Embryo Microinjection and Electroporation in the Chordate Ciona intestinalis
Summary
We present transient transgenesis and gene knockdown in Ciona intestinalis, a chordate sister group to vertebrates, using microinjection and electroporation techniques. Such methods facilitate functional genomics in this simple invertebrate that features rudimentary characteristics of vertebrates, including notochord and head sensory epithelia, and many orthologs of human disease associated genes.
Abstract
Simple model organisms are instrumental for in vivo studies of developmental and cellular differentiation processes. Currently, the evolutionary distance to man of conventional invertebrate model systems and the complexity of genomes in vertebrates are critical challenges to modeling human normal and pathological conditions. The chordate Ciona intestinalis is an invertebrate chordate that emerged from a common ancestor with the vertebrates and may represent features at the interface between invertebrates and vertebrates. A common body plan with much simpler cellular and genomic composition should unveil gene regulatory network (GRN) links and functional genomics readouts explaining phenomena in the vertebrate condition. The compact genome of Ciona, a fixed embryonic lineage with few divisions and large cells, combined with versatile community tools foster efficient gene functional analyses in this organism. Here, we present several crucial methods for this promising model organism, which belongs to the closest sister group to vertebrates. We present protocols for transient transgenesis by electroporation, along with microinjection-mediated gene knockdown, which together provide the means to study gene function and genomic regulatory elements. We extend our protocols to provide information on how community databases are utilized for in silico design of gene regulatory or gene functional experiments. An example study demonstrates how novel information can be gained on the interplay, and its quantification, of selected neural factors conserved between Ciona and man. Furthermore, we show examples of differential subcellular localization in embryonic cells, following DNA electroporation in Ciona zygotes. Finally, we discuss the potential of these protocols to be adapted for tissue specific gene interference with emerging gene editing methods. The in vivo approaches in Ciona overcome major shortcomings of classical model organisms in the quest of unraveling conserved mechanisms in the chordate developmental program, relevant to stem cell research, drug discovery, and subsequent clinical application.
Introduction
Recently, genome sequence and transcribed gene repertoires have become accessible in a number of model organisms. Determining gene functional links, however, and how these connections are hardwired in the DNA to execute transcriptional activity throughout embryonic time and space, remain a major challenge, especially in vertebrates. The complexity of regulatory interactions and the complexity of genomes1,2, notably in vertebrates, slow down efficient functional analyses, in particular at the DNA level in vivo. Indeed, no GRN is currently analyzed from fertilization to the final, terminally differentiated state of the developing organism.
The ascidian C. intestinalis represents a model organism where such complete GRN analyses may be possible. It has an unduplicated genome typical of invertebrates with little gene redundancy. Vertebrates, in contrast have undergone two rounds of genome duplications that have generated larger gene families and functional diversification but also redundancy between paralogs. The compact cis-regulatory sequences (the control units of GRNs) of C. intestinalis, and an invariable embryonic lineage with few cells are reminiscent of C. elegans. Furthermore, its unique phylogenetic position as an invertebrate sister group to vertebrates within chordates allows developmental observation, at cellular resolution, of the forming chordate body plan3-6.
The key issue that ensures the future success of functional genomics in model organisms is the generation of large numbers of embryos for quantitative in vivo perturbations. The development of the microinjection technique in Ciona eggs7, and the possibility to quickly obtain many transiently transgenic animals by in vivo electroporation in hundreds of Ciona zygotes8,9 has been a breakthrough in Ciona functional genomics. Here, we first present the more laborious microinjection technique for targeting individual genes that remains the method of choice for morpholino (MO)-mediated gene knockdown, notably of maternal transcripts. MO oligos typically target the initiator ATG or the splice sites of the RNA and interfere with protein translation. We then show how electroporation in fertilized eggs of Ciona allows for large numbers of embryos to be analyzed in a quantitative fashion, thus opening avenues of efficient analysis of enhancers in vivo and of tissue-specific gain- or loss-of function approaches, with the possibility for simultaneous manipulations and analyses. We further demonstrate how Ciona community tools are utilized for in silico prediction of regulatory regions and efficient cloning of full open reading frames (ORFs) in expression vectors. Finally, we demonstrate differential subcellular localizations of fluorescently tagged or labeled proteins upon overexpression by electroporation.
Protocol
1. Preparations for Microinjection and Electroporation in Ciona Eggs and Zygotes
- Prepare 10 L artificial seawater with HEPES (ASWH) for culturing of embryos. Dissolve 420 mM NaCl, 9 mM KCl, 10 mM CaCl2·2H2O, 24.5 mM MgCl2·6H2O, 25.5 mM MgSO4·7H2O, and 2.15 mM NaHCO3 in double distilled water (ddH2O) by stirring overnight. Add 5 mM HEPES pH 8.0, and verify/adjust the pH to 8.0. Store the ASWH at 4 °C. Warm it up to 18 °C and filter it with filter paper before use.
- Prepare 20x inactivated dechorionation solution: 20% sodium thioglycolate, 1% pronase in ASWH. Store 500 µl aliquots at -20 °C, and dilute to a final volume of 10 ml ASWH before use.
- Prepare 15 to 20 culture dishes for the eggs/embryos. Pour melted 1% agarose in ASWH into 3.5, 5, 9 or 15 cm petri dishes, coating them with a 1-2 mm thin layer. Let the agarose solidify and cover the plates with ASWH. Stock them at 4 °C for 1-2 weeks maximum. Replace the ASWH before use.
- Prepare special injection dishes.
- Pour a 5-7 mm thick layer of melted 1% agarose into a 5 cm Petri dish, and place a mold on the agarose (such as a zip lock closing glued to a plastic cover slip) to produce an indentation upon solidification of the agarose that can hold the eggs. Prepare 4 to 5 injection dishes and cover with ASWH. Store at 4 °C and replace with fresh ASWH before use. Use only fresh plates for injection (maximum one day old).
- Prepare β-galactosidase (LacZ) staining buffer: 1 mM MgCl2·6H2O, 3 mM K4[Fe(CN)6]·3H2O, 3 mM K3[Fe(CN)6] in phosphate-buffered saline (PBS) with Tween (PBT) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.05% Tween). Store in the dark at 4 °C. Prepare LacZ substrate 5-Bromo-4-chloro-3-indolyl ß-D-Galactopyranoside (X-gal, 10 U/ml) in aliquots of 40 mg/ml in dimethylformamide (DMF) and store stocks at -20 °C until use.
