High Throughput, Real-time, Dual-readout Testing of Intracellular Antimicrobial Activity and Eukaryotic Cell Cytotoxicity
Summary
A high throughput, real-time assay was developed to simultaneously identify (1) eukaryotic cell-penetrant antimicrobials targeting an intracellular bacterial pathogen, and (2) assess eukaryotic cell cytotoxicity. A variation on the same technology was thereafter combined with digital dispensing technology to enable facile, high-resolution, dose-response, and two- and three-dimensional synergy studies.
Abstract
Traditional measures of intracellular antimicrobial activity and eukaryotic cell cytotoxicity rely on endpoint assays. Such endpoint assays require several additional experimental steps prior to readout, such as cell lysis, colony forming unit determination, or reagent addition. When performing thousands of assays, for example, during high-throughput screening, the downstream effort required for these types of assays is considerable. Therefore, to facilitate high-throughput antimicrobial discovery, we developed a real-time assay to simultaneously identify inhibitors of intracellular bacterial growth and assess eukaryotic cell cytotoxicity. Specifically, real-time intracellular bacterial growth detection was enabled by marking bacterial screening strains with either a bacterial lux operon (1st generation assay) or fluorescent protein reporters (2nd generation, orthogonal assay). A non-toxic, cell membrane-impermeant, nucleic acid-binding dye was also added during initial infection of macrophages. These dyes are excluded from viable cells. However, non-viable host cells lose membrane integrity permitting entry and fluorescent labeling of nuclear DNA (deoxyribonucleic acid). Notably, DNA binding is associated with a large increase in fluorescent quantum yield that provides a solution-based readout of host cell death. We have used this combined assay to perform a high-throughput screen in microplate format, and to assess intracellular growth and cytotoxicity by microscopy. Notably, antimicrobials may demonstrate synergy in which the combined effect of two or more antimicrobials when applied together is greater than when applied separately. Testing for in vitro synergy against intracellular pathogens is normally a prodigious task as combinatorial permutations of antibiotics at different concentrations must be assessed. However, we found that our real-time assay combined with automated, digital dispensing technology permitted facile synergy testing. Using these approaches, we were able to systematically survey action of a large number of antimicrobials alone and in combination against the intracellular pathogen, Legionella pneumophila.
Introduction
Pathogens that grow or reside temporarily in intracellular compartments are difficult to therapeutically eradicate. Obligate or relatively obligate intracellular pathogens such as Legionella pneumophila, Coxiella burnetii, Brucella spp., Francisella tularensis, and Mycobacterium spp. often require prolonged courses of antimicrobial therapy for cure that may range from months to even years. Furthermore, extracellular pathogens may transiently occupy intracellular niches and in this way escape clearance by normal courses of antimicrobial therapy and later emerge to start new rounds of virulent infection. Staphylococcus aureus1 and uropathogenic Enterobacteriaceae2,3 infections are two increasingly recognized examples. Therefore, a fundamental drug discovery goal is to identify novel antimicrobials that penetrate into intracellular compartments. Optimal therapy to quickly eradicate intracellular organisms and prevent development of resistance through sub-inhibitory antimicrobial exposure is especially desirable.
To this end, we developed a high-throughput screening technology to identify intracellular-penetrant antimicrobials targeting the intracellular growth of the model pathogen, Legionella pneumophila.4 Previous clinical observations indicate that standard antimicrobial susceptibility testing did not accurately predict in vivo therapeutic efficacy against this organism.5 Specifically, this was because major classes of antimicrobials such as β-lactams and aminoglycosides, although highly effective against axenically grown Legionella, do not sufficiently penetrate into the intracellular compartments where Legionella resides.5,6 Later evidence suggested that technically more complex intracellular growth assays effectively predicted clinical efficacy.7 Unfortunately, these assays were extremely laborious endpoints assays, requiring infected macrophages, treated with antimicrobials, to be lysed at different times points for colony forming unit enumeration. Such assays are impractical to do on a large scale and are unsuitable for high-throughput drug discovery.
Therefore, we developed technology for real-time determination of intracellular bacterial growth.6 This was accomplished through use of a bacterial strain modified through integration of either a bacterial luciferase operon8 (first generation assay, described previously)4 or fluorescent protein9 reporters (second generation, orthogonal assay, described here) into the bacterial chromosome. In this way, luminescent or fluorescent signal provides a surrogate, real-time readout of bacterial number.
