Transcriptome Profiling of In-Vivo Produced Bovine Pre-implantation Embryos Using Two-color Microarray Platform
Summary
Microarray technology allows quantitative measurement and gene expression profiling of transcripts on a genome-wide basis. Therefore, this protocol provides an optimized technical procedure in a two-color custom made bovine array using Day 7 bovine embryos to demonstrate the feasibility of using low amount of total RNA.
Abstract
Early embryonic loss is a large contributor to infertility in cattle. Moreover, bovine becomes an interesting model to study human preimplantation embryo development due to their similar developmental process. Although genetic factors are known to affect early embryonic development, the discovery of such factors has been a serious challenge. Microarray technology allows quantitative measurement and gene expression profiling of transcript levels on a genome-wide basis. One of the main decisions that have to be made when planning a microarray experiment is whether to use a one- or two-color approach. Two-color design increases technical replication, minimizes variability, improves sensitivity and accuracy as well as allows having loop designs, defining the common reference samples. Although microarray is a powerful biological tool, there are potential pitfalls that can attenuate its power. Hence, in this technical paper we demonstrate an optimized protocol for RNA extraction, amplification, labeling, hybridization of the labeled amplified RNA to the array, array scanning and data analysis using the two-color analysis strategy.
Introduction
Early embryonic loss in high-producing dairy cows is one of the main challenges in the dairy industry1,2. Bovine has become an interesting model to study human preimplantation embryo development due to their similar developmental process3,4. However, more research is required to have better understanding about genes involved in bovine early embryonic development.
After twenty years since the first microarray technology developed in 19955, the development of more sophisticated probe fabrication technology reduced printing errors and variability of array chips within and between different microarray platforms6. Improved microarray technology resulted in a widely application of this technology in clinical research7 and more recently, in early embryo quality assessment8.
The large amount of required material for microarray technology is the main reason why the microarray technology initially failed to enter a number of research fields such as early embryonic development. More recently, RNA amplification methods have been improved to linearly amplify RNA up to micrograms level from sub-nanogram starting RNA material9. There are several commercial RNA amplification kits available on the market; however, the more popular well developed kits are related to Ribo-Single Primer Isothermal Amplification10 and T7 promoter driven11 methods. The most popular antisense RNA amplification uses in vitro transcription with an oligo dT primer linking to a T7 promoter at 5' end12. This technology allows maintaining the most of representative anti-sense transcripts after linear amplification for arrays hybridization13. This method has been adapted to amplify picogram level of total RNA extracted from bovine embryo8.
Universal Linkage System (ULS) is the labeling method which directly incorporates the DNA or amplified RNA with platinum-linked fluorescent dye either Cyanine 547 or Cyanine 647, by forming a coordinative bond on N7 position of guanine14. This method was adapted in embryos research to generate more stable amplified aRNA without modification compared to the aminoallyl modified aRNA generated by enzymatic method15. Both single dye and two dyes labeling methods have been adapted using Universal Linkage System in microarray. A large microarray comparisons study was that there is a good correlation of data quality between the one- and two-color array platforms6.
Recently, both T7 promoter driven antisense RNA amplification and ULS labeling methods have been developed to provide a more reliable protocol to generate a sufficient amount of high quality labeled aRNA materials for microarray hybridization8,16. Therefore, this study provides a protocol to demonstrate some of the important steps from RNA extraction to data analysis involved in two-color microarray using Day 7 bovine embryos as an example.
Protocol
The animal part of this study was conducted at the Metabolic Research Unit of the University of Alberta, Edmonton, Canada, with all animal experimental procedures approved (Protocol # AUP00000131) by the University of Alberta Animal Care and Use Committee, and animals cared for according to the Canadian Council of Animal Care guidelines (1993).
1. Embryo Production, Isolation of Total RNA and DNase Treatment
- For animal experimental protocols and embryo collection refer in our previous publications17, 18.
- Pool and snap freeze similar stage embryos from each cow in liquid nitrogen and then keep at -80 °C until RNA extraction.
- Extract total RNA via a column-based RNA extraction kit which enables to extract RNA from pico-scale samples (kit information is available in Material List).
NOTE: Recommended kit contains: Conditioning Buffer, Extraction Buffer, 70% Ethanol, Wash Buffer 1, Wash Buffer 2, Elution Buffer, RNA purification columns with collection tubes and microcentrifuge tubes. - Add 10 µL Extraction Buffer to tube containing embryos and incubate for 30 min at 42 °C. Centrifuge tube at 800 x g for 2 min to collect cell extract into the microcentrifuge tube.