- Soak Pasteur pipettes in tap water overnight to prevent the embryos from sticking.
- Prepare injection needles.
- Set up a micropipette puller at conditions such as heat 425, pull 75, velocity 75 and time 200 (from a filament ramp test of 415).
- Pull 4 to 5 filament-containing glass capillaries to produce 8 to 10 long, fine needles normally closed at the tip. Store the needles in a dust free needle box, attached to double adhesive tape on an elevated support, to avoid breaking the tips and which is convenient for needle filling.
NOTE: Test the obtained needles by injecting a few eggs with green vital dye (as described in section 6) and adjust the needle pulling conditions to obtain a tip shape that allows for fine-tuned and repetitive injection as described in 6.6.
- Prepare injection solution. Prepare 4 µl of injection solution, e.g., composed of 1 µl of 2 mM MO, 2 µl of 10 µg/µl green vital dye and 1 µl ddH2O. Add a final concentration of 0.05-0.5 µg/µl capped mRNA or 20 ng/µl of plasmid DNA according to the experimental question. Mix well. Keep on ice if the solution contains mRNA.
2. Collection of Gametes
- Dissect Ciona adults in an 18 °C cooled embryo room. Use small scissors and start from the end that opposes the siphons on the side of the exhaling (shorter) siphon under which the oviduct and the sperm duct run.
- Expose the oviduct and delicately make an incision in the oviduct using the tip of the scissors. Drop the outflowing eggs directly into a 6-well plate containing ASWH by gently pressing the oviduct with the closed scissors stripping off the eggs. Transfer the remaining eggs with a Pasteur pipette pre-rinsed with ASWH.
- Deposit the eggs from different animals in different wells. Observe the egg quality under the dissecting scope.
NOTE: Well developed eggs are round and pink to slightly yellow in color. They are surrounded by a non-cellular vitelline coat, the chorion that forms a well-defined perivitelline space around the egg. The chorion is associated with test cells at its inside and star-shaped follicle cells at its outside. Eggs of low quality often lack the perivitelline space or the follicle cells. They appear less round, more granular or different in color. Immature oocytes are smaller and have a silver white color. - Cut the sperm duct with the scissors and collect concentrated sperm into a 1.5 ml tube using a separate Pasteur pipette. Pool the sperm from different animals (at least two) into the same tube. Store sperm at 4 °C for several days.
3. Fertilization
- Collect sperm and eggs at 18 °C as described in step 2.
- Activate the sperm.
- Prepare 1 ml ASWH in a 1.5 ml tube and mix with 50 µl of 1 M Tris pH 9.5. Then add 20 µl of concentrated sperm and gently mix by inverting the tube.
- Verify the sperm activation in a drop of sperm placed onto a cover of a small Petri dish or a glass slide and observe under the dissecting scope. The spermatozoids should be swimming frantically.
- Add 100-200 µl of the activated sperm solution to each well of eggs (containing between 100 and 1,000 eggs), mix well by pipetting up and down, so the eggs are floating in the medium. Wait for 10 min without moving the embryos.
4. Dechorionation of Eggs and Embryos
- Dilute a 500 µl aliquot of 20x dechorionation solution to 10 ml in ASWH. Activate the dechorionation solution by adding drop-wise 200 µl of 2.5 M NaOH to the 10 ml 1x dechorionation solution. A milky precipitate will form. Gently mix by inverting the tube.
- Collect the eggs or zygotes (after the 10 min wait in step 3.3) into glass tubes using a Pasteur pipette. Pool the eggs from 2 to 3 animals in one tube (around 1,000-5,000 eggs/tube). Sediment with a hand centrifuge by spinning at high speed (1,200 x g) for about 20 sec to form an embryo pellet at the bottom of the tube. Slowly stop the centrifuge and remove the ASWH with a Pasteur pipette.
- Add 4 ml of activated dechorionation solution to the pelleted eggs/zygotes in each tube. Suspend the eggs/zygotes by gently pipetting them up and down using a tap water-treated Pasteur pipette topped with a small rubber pear. The solution should turn yellowish after 1-3 min.
- Follow the dechorionation by removing a small aliquot of the dechorionating egg/zygote suspension using the Pasteur pipette. Deposit a drop on a slide and observe under the dissecting scope. Keep pipetting eggs/zygotes up and down and check every 20-30 sec.
NOTE: First follicle cells detach, then the chorion turns yellow and opaque, and finally it detaches from the zygotes. Pink dechorionated eggs/zygotes will sink to the bottom of the glass tube. The dechorionation should not take more than max. 5 min. - Fill the tube with ASWH once 50% of eggs/zygotes are dechorionated.
NOTE: Very gently centrifuge (8 x g) for about 10-15 sec corresponding to a very low speed just enough to sediment the dechorionated eggs/embryos. Slowly stop the centrifuge as to not perturb the pellet of eggs/zygotes at the bottom of the tube. - Remove nearly all the liquid from the glass tube including the floating material with incompletely dechorionated eggs/zygotes and replace with ASWH. Slowly pipet up and down to wash and gently centrifuge again as in step 4.2. Alternatively, wait for zygotes to settle down by gravity. Wash once more until no chorion debris is left.
- Transfer eggs/zygotes to freshly rinsed culture dishes using a Pasteur pipette. Keep the zygotes (not eggs) at low density (around 200 embryos per 10 cm dish) to avoid their sticking together. Culture the embryos to the desired stage at temperatures between 13 and 20 °C. Alternatively, electroporate the zygotes (Step 5) or inject the unfertilized eggs (Step 6).
5. Electroporation
- Fertilize the eggs and dechorionate the zygotes at 18 °C as in sections 3 and 4. Provide between 50 and 400 eggs per electroporation sample.
- Prepare 50 µl plasmid DNA in a 1.5 ml tube (max. 100 µg plasmid DNA in 50 µl ddH2O) and add 200 µl 0.95 M mannitol. Mix well by vortexing.
- Dispatch and gravity sediment the dechorionated zygotes in siliconized 1.5 ml tubes. Remove the ASWH down to the 100 µl mark on the tube.
- Proceed, consecutively, for each zygote tube as follows: add DNA/mannitol solution using a Pasteur pipette. Gently mix with the zygotes and instantly take up the DNA/mannitol/zygote suspension and transfer to a 4 mm electroporation cuvette.
- Place the cuvette immediately into the electroporation holder and give a single pulse of 16 msec at 50 V.