However, these attributes do not address a major confounder in intracellular infection assays, off-target effects on host cells. In particular, the death of the host cell inherently limits intracellular growth and leads to false positive identification of antimicrobial effect. As many compounds in screening libraries are eukaryotic cell toxic, such false positives would overwhelm true antimicrobials, necessitating a large number of follow-up, endpoint cytotoxicity assays for resolution.
Thus, it was of great interest to be able to assess eukaryotic cell viability and intracellular growth simultaneously. Notably, a characteristic of non-viable eukaryotic cells is loss of cell membrane integrity. Probes that test the permeability of the cell membrane may therefore be used to assess cell viability. We previously characterized the ability of a series of putatively cell membrane-impermeant, fluorescent, DNA-binding dyes to access and stain nuclear DNA of dead cells.4 On binding nuclear DNA, these dyes display a large increase in quantum fluorescent yield resulting in increased signal over background solution fluorescence. As such, these dyes provided a quantitative readout of eukaryotic cell death.4 Notably, we found that several were non-toxic themselves during prolonged co-incubation with J774 macrophages. When added during initial infection, they provided a real-time, fluorescent readout of eukaryotic cell death that can be measured by a microplate fluorimeter or observed microscopically.
Therefore, by combining use of a bacterial reporter and non-toxic, membrane-impermeant, DNA-binding dyes, we were able to develop a simple, non-destructive, real-time assay to measure both bacterial load and eukaryotic cell cytotoxicity simultaneously. This assay has allowed us to screen in 384-well plate format ~10,000 known bioactives including ~250 antimicrobials and >240,000 small molecules with functionally uncharacterized activity for the ability to inhibit intracellular growth of Legionella pneumophila, while at the same time generating eukaryotic cell cytotoxicity data for each compound.6 Our analysis of known antimicrobials against intracellular growth of Legionella was the most comprehensive exploration of this type to date.6
Based on the efficiency of our assay format, we also subsequently explored the potentially synergistic effects of known antimicrobials when used in combination. One of the most common synergy tests, the so-called checkerboard assay, is standardly performed by assessing combinatorial effects of two-fold serial dilutions of two or more antimicrobials.10 In these assays, synergy is defined by the observation of greater effect when two or more antimicrobials are applied together than the sum of the effects of each applied separately. Of note, heretofore, only focused and selective synergy testing was performed against intracellular Legionella pneumophila because of the great effort involved in traditional endpoint assays multiplied by the combinatorial permutations required.
To facilitate synergy testing, we made use of our real-time intracellular growth/eukaryotic cytotoxicity assay in combination with automated digital dispensing technology6. This automation permitted us to dispense serial dilutions of compounds dissolved in DMSO or aqueous solution alone or in combination in 384-well format.11 Furthermore, such robust liquid handling technology permitted us to easily perform higher resolution, square-root-of-two (rather than the standard, lower resolution, doubling) dilution combinations to achieve higher levels of specificity in our two-dimensional, checkerboard synergy analysis. This resolution was especially valuable in addressing concerns in the synergy field about reproducibility when using two-fold dilution series12. Lastly, our assay was quantitative and also therefore measured gradations of inhibition. As a result, the assay captured the entirety of inhibitory information, expressible in isocontour isobolograms in which isocontours connect combinatorial concentrations with similar levels of growth inhibition.6 This plotting strategy allowed visualization of combinatorial dose-response curves. To illustrate our methodology, we describe our protocol for performing these assays and show representative results.
Protocol
1. Real Time Intracellular Growth and Eukaryotic Cell Cytotoxicity Assay
- Preparing Host Cells (J774A.1 Cells)
- Culture J774A.1 Mus musculus macrophage-like cells in suspension in RPMI 1640 with 9% iron-supplemented calf serum. Initially passage in tissue culture flasks. After cells have become confluent in a 75 cm2 tissue culture flask in 15 ml of medium, split by scraping and dilution to 65 ml with the same type of medium, of which 15 ml is returned to the tissue culture flask and 50 ml is transferred to a 250 ml bacterial shaker flask.
- For scale-up, culture in suspension in 250 ml and/or 1,000 ml bacterial flasks filled to one fifth volume with tissue culture medium. Aerate by setting rotation speed at approximately 120 revolutions per minute. For consistent growth, incubate at exactly 5% CO2 and 37 °C.
- Harvest cells when they reach density in the range 2.5 x 106 cells/ml to 5 x 106 cells/ml. Ensure dead cell percentage (most easily assayed by trypan blue staining) does not exceed 25%, as dead cells will increase background noise in cytotoxicity assays.