- Pipette 250 µL Conditioning Buffer onto the purification column filter membrane. Incubate the RNA purification column with Conditioning Buffer for 5 min at room temperature. Centrifuge the purification column in the collection tube at 16,000 x g for 1 min.
- Pipette 10 µL of 70% Ethanol into the mixture of RNA Extraction Buffer and embryos. Mix well by pipetting up and down. Pipette mixture into the preconditioned purification column.
- To bind RNA, centrifuge the purification column for 2 min at 100 x g and immediately follow by a centrifugation at 16,000 x g for 30 s to remove flow through.
- Pipette 100 µL of Wash Buffer 1 into the purification column and centrifuge for 1 min at 8,000 x g.
NOTE: DNase treatment is recommended if performing reverse transcription or amplification after RNA isolation. - Digest genomic DNA right after step 1.8.
NOTE: Recommended DNase kit is available in Material List. Recommended kit contains: DNase I stock solution and Buffer. - Prepare DNase incubation mixture by mixing 5 µL DNase I stock solution with 35 µL Buffer. Mix by gently inverting.
- Pipette the 40 µL DNase incubation mixture directly into the purification column membrane. Incubate at room temperature for 15 min.
- Pipette 40 µL Wash Buffer 1 into the purification column membrane and then centrifuge at 8,000 x g for 15 s.
- Pipette 100 µL Wash Buffer 2 into the purification column and centrifuge for 1 min at 8,000 x g.
- Pipette another 100 µL Wash Buffer 2 into the purification column and centrifuge for 2 minutes at 16,000 x g.
- Transfer the purification column to a new 0.5 mL microcentrifuge tube. Pipette 11 µL of Elution Buffer directly onto the membrane of the purification column. To ensure maximum absorption of Elution Buffer into the membrane, gently touch the tip of the pipette to the surface of the membrane while dispensing the elution buffer.
- Incubate the column for 1 min at room temperature. Centrifuge the column for 1 min at 1,000 x g to distribute Elution Buffer in the column, and then spin for 1 min at 16,000 x g to elute RNA.
- Use bioanalyzer instrument according to the manufacturer's instructions to evaluate the RNA quality and quantity19.
- Store the RNA sample at -80 °C until use.
2. RNA Amplification and Labeling for Microarray Analysis
NOTE: For the first time user working with RNA amplification and labeling procedures, use five ng of spike-in RNA along with the actual aRNA samples to monitor the quality control measurement of the RNA amplification and hybridization. The spike-in RNA mix contains ten in vitro synthesized, polyadenylated transcripts in predetermined ratios. When the procedure performed properly, the labeled transcripts specifically hybridize only to complementary control probes in arrays and the data can be scanned to track quality control assurance with minimal self- or cross-hybridization. More details description regarding the spiked-in control intensities and normalization procedure after scanning the array and data analysis is refer to previous publications20, 21. The following procedure is given to the routine microarray users.
- Prepare 100 pg high quality RNA sample (RNA Integrity number (RIN) value at least > 7.0) in a total volume of 10 – 11 µL.
- Amplify the RNA samples with an amplification kit being able to work with pico-scale samples (as little as 100 pg).
NOTE: Recommended amplification kit information is available in Material List and follows the manufacturer's instructions without any modification. - Use a fluorescence-based quantitation to evaluate the profile of the size range of the amplified aRNA.
- Use the spectrophotometer instrument (based on the absorbance at 260 and 280 nm) to measure the concentration of the amplified aRNA.
- Store the amplified aRNA at -80 °C until ready for labeling.
- Use 2 µg of amplified RNA for fluorescent labeling according to ULS aRNA labeling kit user guide.
NOTE: Since Cyanine 547 and Cyanine 647 dye are sensitive to photo-degradation and Cyanine 647 dye is easily degraded by ozone, the labeling procedure takes place in a minimal amount of light and under an ozone-free environment. - Turn on the ozone box (Figure 1A) until the level of ozone is 0.001 ppm.
- Do the reaction setup inside the ozone box (Figure 1B) by adding 2 µL of Cyanine 547 or Cyanine 647, 2 µL of labeling buffer, and then adjust to a final volume of 20 µL with RNase-free water under the safelight. Add darkroom filter to the light source.
- Mix gently and incubate the tubes in a thermocycler at 85 °C for 15 min. Place samples on ice for at least 1 min. Spin down to collect content of tubes before proceeding with RNA extraction kit, except steps related to DNase treatment (from 1.9 to 1.11), to clean up the Cy-dye-labeled amplified RNA.