- Remove the zygotes from the cuvette using the same Pasteur pipette and spread them out on a culture dish containing fresh and filtered ASWH. Pipette the zygotes before they start cleaving around 1 hour post fertilization (hpf) at 18 °C.
- Rinse the pipette between the samples in a beaker of ASWH.
- Culture the zygotes at 15-20 °C at low enough density of around 200 embryos per 10 cm dish to avoid their sticking together.
6. Microinjection
- Prepare Dechorionated Eggs for the Injection.
- Dechorionate the eggs separately, from 2-4 animals, as described in section 4. Keep the unfertilized and dechorionated eggs in separate small agarose coated culture dishes. Wash the eggs several times in the dishes to remove the debris.
- Test fertilize small batches of the dechorionated eggs (around 50) from each individual. Use a separate 5 cm dish for each batch and apply 2 µl activated sperm (step 3.2). Mix well by swirling the dishes and spread out the eggs in the dish by moving the dishes sideways. Keep the sperm and eggs together for about 10 min without moving.
- Eliminate a maximum of sperm as follows: transfer the fertilized eggs to a fresh culture dish or rinse twice with fresh ASWH. Leave the cleaving embryos unperturbed till the 32-cell stage (at 18 °C for about 3 hours post fertilization). Choose eggs from the best developing batch (best percentage of fertilization and most regular cleavage pattern) for the microinjection.
- Set up the Injection Needles.
- Backfill 4 injection needles in a vertical position to allow for gravity flow along the filament. Deposit 0.5 µl injection solution (step 1.8) containing green vital dye on the back end of the needles that are positioned needle tip down. Wait for the liquid to fill the needle tip at the bottom.
- Reposition the needles horizontally and backfill them with mineral oil using a fine and long metal or plastic pipette tip. Slowly overlay the injection solution with mineral oil expelling all the air bubbles while filling the needle.
- Setup the Micromanipulator in an 18 °C Cooled Room.
- Connect the plastic tubing of the needle holder to a 10 ml glass syringe filled with mineral oil. Backfill the tubing and the needle holder with oil and expel any air bubbles.
- Insert the needle in the needle holder and position the holder on the micromanipulator.
- Adjust the needle holder movement along a straight line at an angle of 45° relative to the surface.
- Orient the dechorionated eggs along the agarose indentation of the injection dish so that the eggs can be injected one by one under the dissection microscope.
- Break the needle tip, if necessary, by gently pushing the needle against the agarose or against a piece of glass cover slip placed on the agarose. Slightly pressure the syringe knob to verify that the needle tip is open (green needle contents will expel).
- Inject the unfertilized eggs one by one by first introducing the needle into the egg and slightly aspirating to break the egg membrane followed by injection. Inject green injection solution into the middle of the egg to a maximum of 1/3 of the cell diameter. (This corresponds to approximately 30 pl for an egg diameter of 140 µm).
- Transfer the injected unfertilized eggs to a fresh culture dish and incubate them at 15-18 °C until fertilization.
- Fertilize the injected eggs with freshly activated sperm as in the test fertilizations (step 6.1.2). Eliminate a maximum of the sperm by transferring the zygotes to a fresh culture dish. Spread out the embryos and do not perturb them during cleavage stages (up to the 32-cell stage).
- Culture the embryos to the desired stage at a temperature comprised between 13 and 20 °C.
- Collect the embryos by swirling them into the middle of the plate and transferring them into siliconized 1.5 ml tubes. Fix and stain, or mount (Steps 7 and 8).
- Compare the obtained phenotypes to the control injected embryos.
NOTE: Inject a second, non-overlapping MO that targets the same transcript to obtain identical phenotypes. Rescue the specific MO phenotype(s) with a modified mRNA encoding your protein of interest but that is not recognized by the MOs. Inject a control MO that gives no phenotype.
7. LacZ Staining
- Collect the embryos at a desired stage in siliconized 1.5 ml tubes and fix them in 0.2% glutaraldehyde in 1 ml ASWH for 15 to 30 min maximum.
- Wash 2x 10 min with 1 ml 1x PBT.
- Rinse once in 500 µl LacZ staining buffer. Replace with X-Gal substrate supplemented 1 ml staining buffer (use 10 µl/ml of 40 mg/ml X-Gal stock). Transfer the embryos into a 12 well plate for better observation of the staining.
- Incubate in the dark. Vary incubation from 0.5 hr to overnight at 4 °C, at RT or at 37 °C depending on the strength of the driver expressing LacZ.
- Wash the embryos up to 4 times in 1 ml 1X PBT to remove staining buffer.
- Post-fix for 10 min at room temperature with 2% paraformaldehyde (PFA) in 1 ml 1x PBT or 0.2% glutaraldehyde in 1 ml 1x PBT for interpretation and visualization of the stained embryos.
8. Mounting of Fluorescently Labelled Wmbryos
- Fix the embryos of a desired developmental stage for 20 min in 2% PFA in 1 ml ASWH.
- Wash the embryos 2x in 1 ml 1x PBT. Avoid any light exposure by keeping the embryos in dark conditions.
- Transfer up to 50 embryos to one slide. Remove a maximum of PBT first with a pipette and carefully with a paper towel.
- Add 20-25 µl mounting medium.
- Add a coverslip by positioning it in an angle (from one side first) and slowly lower it with a pointed object (needle, forceps, pipette tip) avoiding bubbles till the sample is covered. Fix the coverslip at its edges using dots of a transparent nail polish.
- Seal the entire coverslip with a nail polish after drying. Store in the dark at -20 °C.