- Plate in white, tissue culture-treated, 384-well microplates at 5 x 104 J774A.1 cells in 30 µl tissue culture medium per well. Incubate microplates overnight to achieve 90% confluence on day of experiment.
- Preparing Luminescent or Fluorescent Legionella pneumophila
- Passage L. pneumophila (Lp02::flaA::lux8) bacteria on appropriate medium, i.e., buffered charcoal yeast extract (BCYE) medium. If using a thymidine auxotrophic strain, supplement medium with 100 µg/ml thymidine. Prepare patches of bacteria for experiments by spreading organisms thickly on a new BCYE plate and incubating one day (if from a previous plate passage) or two to three days (if from frozen stock) to obtain confluent growth.
- Prepare BCYE plates by dissolving 10 g yeast extract, 10 g N-(2-acetamido)-2-aminoethanesulfonic acid, 0.35 g K2HPO4, and 1 g monobasic potassium α-ketoglutarate in 950 ml deionized water. Adjust to pH 6.9 with Potassium hydroxide (≥2.5 ml of 11.9 molar solution).
- Add 15 g agar and 2 g activated charcoal; and bring to 1 L final volume. Add a magnetic stir bar and autoclave. Cool medium to 55 °C and add filter-sterilized solutions containing 0.4 g L-cysteine and 0.42 g Ammonium iron(III) citrate dissolved in deionized water. Use magnet stir plate to mix prior to pouring Petri plates.
- For testing of L. pneumophila growth in axenic growth medium, prepare ACES-yeast extract broth (AYE) by combining all chemical reagents used for BCYE except for charcoal and agar. Filter sterilize and use immediately, or freeze for later experiments.
- Resuspend organisms in the same tissue culture medium used for J774A.1 cells with 100 µg/ml thymidine supplementation as appropriate.
- Passage L. pneumophila (Lp02::flaA::lux8) bacteria on appropriate medium, i.e., buffered charcoal yeast extract (BCYE) medium. If using a thymidine auxotrophic strain, supplement medium with 100 µg/ml thymidine. Prepare patches of bacteria for experiments by spreading organisms thickly on a new BCYE plate and incubating one day (if from a previous plate passage) or two to three days (if from frozen stock) to obtain confluent growth.
- Macrophage Infection
- Add test compounds of interest (screening compounds, and positive and negative controls for bacterial growth inhibition and eukaryotic cell lysis). Preferably, dissolve stock solutions at ≥500x in DMSO (dimethyl sulfoxide) or an aqueous solution to allow sufficient dilution of vehicle.
- Although stock solutions can be stored frozen, avoid freeze-thaw cycles for labile compounds and antimicrobials.
- Dilute luminescent bacteria to target of 2.5 x 106 CFU (colony forming unit)/m and fluorescent reporter-labeled bacteria to 1.0 x 107 CFU/ml in tissue culture medium and add appropriate non-toxic, membrane-impermeant, nucleic acid-binding dye at 2.5x final assay concentration.
- Add 20 µl of mixture to each J774A.1 culture well, final assay volume 50 µl, final bacterial concentration 1 x 106 CFU/ml (lux operon reporter) or 4 x 10 106 CFU/ml (fluorescent protein reporter).
- Incubate at 37 °C for 1-3 days in 5% CO2 at 100% relative humidity to prevent evaporative edge effects.
- Add test compounds of interest (screening compounds, and positive and negative controls for bacterial growth inhibition and eukaryotic cell lysis). Preferably, dissolve stock solutions at ≥500x in DMSO (dimethyl sulfoxide) or an aqueous solution to allow sufficient dilution of vehicle.
- Assay Readout
- Read bacterial growth and eukaryotic cell toxicity on a microplate luminometer and fluorimeter as appropriate for reporters being used.
- To avoid temperature-associated edge effects, thermally equilibrate microplates prior to luminescence reading by placement in a single layer on a lab bench with lids ajar for approximately 20 min (or in a biological safety cabinet with lids off for approximately 10 min).
- Return microplates to incubator if real-time readout at later time points is desired.
- Read bacterial growth and eukaryotic cell toxicity on a microplate luminometer and fluorimeter as appropriate for reporters being used.
2. Data Analysis
- Normalize data to positive and negative controls to calculate percent or fold-inhibition for intracellular bacterial growth and percent cytotoxicity for eukaryotic cells.
