derived antibody used in our assay. We have not identified a commercially available nonrabbit-derived anti-activated Caspase 3 antibody that specifically labels apoptotic cells by whole-mount immunofluorescence in zebrafish embryos. Thus, cell-type specific antibodies that are raised in rabbits will not be compatible with the Casp3 assay for colabeling experiments. One alternative is to perform a TUNEL assay which requires a mouse-derived antibody to label apoptotic cells6. However, we have found that the whole-mount TUNEL assay performed on zebrafish embryos is very sensitive to a number of variables in the reaction, including the concentration of TdT enzyme, duration of embryo incubation in TdT enzyme, and the number of embryos present per tube (our unpublished observations). Thus, it is challenging to find a balance of enzyme that gives consistent interpretation of results. Therefore, a preferable alternative is to analyze apoptotic cells in transgenic zebrafish embryos that express a fluorophore (with commercially available robust antibodies) like GFP in a specific cell type. Then, colabeling experiments can be performed using a primary mouse anti-Gfp antibody followed by a green-fluorescing anti-mouse secondary antibody. See Figure 5 for an example.
While the Casp3 protocol gives robust, reliable staining of apoptotic cells in embryos from the 4-cell stage through 32 hpf, it has limited application beyond 32 hpf due to the inability of the antibodies to penetrate the skin of older embryos. Additional techniques to permeabilize the embryos, including treatment with collagenase, trypsin, acetone, or ethanol, could potentially be optimized to adapt this assay to the analysis of older embryos. Staining with AO, however, can be performed on live zebrafish up to at least 5 days-old9. Thus, the AO assay circumvents the embryonic stage restriction and represents a rapid assay to analyze apoptotic cells (1-2 hr). By contrast, the Casp3 assay requires overnight fixation of embryos and takes 2-4 days to complete. The disadvantages of the AO assay include that efficient photographic documentation of embryos can be challenging with multiple samples since embryos are alive and apoptotic cells are rapidly formed and cleared. This can be especially problematic if embryos need to be mounted to obtain a specific orientation for documentation. Additionally, we have found that AO staining diminishes after fixation with PFA, so it is not compatible with colabeling experiments involving immunofluorescence.
Quantitative analysis of activated-Caspase 3 can be performed utilizing software programs that measure fluorescence intensity of documented images. We have quantified IR-induced apoptosis in the neural tissue, a method we have described in detail elsewhere10. Depending on the experiment, quantitative analysis of activated-Caspase 3 may require further manipulation of embryos, such as flat-mounting, to obtain proper orientation for optimal tissue analysis. In addition, the linearity of the analysis will depend on the thickness of the tissue being analyzed. For instance, we analyzed IR-induced apoptosis in neural tissue by measuring active-Caspase 3 immunofluorescence in the shallow neural tissue of the spinal cord rather than the neuron-dense tissue of the brain4,10.
In our experience, the Casp3 assay is the most robust and reliable assay available to analyze apoptosis in zebrafish embryos between the 4-cell stage and 32 hpf when analysis requires ample time for documentation. When appropriate antibodies or transgenic lines are available to identify specific cells, this assay is also highly amenable to colabeling to identify the specific cell types undergoing apoptosis.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Funding was provided by the Huntsman Cancer Foundation (CJ, CT), American Cancer Society grant #RSG-13-025-01-CSM (RS) and NIH T32 GM007464-36 (SS).