Representative Results
Microinjection for gene regulatory network studies
Embryos of the ascidian Ciona intestinalis are well suited for gene functional or gene regulatory studies at cell lineage level and with cellular resolution. mRNAs, MOs or labels can be microinjected into the unfertilized egg or into individual blastomeres following fertilization. MO mediated gene knockdown using the microinjection technique is described in step 6. MOs specifically target the mRNA of selected genes and prevent their translation into protein. MO mediated loss-of-function (LOF) of developmental genes changes the expression of a broad array of downstream genes, overall revealing a glimpse into the embryonic GRN10. Such analyses in Ciona provided the first whole embryo regulatory blueprint in a metazoan11. Figure 1 displays an example of how microinjection was utilized to study the upstream regulation of the expression of the conserved Ci-Myelin Transcription Factor (Ci-MYT) at the gastrula stage of Ciona embryos. Monitored by in situ hybridization (ISH), endogenous expression (Figure 1a, schematized in Figure 1a') is observed in the 6-row neural plate precursors, of notably the brain (rows III/IV, in red) and the spinal cord (row I, in green). In addition, Ci-MYT upstream regulation was analyzed by MO injection targeting several factors that are expressed in or adjacent to neural precursors prior to Ci-MYT expression onset (Figure 1b-g). MO knockdown of early embryonic factors, such as the Forkhead box A-a (FoxA-a) transcription factor and the fibroblast growth factor 9/16/20 (FGF9/16/20) (Figure 1b or 1c, respectively) eliminates overall Ci-MYT expression. Other transcription factors only partially affect Ci-MYT expression resulting in MO-mediated downregulation in a subset of brain precursors (blue arrow heads in Figure 1d, f and g). Conversely, Ci-MYT expression is ectopically activated upon downregulation of Nodal (Figure 1e, red arrow heads), suggesting that Nodal normally represses Ci-MYT expression in these lateral nerve cord precursors.
Electroporation for efficient gene functional and enhancer studies
Electroporation of plasmid DNA into Ciona zygotes is an efficient method for transient transgenesis and subsequent observation of phenotypic changes in vivo. Unlike microinjection of mRNA, electroporation allows for tissue specific overexpression, in which gene coding regions (ORFs, open reading frames) are expressed under control of tissue specific drivers. These normally constitute cis-regulatory regions of well-known genes (such as the brachyury enhancer for expression in notochord precursors8). Conversely, electroporation is instrumental for the efficient analysis of novel regulatory regions where reporter genes (LacZ or green fluorescent protein, GFP) reveal their activity in vivo. The workflow for the identification and subsequent electroporation mediated analysis of such a novel regulatory region (for Ci-MYT) is shown in Figure 2.
A vast repertoire of Ciona community data, readily accessible in ascidian specific genome browsers12,13, like the Ascidian Network for In situ Expression and Embryological Data (ANISEED)23, is inspected for the in silico identification of regulatory regions (Figure 2A). Subsequent polymerase chain reaction (PCR) based cloning of these regions into expression vectors allows for electroporation mediated testing in vivo. (Figure 2B). The genome browser screen-shot in Figure 2A shows the upstream region of the KH transcript model for Ci-MYT (first three exons, in orange, and two introns are visible). Different genome browser tracks annotate functional genome data predicted for this region, such as chromatin immunoprecipitation (ChIP) data14 (here shown for ZicL, Ci-zinc finger of the cerebellum L), sequence conservation between two related Ciona species15,24 or nucleosome occupancy16,17 (from top to bottom in Figure 2A). Generally, exonic protein coding regions are highly conserved (alignment track in Figure 2A). Additional high conservation peaks appear in non-coding regions upstream and in the first intron. Two of these are predicted to be nucleosome free according to a sequence based algorithm16,17 (red frames in Figure 2A) suggesting accessibility to transcription factors. Indeed, Chip-on-ChIP signals14 for Ci-ZicL transcription factor binding (green bars in Figure 2A) are enriched in these two conserved regions. Furthermore, using an interface25 that searches for potential transcription factor binding sites, we identified ZIC sites located within the genome sequences marked by Ci-ZicL ChIP clusters (orange arrows and sequences with underlined ZIC-core, Figure 2A). The binding of Ci-ZicL transcription factor in conserved and predictably nucleosome free regulatory regions of Ci-MYT is consistent with the downregulation of Ci-MYT expression observed in MO-injection mediated knockdown experiments targeting ZicL (Figure 1d).
After determining potential cis-regulatory regions in silico for Ci-MYT expression, these are PCR amplified from Ciona genomic DNA and inserted by recombination cloning into LacZ reporter plasmids19. The constructs are then electroporated into fertilized Ciona eggs (Figure 2B) and analyzed for their activity by LacZ staining (Figure 3).
Figure 3 shows that by such an in silico approach it was possible to identify two separate cis-regulatory sequences that recapitulate Ci-MYT mRNA expression domains (compare Figure 1a). Generally, transiently transgenic embryos electroporated with plasmid DNA show mosaic expression in those cells only that have incorporated the DNA. Mosaic LacZ expression is thus driven by pmyt-R3 and can be found in all the precursors that also express Ci-MYT mRNA i.e. neural plate precursors of the posterior row I and the anterior rows III/IV at gastrula and early neurula stages (Figure 3a, b, purple and red arrow heads, respectively). In contrast, pmyt-R5 is active only in posterior (row I) neural plate precursors and starting at the later neurula stage (Figure 3c, red arrow heads). The two regions thus encode two separate spatial and temporal aspects of Ci-myt gene regulation. Interestingly, both regulatory regions also stain additional precursors not seen in the ISH, notably in the anterior and lateral neural plate and in muscle (Figure 3a, b, pink, white, and yellow arrow heads, respectively). Such wider expression may point to repressive elements that are not contained in the isolated regulatory fragments (such as for Nodal that represses lateral neural fate, see Figure 1e) or to low expression levels detected with accumulating LacZ enzymatic signal.
In addition to enhancer studies, electroporation in Ciona also facilitates the functional analyses of Ciona coding genes by their overexpression. A large collection of Ciona full length ORFs is available to the community, and was furthermore annotated for human orthologs, including disease-associated genes18. Individual genes or groups of genes from this recombination-cloning compatible full ORF cDNA library (Figure 2B) are easily transferred into destination vectors containing appropriate drivers to be expressed in embryos. Resulting expression clones can furthermore be co-electroporated with cis-regulatory reporter constructs to test their putative role in enhancer activation. Figure 2B illustrates the isolation of full ORF clones for the transcription factors Ci-ZicL and Ci-GATAa and their recombination into adapted destination vectors that may contain translational tags19 (e.g. green fluorescent Venus- or viral hemagglutinin (HA)-tag) and a tissue specific driver such as the Friend Of Gata regulatory region (pfog driver) for early pan-ectodermal expression15 used in the present study. The effect of Ci-ZicL overexpression in ectodermal and neuroectodermal tissues was analyzed by co-electroporation of pmyt-R3>LacZ and pmyt-R5>LacZ reporters (Figure 3b', b'' and c', c'', respectively). Our electroporation analyses suggest that Ci-ZicL may activate Ci-MYT expression through pmyt-R3 and pmyt-R5 enhancer regions as both reporter constructs showed ectopic LacZ expression in ectodermal regions (green arrow heads in Figure 3b', b'' and c', c'',). As shown in Figure 3d, electroporation of plasmid DNA in hundreds of Ciona eggs allows for statistically relevant quantification of expression changes, illustrated for ectopic pmyt-R3>LacZ expression upon concentration dependent, pan-ectodermal Ci-ZicL expression.