- To assess assay robustness, calculate statistical separation between positive and negative controls for both bacterial growth inhibition and cytotoxicity using Z-factor (Z') analysis where Z' = 1 – 3(σp + σn)/|µp + µn|. In this equation, σp and σn are the standard deviations for the positive and negative control wells, respectively; µp and µn are the mean values for the positive and negative control wells, respectively.
- For high-throughput screening assays, calculate standard z-scores (or other alternative, quantitative statistical measures) to assess effects of test compounds of interest. Alternatively, calculate a simple fold-reduction or log-fold reduction as a potentially physiologically more relevant measure of effect magnitude for geometrically replicating bacteria.
3. Single and Multi-dimensional (i.e., Synergy) Dose-response Testing and Data Interpretation
- Prepare macrophages and bacteria as described in protocol section 1.
- Just prior to infection or axenic incubation, add antimicrobials of experimental interest in a serial, doubling dilution series, with the goal of spanning on the high end a concentration that completely eliminates growth (i.e., the minimal inhibitory concentration or MIC) and on the low end a concentration that shows no obvious activity.
- Use an automated liquid handling system to facilitate single dose-response and combinatorial synergy testing setup through direct, automated addition of antimicrobial dilutions according to manufacturer's protocol.
- Consider use of sub-doubling dilutions, for example,
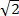 -fold dilution series, and assay replicates to increase accuracy and reproducibility of data.
-fold dilution series, and assay replicates to increase accuracy and reproducibility of data. - For macrophage infection, dilute luminescent bacteria to target of 2.5 x 106 CFU (colony forming unit)/ml in tissue culture medium. Add 20 µl of mixture to each J774A.1 culture well, final assay volume 50 µl, final bacterial concentration 1 x 106 CFU/ml; and incubate at 37 °C in 5% CO2 at 100% relative humidity.
- For axenic growth testing, dilute luminescent bacteria to final concentration of 1 x 106 CFU/ml in AYE medium, add 50 µl to each well of a 384-well plate and incubate at 37 °C in 5% CO2 at 100% relative humidity.
- Calculate fractional inhibitor concentration index for antimicrobial combinations that inhibit bacterial growth. For each drug in the combination (a, b, . . n) that results in complete or >99% inhibition of bacterial growth, calculate the individual fractional inhibitory concentration (FICa, b, . . n.) as follows: FICa = the concentration of compound "a" divided by the minimal inhibitor concentration of "a" by itself.. Using the same formula, calculate FICb, etc. Then calculate the combinatorial FIC index or
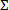 FIC = FICa + FICb + . . .FICn for each inhibitory combination.
FIC = FICa + FICb + . . .FICn for each inhibitory combination.
- Use the combinatorial index that deviates furthest from 1.0 to assess synergy. Consider a cutoff of ≤0.5 as a conservative indication of a synergistic effect and ≥4, an antagonist effect.12
- Plot isobolograms, isocontour isobolograms, and/or isosurface isobolograms6 as alternative graphical representations of combinatorial effects.
Representative Results
Microplate intracellular growth assay
Figure 1 diagrams the assay steps. The automated steps shown can be performed manually. However, throughput is greatly facilitated using liquid handling systems.
Figure 2 shows representative results from a 384-well microplate, dual-readout, real-time intracellular growth and eukaryotic cell cytotoxicity assay using a Legionella strain (Lp02) marked with either a lux operon (Figures 2A, 2B) or mNeptune2 fluorescent protein (Figure 2C, 2D) reporter. Positive and negative control compounds (DMSO, antibiotics, saponin) were added to plates through use of a pin transfer robot to simulate performance in a high-throughput screening setting. Significant Legionella growth (Figures 2A, 2C) can be easily distinguished in comparison to saponin lysis and antibiotic-treated controls at 24 to 72 hr for luminescent bacteria and 48 to 72 hr for mNeptune2-labeled bacteria, respectively. Legionella typically lyses host cells after replicating for 48 to 72 hr. This can be appreciated in Figures 1B and 1D, which show increased fluorescence of SYTOX Green (a representative, impermeant, nucleic acid-binding dye and marker of host cell death) over time. Cytotoxicity eventually reaches the maximal amount observed in the detergent (saponin), host cell lysis, positive control. mNeptune2-labeled organisms were added at a four-fold higher inoculum than lux operon-labeled bacteria, likely accounting for more rapid host cell killing and earlier associated rise in fluorescence signal in mNeptune2 experiments. As expected, effective antimicrobial treatment (levofloxacin or azithromycin) prevented both bacterial growth and host cell lysis. Conversely, cytotoxic compounds that limited intracellular growth through destruction of the host cell (e.g., saponin) could easily be identified by a combination of low luminescence or mNeptune2 fluorescence, and high cytotoxicity-associated signal. Furthermore, observation of combined decrease in both luminescence and mNeptune2 fluorescence (bacterial growth), and cytotoxicity provided reasonable confidence that a compound is a true intracellular growth inhibitor (e.g., azithromycin and levofloxacin positive controls) rather than a false positive resulting from spurious interference with bacterial reporter signal.