- Use the spectrophotometer instrument to measure and determine the concentration of labeled amplified RNA and keep the labeled aRNA at -80 °C maximum for 3 days before use.
3. Microarray Hybridization and Washing
NOTE: The following important steps are adapted according to Two-color Microarray-based Gene Expression Analysis Protocol (version 6.5, May 2010).
- Add equal amount of 825 ng from each Cyanine 547- and Cyanine 647-labeled aRNA and mix with 25x fragmentation and 10x blocking buffers.
- Heat the mixture at 60 °C for 15 min and cool on ice for 1 min.
- Add equal amount of 2x hybridization buffer and mix well.
- Centrifuge at 13,000 rpm, then the mixture is ready for hybridization.
- Load 100 µL from the mixture onto the array slide and seal the slide inside the hybridization chamber.
- Load the assembled chamber into the hybridization oven for 17 h at 65 °C rotating at 10 rpm in oven.
- Wash the arrays according to protocol with ozone protection treatment by dipping into stabilization and drying solution for 30 s.
- Remove the arrays from the solution and make sure the array surface is dry with no dust particles
4. Scanning Microarray Slide
- Keep the arrays in dark place and turn on scanner until it is ready to use (15 min wait time).
- Load the array barcode on left, into the scanner and start scanning. Do the spot analysis according to the software user guide and save the data as (gpr) file format.
5. Statistical Analysis
- Download the FlexArray software http://genomequebec.mcgill.ca/FlexArray/license.php Click on “import raw data” and load the gpr files into FlexArray.
- Perform the normalization and statistical analysis if necessary by following the instruction. Note: Calculate the threshold for positive spot selection in this study as it was described previously22.
- Select the drop down manual from the plot viewer of the FlexArray to view all the hybridized spots and background signals. Note: A similar picture of spiked in controls after hybridization to the microarray can be seen in previous study8.
- Select a Loess normalization within array following by a quantile between arrays normalization process from the FlexArray drop down manual accordingly.
- Export all the normalized data from FlexArray into an Excel file for further usage.
6. Bioinformatics Analysis
The original annotation of the probe sequences from Bovine Embryo-specific Transcripts (BESTv1) microarray has been described previously8. In present study, 20,403 unique Gene Symbols (GS) were obtained (Supplement File1) through the EmbryoGENE Laboratory Information Management System (LIMS) ELMA (http://elma.embryo-gene.ca). A list of 6,765 unique genes (Supplement File2) was selected and corresponded to the all positive signals after microarray normalization process (Step 5.5) from bovine Day 7 embryos gene expression profile. Positive signal threshold was calculated based on pervious publication16 from the signal intensity of an A value.
- To perform functional analysis with PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System23, 24, open the following link (http://www.pantherdb.org/about.jsp).
- Upload a list of 6,765 embryo-specific GS to identify specific biological process during embryonic development using statistical overrepresentation test and use default setting of Bos taurus (all genes in database) as reference list25.
NOTE: This allows the identification of developmental-related processes from the GO terms that are statistically over- or under-expressed using a binomial test. - Set P-value threshold at < 0.05 as software default. Data can be downloaded and exported into Excel file for further selection16.
Representative Results
A representative result of total RNA and amplified aRNA from Day 7 bovine embryos is shown in Figure 3 and summarized in Table 1.
RNA integrity and profile can be assessed after RNA extraction. Quality assessment of RNA could be done by bioanalyzer instrument (Figure 3A) and only those samples with RIN value higher than 7.0 are qualified to be used for amplification (Table 1).
The quality and quantity of amplified RNA should be evaluated after amplification. The similarity of amplified RNA profile and appropriate size range are the main factors for quality control (Figure 3B). After the two-round amplification, the amplified RNA fragments are very similar in size, ranging between 200 and 800 bp (Figure 3B) and all aRNAs of similar size range are selected for further fluorescence labeling. Also, a successful amplification of the original total RNA will yield at least more than a thousand fold increase of aRNA (Table 1).
Fluorescence labeling efficiency can be calculated according to degree of labeling ratio. The formula is available in the ULS aRNA labeling kit user guide. The user guide suggests the degree of labeling ratio should be 1.0 – 3.6%, indicating that an average of 1 – 3.6 ULS molecules per 100 nucleotides.
The image files after hybridization and scanning can be viewed from FlexArray. A good quality scanned image after microarray hybridization and washing is shown in Figure 4A. If labeled materials are lower or higher than this range, signals will be too low to detect or high background levels will be detected after scanning (Figure 4B).