Electroporation for in vivo subcellular localization
Figure 4 depicts the subcellular localization of tagged transcription factors when expressed upon electroporation in ectodermal blastomeres using adapted expression constructs (schematized in Figure 2B). By C-terminally tagging transcription factors (such as Ci-GATAa) with a Venus tag (Figure 4b) or a HA-tag (Figure 4c) and overexpressing them in ectodermal tissues (pfog) we could observe that in vivo,'Ci-GATAa is mostly localized to nuclei, as expected for a transcription factor, whereas Venus expression is ubiquitous, including the cytoplasm. Similar experiments may be performed to study the dynamics of subcellular localization over time, of individual or combinations of transcription factors, possibly revealing their co-localization with different tags.
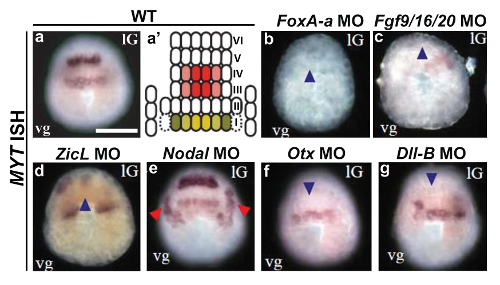
Figure 1: Assessing Ci-MYT regulation by microinjection in Ciona eggs. Ci-MYT mRNA expression in WT and Morpholino (MO) injected embryos. (a) WT expression pattern of Ci-MYT in the neural plate. (a') Schematic representation of stained precursors in the neural plate (NP). Row I/II: posterior neural fate; row III/IV: anterior neural fate; row V/VI: palp fate. (b–g) Ci-MYT expression upon microinjection of various MOs for knocking down indicated genes that participate in the early Ciona GRN (pictures adapted from Imai et al., 2006)11. Ci-MYT, Ci-myelin transcription factor; WT, wild type; blue arrowheads: loss of Ci-MYT signal; red arrowheads: ectopic Ci-MYT signal. Scale bar = 100 µm refers to all images. Please click here to view a larger version of this figure.
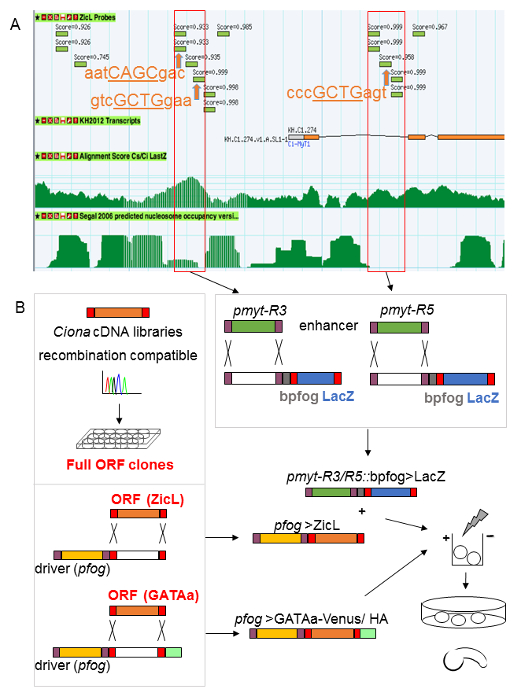
Figure 2: Workflow for in silico search of enhancer regions and recombination cloning for transient transgenesis by electroporation in Ciona embryos. (A) A snapshot from the ANISEED23 ascidian genome browser identifies upstream regulatory sequences of Ci–myt. Red frames mark two potential cis-regulatory regions, pmyt-R3 (scaffold_196:76883..77079) and pmyt-R5 (scaffold_196:75856..76041) that were selected for analysis by electroporation. Names of the tracks: ZicL probes: ChIP-on-Chip data14; KH2012 Transcripts: Ciona intestinalis transcript models; Alignment Score Cs/Ci LastZ: sequence conservation between Ciona savignyi (Cs)/Ciona intestinalis (Ci)15,24; Segal 2006 predicted nucleosome occupancy version 1: chick trained computational algorithm by Segal et al., 200616 applied to Ci as in Khoueiry et al., 201017. Green bars: Ci-ZicL ChIP peaks14; orange arrows: predicted ZIC sites; GCTG: core ZIC binding site. (B) Scheme for generating overexpression constructs from a Ciona full length ORF cDNA library, recombined into destination vectors containing tissue specific drivers (pfog) and protein tags subsequently co-electroporated with reporter constructs (such as pmyt-R3>LacZ or pmyt-R5>LacZ) for in vivo readout. Ci-MYT, Ci-myelin transcription factor; Ci-ZicL, Ci-Zinc finger of the cerebellum L transcription factor; pfog, regulatory region of Ci-Friend of GATA. Please click here to view a larger version of this figure.