Table 1–3 shows representative Z' for three different bacterial reporters (lux operon, mNeptune2, tdTomato) and corresponding fluorescence readings relative to controls. A Z' >0.5 indicates a statistically robust assay appropriate for high-throughput settings; however, lower Z' may be suitable depending on experimental goals, i.e., the ability to reliably distinguish more or less subtle effects of test compounds or perturbations. Based on results from Figure 2, mNeptune2 was not as sensitive a reporter as the bacterial luciferase operon. Therefore, as expected, a robust difference between mNeptune2 signal in negative (DMSO) and positive (levofloxacin, azithromycin, saponin) growth inhibitor controls was not observed until day 2 or day 3 of incubation, in contrast to early, reasonably robust signal of lux operon reporter bacteria on day 1 after infection. In contrast, tdTomato, an alternative bacterial fluorescent reporter, showed suboptimal Z' throughout the infectious course. Suboptimal fluorescent characteristics resulted in part from encroachment of the fluorescence excitation and emission tail into the range used for optimal readout of tdTomato signal. This encroachment can be appreciated from the positive Z' found in the tdTomato saponin versus levofloxacin comparison (implying a significant difference in bacterial numbers under two conditions where bacteria should not replicate). This result can be explained by saponin lysis causing a strong fluorescence signal, whose tail is detected as tdTomato emission, and therefore leading to a spurious difference between the saponin and antibiotic controls. Therefore, while the bacterial lux and mNeptune2 reporters in combination with fluorescence appear usable for dual-readout, solution-based, real-time assays, the tdTomato and fluorescence combination, at least without further mathematical signal deconvolution, does not.
Figure 3 shows previously published examples of dose-response curves of known antimicrobials using lux operon, reporter-labeled organisms.6 Notably, Legionella does not grow in tissue culture medium. Therefore, replication of bacteria in macrophage infection assays solely represents intracellular growth. However, Legionella will grow axenically in a specialized, low sodium, liquid ACES (N-(2-acetamido)-2-aminoethanesulfonic acid) yeast extract medium.20 Here effects on intracellular and axenic growth were compared. Using these two growth systems, we observed that polar antimicrobials such as β-lactams (meropenem, ceftriaxone) and aminoglycosides (data not shown) are poor inhibitors of intracellular growth, in contrast to their potent effects on axenic growth. Presumably poor efficacy in macrophage infection assays is based on an inability to access the intracellular Legionella replicative niche. In contrast, eukaryotic cell-penetrant antimicrobials such as quinolones (levofloxacin) and azithromycin, demonstrated potent effects on both intracellular and axenic bacteria.
Additionally, use of the dual-readout assay allowed dose-response curves for both bacterial inhibition and eukaryotic cytotoxicity to be determined in the same experiment. Specifically, IC50 (concentration for 50% bacterial growth inhibition) and CC50 (concentration that induces 50% eukaryotic cell death) can be determined, and selectivity, CC50/IC50, calculated. All three measures are important criteria for compound progression in drug discovery efforts. Figure 4 shows an example of such dual, dose-response curves in response to the antibiotic doxycycline, data obtained from the same screening wells over time. In Figure 4A, on day 1 of infection, about a hundred-fold bacterial replication is evident in the zero antibiotic test wells compared with the highest levels of inhibition observed with antibiotic. There was minimal associated eukaryotic cell toxicity except at very high doxycycline concentrations (≥10 µg/ml). On day 2 (Figure 4B), bacterial lux signal was approximately 10-fold greater than day 1 with significant bacterial replication-associated cytotoxicity observed at low antimicrobial concentrations. As expected, bacterial replication-induced host cell death tracks closely with bacterial numbers (lux signal) throughout the antimicrobial dose-response curve. At higher antimicrobial concentrations cytotoxicity is reduced to baseline, until very high concentrations, where doxycycline again appears toxic as observed on day 1. The apparent shifting of the luminescence dose-response towards slightly higher antimicrobial concentrations compared to fluorescence-based cytotoxicity measurements is somewhat exaggerated when plotting luminescence and cytotoxicity on log and linear scales, respectively, as shown. IC50 for bacterial growth was approximately 100 ng/ml while CC50 was approximately 10 µg/ml, consistent with the CC50 described previously for doxycycline treatment of the leukocyte lineage, HL-60 cell line.21 Therefore, overall selectivity determined in this dual-readout experiment was approximately 100.