In current study, positive signal threshold was set at 7.6 (probe signal higher than 7.6 or background < 7.6 considered to be positive). Approximately 46% of the probe sets representing 20826 positive spots are indicated in red color in bovine Day 7 embryos (Figure 5). After removing unannotated and duplicate gene symbols, only 6,765 unique gene symbols is left for PANTHER analysis.
These 6,765 genes represent the unique RNA transcripts expressed in the Day 7 bovine blastocyst. In order to find the biological process specific to bovine blastocyst, PANTHER Overrepresentation Test is performed. The top 10 Gene Ontology (GO) term IDs with the highest fold enrichment index is selected (Figure 6). In general, these specific GO terms largely represent the active cellular division process such as mitosis and chromosome segregation, regulation of cell cycle and initiation of cytoplasmic and mitochondrial translation.
Figure 1: Ozone-free Box (A) and Array Labeling Reaction (B). A) Ozone Free Box consists of a catalytic converter unit which recycles the air and destroys ozone and a highly sensitive ozone sensor to monitor the ozone level inside the box during microarray performance. B) Labeling procedure and array hybridization perform inside the box to avoid the fluorescent Cyanine 647 dye degradation by ozone. Please click here to view a larger version of this figure.
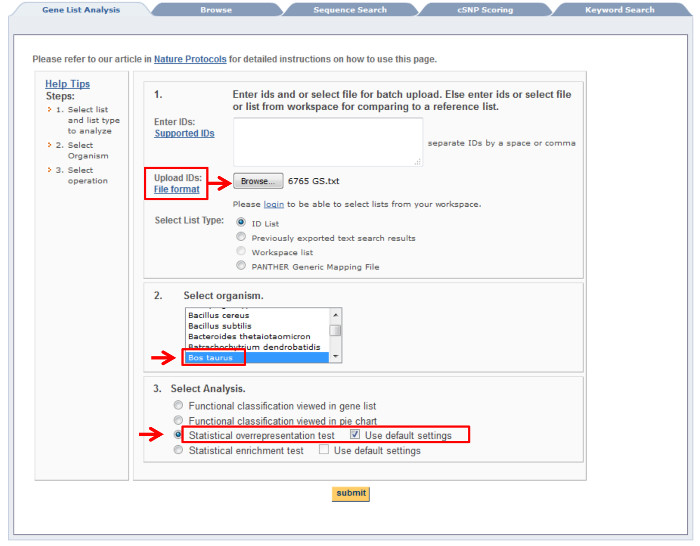
Figure 2: PANTHER Gene List Analysis. Red arrows indicate the area need to be filled. Please click here to view a larger version of this figure.
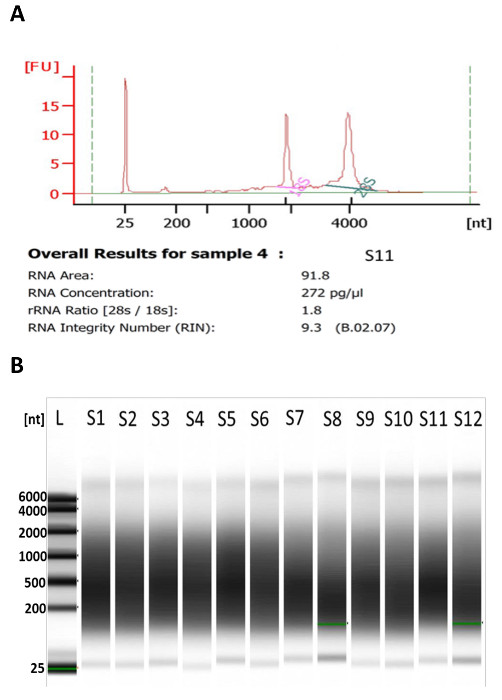
Figure 3: A Representative Bioanalyzer Electropherogram of Total RNA (A) and the Gel Image of Amplified RNA (B). A) Showing overall result with RIN value, RNA concentration and rRNA ratio. B) The Profile of amplified RNA after two-round amplification. Amplified RNA fragments are expected to be in a range between 200 and 800 bp. Please click here to view a larger version of this figure.