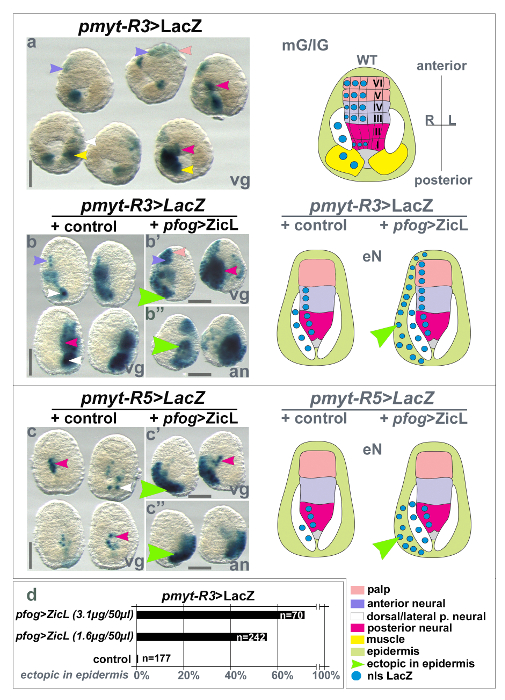
Figure 3: Electroporated Ciona embryos showing Ci-ZicL inducible spatio-temporal cis-regulation of Ci-MYT. LacZ stained embryos at mid-gastrula (mG)/late-gastrula (lG) or early neurula (eN) stage are shown that were co-electroporated with either pmyt-R3>LacZ or pmyt-R5>LacZ and a control construct (pfog>mCherry) or pfog>ZicL. The pfog driver19 mediates ectopic (pan-animal) ZicL expression in ectoderm and neuroectoderm and causes ectopic LacZ stain in these cells. (a) pmyt-R3 LacZ activity in mG/lG stage embryos (small arrowheads, left panel) or in correspondingly colored schematic territories (blue circles, right panel); vegetal view (vg). Row I/II: posterior neural fate; Row III/IV: anterior neural fate; Row V/VI: palp fate. (b) pmyt-R3 activity at eN stages in tissues as in a. (b') Ectopic pmyt-R3 activity upon Ci-ZicL co-electroporation is indicated by green arrowheads. (b'') Same embryos as in b', oriented animal pole up (an). (c) pmyt-R5 LacZ activity at eN stages (c') Ectopic pmyt-R5 activity upon Ci-ZicL co-electroporation is indicated by green arrowheads. (c'') Same embryos as in c' but oriented animal pole up (an). (d) Quantification of representative experiments for Ci-ZicL over-activating pmyt-R3 at low concentrations. One biological repeat is represented. Ci-MYT, Ci-myelin transcription factor; Ci-ZicL, Ci-Zinc finger of the cerebellum L transcription factor; Ci-FOG, Ci-Friend of GATA; WT, wild type; mG, mid-gastrula; lG, late-gastrula; eN, early neurula; an, animal view; vg, vegetal view; nls LacZ, nuclear localization signal for LacZ. Scale bar = 100 µm. Please click here to view a larger version of this figure.

Figure 4: Differential subcellular localization of proteins upon electroporation. Localization of Venus-tagged or Hemagglutinin- (HA)-tagged proteins overexpressed in ectodermal cells using the pfog driver19. (a–c) DIC images. (a', b') Corresponding fluorescence images. (c') Corresponding fluorescence image upon immune histological labelling with TRITC. Scale bar = 100 µm refers to all images. DIC, Differential Interference Contrast. Please click here to view a larger version of this figure.
Discussion
There are two major techniques to generate transgenic Ciona embryos: microinjection of RNA or DNA and electroporation of DNA. These techniques for gene perturbation in Ciona embryos were first used in the 1990s7,8 and from then on successively improved, and now utilized in Ciona (and other ascidian genera)20 worldwide.
Critical steps in the protocol
Successful transgenesis into Ciona gametes that are easily collected from adult animals depends on a number of critical factors. The quality of gametes depends on the spawning season (generally from May to December in the Northern Atlantic, changing with water temperature and oxygenation), on the sea water quality in the aquariums (at correct salinity, adults stay healthy for 2-4 weeks), and finally on correct handling of gametes upon extraction from the adults, as detailed below for the various steps in the protocol.
During preparation steps, incubating Pasteur pipettes with tap water will prevent embryos from sticking and sea water pre-incubation of agarose-coated petri dishes will remove toxic substances released from the agarose. Generally, dishes may be prepared up to 2 weeks before use, stored at 4 °C to avoid bacterial growth and rinsed freshly before use. One day old, small dishes seem best for injections.
Efficiency and length of dechorionation is a crucial step in the protocol as prolonged treatment of eggs/embryos will harm development. Careful preparation (no shaking) and correct storage (at 4 °C for up to one week) preserve the quality of the dechorionation solution. Proteases in the solution lose efficiency if either mixed too violently at preparation or over time, notably if kept at room temperature for longer hours/days. We have optimized this critical step by using fresh dechorionation solution conveniently prepared from frozen aliquots of 20x stock solution.
Correct handling of fragile dechorionated eggs/embryos is essential and shearing or stabbing of embryos while pipetting will be detrimental. When pipetting, embryos are maintained as much as possible in suspension (by gently 'blowing' liquid to swirl them up followed by smooth but quick pipetting), while holding the glass pipette in vertical rather than horizontal position. Such care applies to all pipetting steps for washing, resuspension and transferring eggs/embryos in and out of tubes, cuvettes or plates, before and after fixation. Upon fixation, special care should also be taken to separate pipetting devices for live versus fixed embryos to avoid any toxicity from remains of fixatives.
Microinjection is rather tricky in Ciona eggs/embryos due to their small size and the resilience of the vitelline membrane and critically depends on an optimized tip size of the injection needle, a perfect angle of injection (needle movement in one line, along the egg radius) and a finely tuned, manual breaking of the vitelline membrane (by suction prior to aspiration for injection). The latter is only possible if air bubbles are entirely eliminated from the injection tubing that contains mineral oil for smoothing the suction/injection pressure. In addition, the smallest pieces of dust or egg debris will clog the needle, and are easily avoided by clean working conditions and rinsing steps for eggs prior to transfer on the injection dish. Post injection, the transfer of eggs onto fresh dishes will avoid toxic effects from dead embryos not having survived the injection procedure.
Troubleshooting
Potential problems in the procedure may be addressed by the following solutions. Low fertilization rates may arise from insufficient sperm activation, low egg cleanliness or large fertilization volumes. Use freshly pooled and less diluted sperm. Verify the sperm movement upon activation. Wash the eggs with ASWH prior to fertilization as described in step 4.2. Reduce the water volume and reduce the amount of eggs per fertilization dish/well. Losing embryos during the preparation steps can be avoided by adding a drop of dechorionation solution to more efficiently centrifuge down the undechorionated eggs/embryos. Dechorionation time should be optimized as partially dechorionated eggs will not sediment and prolonged dechorionation is toxic, causing the embryos to explode.
Asynchronous development may arise from residual sperm released during dissection. Keep the egg batches separated from different animals to avoid uncontrolled cross-fertilization. Abnormal development can have various causes. At the beginning and at the end of the season, after keeping the embryo from different individuals separated, the best batches may be selected. Avoid perturbing the embryos for the 10 min that follow fertilization to avoid interfering with their ooplasmic segregation. Ensure that the embryos didn't start dividing while being transferred as pipetting separates the sister blastomeres and results in partial embryos. Use less sperm for dechorionated eggs to avoid polyspermy. Use freshly prepared but rinsed agarose dishes. Ensure that the pipetting device is fixative free. Keep control embryos that were not dechorionated and not electroporated to detect at which step development was disrupted. Supplement the embryos with antibiotics if contamination and decomposition occurs.