Materials
| 1.5 ml tubes | Denville Scientific | c2170 | Nonhazardous. Any similar 1.5 m plastic tubes will suffice. |
| 10x Phosphate Buffered Saline (PBS) | Sigma Aldrich | P5493 | Nonhazardous. |
| a-activated-human-Caspase-3 antibody | Fisher Scientific | BDB559565 | May be harmful if inhaled or swallowed. |
| a-GFP monoclonal antibody (mouse) | Life Technologies | 33-2600 | May be harmful if inhaled or swallowed. |
| a-HuC/HuD human neuronal protein, mouse IgG2b, monoclonal 16A11 – UNCONJ | Life Technologies | A21271 | May be harmful if inhaled or swallowed. |
| Albumin, Bovine BSA Heat Shock Isolation | Amresco | CAS 9048-46-8 | Nonhazardous. Can also be purchased through Bioexpress: 0332-1006 |
| Alexa Fluor 488 Donkey anti-mouse IgG | Life Technologies | A11029 | Use at 1:200 dilution. May be harmful if inhaled or swallowed. |
| Alexa Fluor 488 Donkey anti-Rabbit IgG | Life Technologies | A21206 | Use at 1:200 dilution. May be harmful if inhaled or swallowed. |
| Alexa Fluor 568 Donkey anti-Rabbit IgG | Life Technologies | A10042 | Use at 1:200 dilution. May be harmful if inhaled or swallowed. |
| Dimethyl Sulfoxide (DMSO) | Sigma Aldrich | D8418 | Flammable. |
| Dumont #5 Inox Forceps (standard tips/straight) | Fine Science Tools | 11251-20 | Sharp! |
| FBS HyClone Fetal Bovine Serum | Thermo-Fisher | SH3091003 | Nonhazardous. |
| Fisher Scientific Ocelot Rotator | Fisher Scientific | 05-450-21 | Any rotator with manual speed control is suitable. |
| Instant Ocean Aquarium Salt | Petco | 1373684 | Nonhazardous. |
| Methanol (acetone-free) | Sigma Aldrich | M1775 | Flammable: toxic by inhalation, ingestion and contact. |
| Methylcellulose | Sigma Aldrich | M0387-1006 | Nonhazardous. |
| Paraformaldehyde | Sigma Aldrich | P6148 | Flammable: toxic by inhalation, ingestion and contact. |
| Probe (Angled 80 Short/15.5cm/0.15mm tip diameter) | Fine Science Tools | 10140-02 | Sharp! |
| Pronase | Roche | 11459643001 | Nonhazardous. |
| Pyrex Spot Plate | Fisher Scientific | 13-748B | Nonhazardous. |
| Transfer Pipets, Samco general purpose | Thermo-Scientific | 204 | Nonhazardous. |
| Triton X-100 | Sigma Aldrich | T8787 | Irritant: harmful by ingestion or contact. |
| Tween-20 | Sigma Aldrich | P7949 | Nonhazardous. Other brands include VWR #97063-872. |
Riferimenti
- Tait, S. W., Green, D. R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621-632 (2010).
- Taylor, R. C., Cullen, S. P., Martin, S. J. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231-241 (2008).
- Eimon, P. M., et al. Delineation of the cell-extrinsic apoptosis pathway in the zebrafish. Cell Death Differ. 13, 1619-1630 (2006).
- Jette, C. A., et al. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ. 15, 1063-1072 (2008).
- Kratz, E., et al. Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ. 13, 1631-1640 (2006).
- Sidi, S., et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. . Cell. 133, 864-877 (2008).
- Zhao, X., et al. Interruption of cenph causes mitotic failure and embryonic death, and its haploinsufficiency suppresses cancer in zebrafish. J. Biol. Chem. 285, 27924-27934 (2010).
- Yager, T. D., Ikegami, R., Rivera-Bennetts, A. K., Zhao, C., Brooker, D. High-resolution imaging at the cellular and subcellular levels in flattened whole mounts of early zebrafish embryos. Biochem. Cell Biol. 75, 535-550 (1997).
- Furutani-Seiki, M., et al. Neural degeneration mutants in the zebrafish, Danio rerio. Development. 123, 229-239 (1996).
- Sorrells, S., et al. Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of p53. PLoS Genet. 8, (2012).
- Gavrieli, Y., Sherman, Y., Ben-Sasson, S. A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119, 493-501 (1992).
- Cole, L. K., Ross, L. S. Apoptosis in the developing zebrafish embryo. Dev. Biol. 240, 123-142 (2001).
- Abrams, J. M., White, K., Fessler, L. I., Steller, H. Programmed cell death during Drosophila embryogenesis. Development. 117, 29-43 (1993).
- Beers Jr, R. F., Hendley, D. D., Steiner, R. F. Inhibition and activation of polynucleotide phosphorylase through the formation of complexes between acridine orange and polynucleotides. Nature. 182, 242-244 (1958).
- Delic, J., Coppey, J., Magdelenat, H., Coppey-Moisan, M. Impossibility of acridine orange intercalation in nuclear DNA of the living cell. Exp. Cell Res. 194, 147-153 (1991).
- Westerfield, M. . The Zebrafish Book. , (1993).
- Shin, J., Park, H. C., Topczewska, J. M., Mawdsley, D. J., Appel, B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 25, 7-14 (2003).
- Park, H. C., et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279-293 (2000).