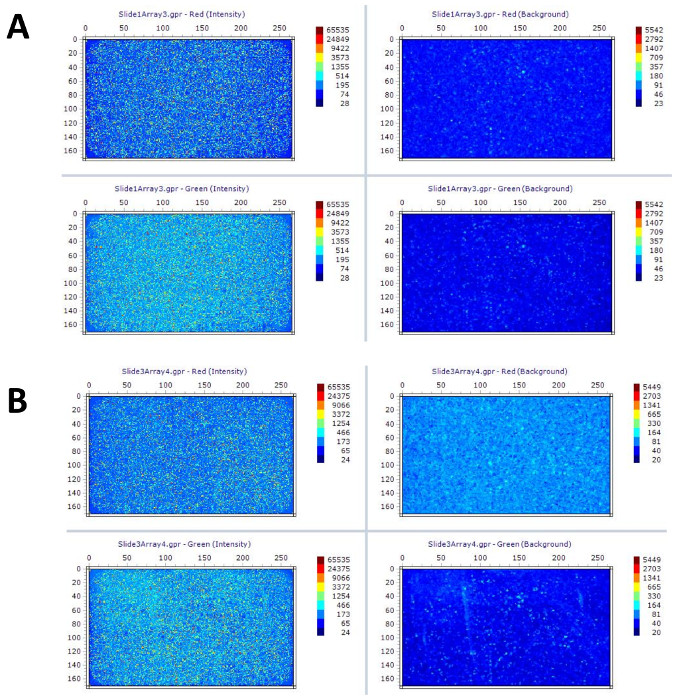
Figure 4: Image of Array Intensity Map. (A) A good image of an array slide showing green and red channel correspond to Cyanine 547 and Cyanine 647 signals after scanning with high spot intensity (indicated on the left with blue green color) and low background (indicated on the right with dark blue color). (B) A bad image of an array slide showing very high background image from red channel (indicated from the upper panel with the similar blue green color from the image of Cyanine 647 spot intensity). Color chart indicates the intensity of signal with numerical values corresponding to different colors. The value of signal intensity is the lowest in the range of dark blue color. Please click here to view a larger version of this figure.
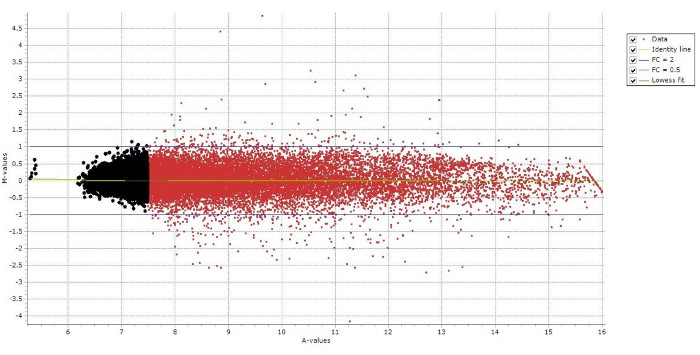
Figure 5: MA Plot for Day 7 Bovine Embryo Gene Expression Profile. 20,826 red spots represented positive signals above background signals. Background spots are highlighted in black color. Please click here to view a larger version of this figure.
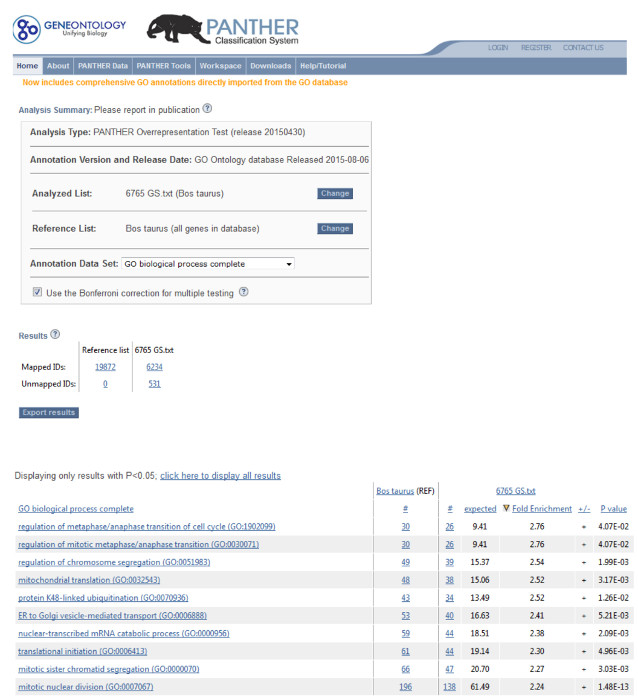
Figure 6: The Top 10 Gene Ontology (GO) Term IDs with the Highest Fold Enrichment Index. These specific GO terms largely represent the active cellular division process such as mitosis and chromosome segregation, regulation of cell cycle and initiation of cytoplasmic and mitochondrial translation. Please click here to view a larger version of this figure.