If the injection device has 'clogged,' the injection needle opening may be verified by expelling some of its stained content. Clogging material may be wiped off by carefully dragging the needle tip through the agarose. Air bubbles in the injection tubing and/or needle should be removed. Change the needle for better control of the injection volume as the needle tip may break or clog along injection. Rinse the eggs and place them in a clean injection dish if debris has accumulated in the dish. Centrifuge the injection solution prior to filling fresh needles in order to sediment any small particles or crystals. Also helpful is practicing injection only with vital dye. Verify the injection technique by fertilizing injected and non-injected eggs. Finally, consulting with an experienced colleague can be helpful to improve injection technique.
To control for off-target effects a second non-overlapping MO and rescue with a modified mRNA that is not recognized by the MOs can faithfully rule out unspecific phenotypic effects in Ciona.
Microinjection: significance and limitations
Microinjection in Ciona was utilized to generate the first blueprint for a whole embryo gene regulatory network (GRN) in a chordate11. However, despite such a remarkable achievement, microinjection in Ciona embryos has limitations, due to the relatively small size (0.14 mm in diameter) of Ciona eggs. Compared to those of other chordate species where foreign DNA/mRNA is introduced by microinjections, Ciona eggs are comparatively difficult to handle and the rate of injected Ciona embryos per experiment remains low (on average 30 to 50 embryos per experiment), impeding quantitative analyses. Nevertheless, for perturbing maternal factors in Ciona, microinjection of MOs is the method of choice also to study their involvement in zygotic onset of development from the 16 to 32 cell stages15. In addition, it is possible though much more difficult to microinject individual blastomeres up to the 16-32 cell stage. Finally, microinjection remains the most efficient technique for generating transgenic embryos (by mRNA or DNA injection) in ascidian species other than Ciona, for which fully optimized electroporation protocols do not exist.
Electroporation: significance and limitations
To overcome the low numbers and other constraints of microinjection, most of the Ciona community has used electroporation of plasmid DNA since it was developed and published as an optimized and standardized protocol by Zeller et al. in 20049. Indeed, thousands of transgenic Ciona embryos can be generated within one hour with up to 500 embryos simultaneously electroporated in one cuvette using a single 50 V pulse of 16 msec. The approach of generating massive numbers of transgenic embryos is still unique amongst chordate model organisms. This had led Ciona to be quickly recognized as a powerful model system for performing in vivo analyses of potential cis-regulatory regions and cellular/subcellular localization, as exemplified here.
Although electroporation in Ciona zygotes has fundamentally revolutionized the chordate GRN research field, the technique nevertheless faces some limits. First, plasmids are transiently introduced into Ciona embryos and are most likely inherited as extrachromosomal arrays, making the stability of electroporated plasmid DNA speculative. Furthermore, it is very difficult, perhaps even impossible, to control the copies of electroporated plasmids that will be taken up per zygote, thus resulting in phenotypic fluctuations. A further problem that makes it difficult to interpret electroporation outcomes is a phenomenon that we call mosaicism. Electroporated plasmid DNA can be taken up at different positions of the one-cell stage embryo; which determines which and how many quarters of the zygote will receive plasmid to be inherited by the descendant blastomeres. Such variations clearly make the interpretation of quantitative effects more difficult. However, most problems that come with electroporation variability are solved by an appropriate amount of biological samples, with counting and statistics of LacZ (or GFP) positive cells, which is easily obtainable in a single batch.
Versatility and potential of electroporation for genetic engineering
Electroporation eventually allows for mid-throughput screening of potential cis-regulatory regions and quantitative analyses of gene function once cloned into reporter or expression plasmids. Due to the compact Ciona genome, identifying and cloning of potential regulatory regions is fairly straightforward3. A strategy for using the wealth of community data is demonstrated in Figure 2A. Cloning of reporter and gene coding plasmids is further facilitated by the generation of Ciona adapted, GATEWAY compatible vector suites19 and a compatible full length ORF library18. Both tools are available to the community and allow for efficient, restriction enzyme-free recombination of regulatory drivers and coding regions, as depicted in Figure 2B.
In recent years, electroporation was successfully adapted to achieve tissue-targeted gene manipulation with plasmids that express gene editing tools like the CRISPR/Cas9 components under the control of tissue specific drivers21,22. Such approaches will quickly advance our understanding of the dynamics of GRNs and the potentially conserved signaling mechanisms, including transcription factor codes involved in tissue formation and the stepwise exit from pluripotency. Furthermore, tissue-specific loss- or gain-of-function approaches (by CRISPR/Cas9 or by overexpression) using electroporation will be instrumental to study Ciona orthologs of human disease-associated genes. Indeed, catalogues for Ciona orthologs of human disease genes and their full length ORF coding clones are readily available for analysis in Ciona18. Due to the genome wide duplications in vertebrates, most human genes possess functionally redundant paralogs that complicate analyses of gene function1. Ciona being a simpler system with the typical chordate tissue arrangement in larvae should uncover fundamental roles of such important, often functionally conserved genes.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the University of Innsbruck, Austria, and the Austrian Academy of Sciences (ÖAW).