| Sample | Number of embryo | Tipo | Embryo morphological quality | RNA concentration (pg/µL) | RIN | Used total RNA for amplification (pg) | Amplified RNA concentration (ng/µL) |
| S1 | 4 | Blastocyst | Grade 1&2 | 101 | 8.6 | 858.5 | 3498.4 |
| S2 | 1 | Blastocyst | Grade 2 | 142 | 7.4 | 1207 | 1682.4 |
| S3 | 2 | Blastocyst | Grade 1 | 135 | 8.8 | 1147.5 | 3185.3 |
| S4 | 3 | Blastocyst | Grade 1 | 177 | 9 | 1504.5 | 2906.4 |
| S5 | 5 | Blastocyst | Grade 1 | 636 | 8.3 | 5406 | 3437.5 |
| S6 | 3 | Blastocyst | Grade 1&2 | 159 | 9.1 | 1351.5 | 1689.8 |
| S7 | 3 | Blastocyst | Grade 1 | 154 | 9 | 1309 | 4507.1 |
| S8 | 4 | Blastocyst | Grade 1 | 304 | 8.5 | 2584 | 3089.3 |
| S9 | 5 | Blastocyst | Grade 1 | 282 | 9.1 | 2397 | 3917.8 |
| S10 | 6 | Blastocyst | Grade 1&2 | 374 | 9.4 | 3179 | 2979 |
| S11 | 5 | Blastocyst | Grade 1 | 272 | 9.3 | 2312 | 2576 |
| S12 | 3 | Blastocyst | Grade 1 | 206 | 9 | 1751 | 2567.9 |
Table 1: Sample and RNA information. The concentration and quality of RNA should be evaluated after extraction. RNA integrity and concentration can be assessed by any bioanalyzer. RNA integrity number should be more than 7.0. The RNA concentration should be examined by spectrophotometer instrument after amplification. Embryo quality was evaluated according to the Manual of the International Embryo Transfer Society (3rd ed., Savoy, IL).
Discussion
The first problem to perform microarray analysis using Day 7 bovine embryos is not getting sufficient amounts of high quality RNA for studying gene expression. Traditional phenol/chloroform RNA extraction and ethanol precipitation method is not recommended for Day 7 embryos, resulting in low yield and possible leftover phenol inhibiting RNA amplification reaction. Instead, a standard column-based method is better to isolate total RNA and then elute the RNA with minimum elution buffer to increase the concentration. An average of 780 pg of total RNA per embryo is obtained in current study which is comparable with other reports using an identical methodology26 and the same RNA extraction kit27, 28 used presently. For RNA extraction, it is recommended to dissolve precipitate prior to use by mixing thoroughly or warm the extraction buffer vial to re-dissolve the extraction buffer prior to use. Moreover, incubation time is also important for RNA yield in extraction step. Incubation time less than a full 30 min at 42 °C reduced RNA yield.
On column DNase digestion is necessary to remove genomic DNA contamination during total RNA purification. The DNase enzyme is very sensitive to vortex and centrifuge. Therefore, DNase and buffer should be mixed by gently inverting.
The quality of the RNA is very important in microarray analysis and samples with an RIN lower than 7 should not be considered for hybridization. In current study, the total RNA profiles obtained by bioanalyzer are similar to those observed previously in studies from bovine blastocysts26, 27. It is worth to be noted that when using pre-hatched embryos, especially RNA prepared from oocytes and embryos before the maternal to embryonic transition, the RIN value is often lower than 7 compared to the total RNA from Day 7 embryos and this is due to a variation in the 28S/18S rRNA ratio27.
The amount of extracted RNA from bovine embryos is not sufficient for platform hybridization, hence, RNA amplification is required. There are several methods available for RNA amplification such as PCR, in vitro transcription (IVT), and Ribo-SPIA (single primer isothermal amplification). Although the latter method is faster and in one day provides sufficient material for arrays, it required at least 5 ng as starting material29. Hence, we chose the IVT method which is able to work with low starting materials12 (as little as 100 pg). Moreover, it has already been tested to amplify total RNA extracted from bovine embryo successfully29. In current study, our amplification method produced ~28 µg amplified RNA per embryo from 598 pg total RNA per embryo. Additionally, after the two-round amplification, our samples showed a similar profile and appropriate size range (between 200 and 800 bp) which are in agreement with previous studies13, 15.