Materials
| Lab with constant temp. (~ 18°C) | For embryo work | ||
| Ciona intestinalis adults | UPMC – Station Biologique, Roscoff, France |
For collecting gametes | |
| NaCl | Roth | 3957.1 | For ASWH |
| KCl | Roth | 6781.1 | For ASWH |
| CaCl2x2H2O | Roth | A119.1 | For ASWH |
| MgCl2x6H2O | Roth | 2189.1 | For ASWH |
| MgS04x7H2O | Roth | P027.2 | For ASWH |
| NaHCO3 | Roth | 6885.1 | For ASWH |
| Hepes | Roth | HN78.3 | For ASWH |
| Sodium thioglycolate | Sigma | MZ-6 | For Dechorionation |
| Pronase | Sigma | T0632-100G | For Dechorionation |
| D-Mannitol | Sigma Aldrich | M9546-250G | For electroporation |
| Plasmid DNA (prep >1µg/µl) | Macherey-Nagel | 740410.100 | For electroporation or microinjection |
| Agarose | any supplier | For coating embryo culture dishes | |
| Plastic petri dishes | VWR | 612-2079 | Coated with 1% Agarose in ASWH, covered with ASWH |
| Small scissors | any supplier | For dissecting gametes | |
| NaOH | Roth | 6771.1 | For activating dechorionation solution |
| Tris | Roth | 5429.3 | 1M pH9.5, for sperm activation |
| Pasteur pipettes | VWR | 612-1701 | Treated by immersion in tap water for at least 12h |
| Hand centrifuge | Hettich Zentrifugen | 1011 | Use glass centrifuge tubes |
| 1.5ml Eppendorf tubes | Sigma | T3406-250EA | |
| 2ml Eppendorf tubes | Sigma | T3531-200EA | |
| Square electroporator | Bio Rad Gene Pulser Xcell | ||
| Electroporation cuvettes 4 mm | Eurogentec | CE-0004-50 | |
| Coverslips | Sto Premium | PR248212600 | |
| Glass slides | VWR | ECN 613-1551 | |
| Glutaraldehyde | Sigma | G6257-10ML | For fixation in LacZ staining |
| Paraformaldehyde | Sigma | P6148-500G | For fixation of fluorescent samples |
| Vectashield | Vector | H-1000 | For mounting of fluorescent samples |
| X-Gal (5-Bromo-4-chloro-3-indolyl ß-D-Galactopyranoside) | Roth | 2315.2 | LacZ substrate; stock in 40mg/ml DMF (Dimethylformamide) |
| K5Fe(CN)6 | Merck | 4984 | For LacZ staining buffer |
| K3Fe(CN)6 | Merck | 4973 | For LacZ staining buffer |
| Na22HPO4 | Roth | 4984.2 | For PBT |
| KH2PO4 | Roth | 3904.2 | For PBT |
| Tween | Sigma | SLBJ5103V | For PBT |
| Mineral Oil | Sigma | M8410 | Embryo culture tested |
| Green Vital Dye | Fast Green Sigma | F7251 | for microinjection; 10µg/µl |
| Micropipette Puller (Flaming/Brown) | Sutter Instruments | Model P-97 | For pulling injection needles |
| Borosilicate glass capillaries GC100TF-10 | Harvard Apparatus Ltd. UK | 30-0038 | 1.0mm O.D. x0.78mm I.D., containing filament, for microinjection |
| Micromanipulator | Narishige | MWS-2 | Three-axis Oil Hydraulic System; for 3D fine movement during injection |
| Injection holder | Narishige | 1M-H1 | For holding injection needles, entire system filled with mineral oil |
| All Glass Syringe | Walter Graf & Co GmbH & Co. KG | No. 95199.10427A4 | 10ml FortunaOptima; filled with mineral oil, for fine tuning of microinjection pressure |
| Morpholinos | GeneTools LLC, Pilomath, OR, USA | Injection at 0.25-0.5mM | |
| Capped mRNA | Ambion mMessage mMashine kit | Injection at 0.05-0.5µg/µl | |
| Plasmid DNA | Injection at 20 ng/µl | ||
| Recombination cloning kit | Gateway BP Clonase Enzyme Mix Invitrogen | 11789-013 | |
| Recombination cloning kit | Gateway LR Clonase Enzyme mix Invitrogen | 11791-019 |
Riferimenti
- Holland, P. W., Garcia-Fernandez, J., Williams, N. A., Sidow, A. Gene duplications and the origins of vertebrate development. Dev Suppl. 120 (1), 125-133 (1994).
- Simmen, M. W., Leitgeb, S., Clark, V. H., Jones, S. J., Bird, A. Gene number in an invertebrate chordate, Ciona intestinalis. Proc Natl Acad Sci U S A. 95 (8), 4437-4440 (1998).
- Dehal, P., et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 298 (5601), 2157-2167 (2002).
- Satoh, N., Satou, Y., Davidson, B., Levine, M. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 19 (7), 376-381 (2003).
- Delsuc, F., Brinkmann, H., Chourrout, D., Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 439 (7079), 965-968 (2006).
- Lemaire, P. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev Biol. 332 (1), 48-60 (2009).
- Hikosaka, A., Satoh, N., Makabe, K. Regulated spatial expression of fusion gene constructs with the 5′ upstream region of Halocynthia roretzi muscle actin gene in Ciona savignyi embryos. Roux’s Arch Dev Biol. 203 (1-2), 104-112 (1993).
- Corbo, J. C., Levine, M., Zeller, R. W. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 124 (3), 589-602 (1997).
- Zeller, W. R. Generation and use of transgenic ascidian embryos. Methods Cell Biol. 74 (74), 13-30 (2004).
- Satou, Y., Imai, K. S., Satoh, N. Action of morpholinos in Ciona embryos. Genesis. 30 (3), 103-106 (2001).
- Imai, K. S., Levine, M., Satoh, N., Satou, Y. Regulatory blueprint for a chordate embryo. Science. 312 (5777), 1183-1187 (2006).
- Tassy, O., et al. The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 20 (10), 1459-1468 (2010).
- Brozovic, M., et al. ANISEED 2015: a digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. , (2015).
- Kubo, A., et al. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development. 137 (10), 1613-1623 (2010).
- Rothbächer, U., Bertrand, V., Lamy, C., Lemaire, P. A combinatorial code of maternal GATA, Ets and β-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 134 (22), 4023-4032 (2007).
- Segal, E., et al. A genomic code for nucleosome positioning. Nature. 442 (7104), 772-778 (2006).
- Khoueiry, P., et al. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol. 20 (9), 792-802 (2010).
- Gilchrist, M. J., et al. A pipeline for the systematic identification of non-redundant full-ORF cDNAs for polymorphic and evolutionary divergent genomes: Application to the ascidian Ciona intestinalis. Dev Biol. 404 (2), 149-163 (2015).
- Roure, A., et al. A multicassette Gateway vector set for high throughput and comparative analyses in ciona and vertebrate embryos. PLoS One. 2 (9), e916 (2007).
- Kumano, G., Takatori, N., Negishi, T., Takada, T., Nishida, H. A maternal factor unique to ascidians silences the germline via binding to P-TEFb and RNAP II regulation. Curr Biol. 21 (15), 1308-1313 (2011).
- Sasaki, H., Yoshida, K., Hozumi, A., Sasakura, Y. CRISPR/Cas9-mediated gene knockout in the ascidian Ciona intestinalis. Dev Growth Differ. 56 (7), 499-510 (2014).
- Stolfi, A., Gandhi, S., Salek, F., Christiaen, L. Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development. 141 (21), 4115-4120 (2014).