In this protocol, we used a non-enzymatic labeling protocol, platinum-linked cyanine (Cyanine 547 and Cyanine 647) dyes. This method allows labeling of amplified or non-amplified RNA. Having amplification step prior to labeling allows using natural nucleotides, leading to higher yields amplified RNA, reduce bias and generate longer fragments compared to enzymatic method. The Cyanine 547 and Cyanine 647 dyes are common fluorescent dyes recommend for two-color array. However, Cyanine 647 dye is sensitive to ozone degradation. Hand held ozone monitor should be used to make sure the level of ozone below 2 ppb. In addition, dust free environment is necessary to avoid dust particles falling into hybridization solution or during scanning.
Microarray experiment can be designed in one- or two-color approach. Each experimental approach has some advantages and disadvantages. Using two-color designs reduces the variability and background noise, allows for direct comparison and for loop designs with defining a common reference sample. Although dye-specific biases can substantially affect results when experiments are performed using two-color designs, these biases can be mitigated by performing dye swaps. Such technical replication adds to experimental costs, but can enhance both accuracy and sensitivity in measuring differential expression31.
Compare to other bioinformatics software (such as GOrilla) PANTHER allows comparing dataset with Bos taurus genome as reference. Our results indicate that during Day 7 bovine embryonic development the following biological components and functions are involved: regulation of metaphase/anaphase transition of cell cycle, regulation of mitotic metaphase/anaphase transition, regulation of chromosome segregation, mitochondrial translation, protein K48-linked ubiquitination, ER to Golgi vesicle-mediated transport, nuclear-transcribed mRNA catabolic process, translational initiation, mitotic sister chromatid segregation, mitotic nuclear division.
The protocols presented in this paper, from fluorescent labeling of the amplified RNA to scanning of arrays should be executed with extra awareness of the above environmental factors to maintain high quality data.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Research supported by Alberta Livestock and Meat Agency, Alberta Innovates – BioSolutions, Alberta Milk, and Livestock Research Branch, Alberta Agriculture and Forestry.
Materials
| PicoPure RNA Isolation Kit | Applied Biosystems | KIT0204 | |
| RNase-Free DNase Set (50) | Qiagen | 79254 | |
| Agilent RNA 6000 Pico Kit | Agilent Technologies | 5067-1513 | |
| Arcturus RiboAmp HS PLUS Kit | Applied Biosystems | KIT0505 | |
| 2100 Bioanalyzer Instruments | Agilent Technologies | G2940CA | |
| RNA Screen Tape | Agilent Technologies | 5067-5576 | |
| ULS Fluorescent Labeling Kit | Kreatech Diagnostics | EA-021 | |
| Custom Gene Expression Microarrays | Agilent Technologies | G2514F | |
| Agilent Gene Expression wash buffer 1 | Agilent Technologies | Part #5188-5325 | |
| Agilent Gene Expression wash buffer 2 | Agilent Technologies | Part #5188-5326 | |
| 2X Hi-RPM Hybridization buffer | Agilent Technologies | Part #5190-0403 | |
| 25X Fragment buffer | Agilent Technologies | Part #5185-5974 | |
| 10X GE Blocking Agent | Agilent Technologies | Part #5188-5281 | |
| Stabilization and drying solution | Agilent Technologies | Part #5185-5979 | |
| Gasket slides enabled by Agilent SureHyb techonolgy | Agilent Technologies | G2524-60012 | Pack of 20 gasket slides, 4 microarrays/slide |
| Two-Color RNA Spike-In Kit | Agilent Technologies | Cat# 5188-5279 | |
| GenePix 4000B array scanner | Molecular Devices | GENEPIX 4000B-U | |
| Ozone Free Box | BioTray | OFB_100x200 | |
| GAL file | Agilent Technologies | – |
Riferimenti
- Royal, M. D., Smith, R. F., Friggens, N. C. Fertility in dairy cows: bridging the gaps. Animal. 2 (08), 1101-1103 (2008).
- Diskin, M. G., Murphy, J. J., Sreenan, J. M. Embryo survival in dairy cows managed under pastoral conditions. Anim. Reprod. Sci. 96 (3-4), 297-311 (2006).
- Wrenzycki, C., et al. Effects of culture system and protein supplementation on mRNA expression in pre-implantation bovine embryos. Hum. Reprod. 16 (5), 893-901 (2001).
- Menezo, Y. J., Herubel, F. Mouse and bovine models for human IVF. Reprod. Biomed. Online. 4 (2), 170-175 (2002).
- Schena, M., Shalon, D., Davis, R. W., Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 270 (5235), 467-470 (1995).
- Patterson, T. A., et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 24 (9), 1140-1150 (2006).
- Rhodes, D. R., Chinnaiyan, A. M. Integrative analysis of the cancer transcriptome. Nat. Genetics. 37, 31-37 (2005).
- Robert, C., et al. Combining resources to obtain a comprehensive survey of the bovine embryo transcriptome through deep sequencing and microarrays. Mol. Reprod. Dev. 78 (9), 651-664 (2011).
- Nygaard, V., Hovig, E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucleic Acids Res. 34 (3), 996-1014 (2006).
- Kurn, N., Chen, P., Heath, J. D., Kopf-Sill, A., Stephens, K. M., Wang, S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem. 51 (10), 1973-1981 (2005).
- Van Gelder, R. N., von Zastrow, M. E., Yool, A., Dement, W. C., Barchas, J. D., Eberwine, J. H. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. 87 (5), 1663-1667 (1990).
- Phillips, J., Eberwine, J. H. Antisense RNA Amplification: A linear amplification method for analyzing the mRNA population from single living cells. Methods. 10 (3), 283-288 (1996).
- Gilbert, I., Scantland, S., Dufort, I., Gordynska, O., Labbe, A., Sirard, M. A., Robert, C. Real-time monitoring of aRNA production during T7 amplification to prevent the loss of sample representation during microarray hybridization sample preparation. Nucleic Acids Res. 37 (8), 65 (2009).
- Gijlswijk, R. P., Talman, E. G., Jansse, P. J., Snoeijers, S. S., Killian, J., Tanke, H. J., Heetebrij, R. J. Universal Linkage System: versatile nucleic acid labeling technique. Expert Re. Mol. Diagn. 1 (1), 81-91 (2001).
- Gilbert, I., Scantland, S., Sylvestre, E. L., Dufort, I., Sirard, M. A., Robert, C. Providing a stable methodological basis for comparing transcript abundance of developing embryos using microarrays. Mol. Hum. Reprod. 16 (8), 601-616 (2010).
- Tsoi, S., et al. Development of a porcine (Sus scofa) embryo-specific microarray: array annotation and validation. BMC Genomics. 13, 370 (2012).
- Salehi, R., et al. Superovulatory response and embryo production in Holstein cows fed diets enriched in oleic, linoleic or α-linolenic acid. Reprod. Fertil. Dev. 26 (1), 218-218 (2013).
- Thangavelu, G., Colazo, M. G., Ambrose, D. J., Oba, M., Okine, E. K., Dyck, M. K. Diets enriched in unsaturated fatty acids enhance early embryonic development in lactating Holstein cows. Theriogenology. 68 (7), 949-957 (2007).
- . Agilent 2100 Bioanalyzer User’s Guide Available from: https://www.agilent.com/cs/library/usermanuals/Public/G2946-90004_Vespucci_UG_eBook_(NoSecPack) (2016)
- Kerr, K. F. Extended analysis of benchmark datasets for Agilent two-color microarray. BMC Bioinformatics. 8, 371 (2007).
- Zhu, Q., Miecznikowski, J. C., Halfon, M. S. A wholly defined Agilent microarray spike-in dataset. Bioinformatics. 27 (9), 1284-1289 (2011).
- Vallee, M., Gravel, C., Palin, M. F., Reghenas, H., Stothard, P., Wishart, D. S., Sirard, M. A. Identification of novel and known oocyte-specific genes using complementary DNA subtraction and microarray analysis in three different species. Biol. Reprod. 73 (1), 63-71 (2005).
- Thomas, P. D., et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13 (9), 2129-2141 (2003).
- Mi, H., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33, 284-288 (2005).
- Mi, H., Muruganujan, A., Casagrande, J. T., Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551-1566 (2013).
- Ross, P. J., Wang, K., Kocabas, A., Cibelli, J. B. Housekeeping gene transcript abundance in bovine fertilized and cloned embryos. Cell Reprogram. 12 (6), 709-717 (2010).
- Gilbert, I., et al. The dynamics of gene products fluctuation during bovine pre-hatching development. Mol. Reprod. Dev. 76, 762-772 (2009).
- Vallee, M., et al. Revealing the bovine embryo transcript profiles during early in vivo embryonic development. Reproduction. 138 (1), 95-105 (2009).
- Dafforn, A., et al. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques. 37 (5), 854-857 (2004).
- Beaujean, N., Jammes, H., Jouneau, A., Dufort, I., Rovert, C., Sirard, M. A. Nuclear Reprogramming. Studying Bovine Early Embryo Transcriptome by Microarray. , (2015).
- Patterson, T. A., et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 24 (9), 1140-1150 (2006).