Figure 5 shows previously published isocontour isobolograms associated with two-dimensional synergy tests in which pairwise combinations of antimicrobials were tested for enhanced effect against intracellular L. pneumophila.6 Although standard isobolograms connect the lowest combinatorial concentrations of antibiotics that completely inhibit growth, we opted, based on available quantitative inhibitory data, to connect points with similar levels of inhibition using isocontours. The right most isocontour in these diagrams corresponds to the standard isobologram line. Concave isobolograms generally indicate synergy, while convex isobolograms generally indicate indifference or antagonism. Examples of synergy (panels A-C) and indifference are shown (D-F), in this case corresponding to FIC index values of <0.5 and ≥1, respectively.6
Of note, pairwise combinations of azithromycin, minocycline, and rifampicin demonstrated synergy. Therefore, these agents were tested together in three-way combination as shown in Figure 66. Here, a surface was drawn to connect the lowest combinatorial concentrations of the three antimicrobials that led to >99% inhibition of bacterial intracellular growth. The observed concave-shaped surface, by itself suggestive of three-dimensional synergy, corresponded to a low FIC index (0.325) indicative of substantial synergy effect.

Table 1: Z' for lux operon-reporter, Legionella infection- comparison of positive and negative controls. Z' correspond to data points shown in Figure 2A, 2B. This table has been modified from Chiaraviglio and Kirby 2014.4 Positive luminescence Z' >0.25 are highlighted with yellow characters on a black background.

Table 2: Z' for mNeptune2-reporter, Legionella infection — comparison of positive and negative controls. Z' correspond to data points shown in Figure 2C, 2D. Positive mNeptune2 fluorescence Z' >0.25 are highlighted with red characters.
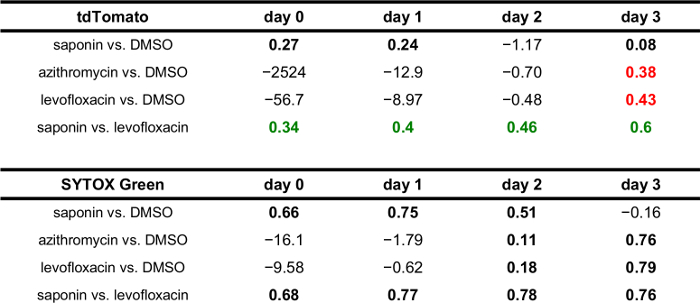
Table 3: Z' for tdTomato-reporter, Legionella infection — comparison of positive and negative controls. Positive tdTomato fluorescence Z' >0.25 are highlighted with red characters. False positive Z' >0.25 related to overlap of SYTOX Green and tdTomato emission are highlighted with green characters.
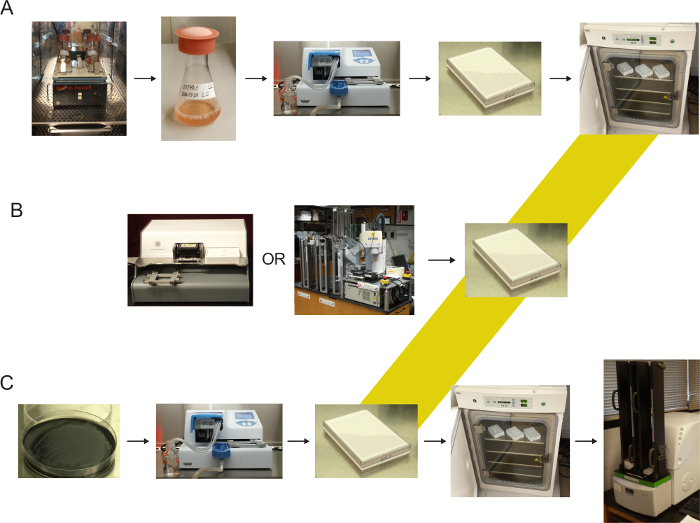
Figure 1: Pictorial representation of dual-reporter assay setup. (A) Replating of J774A.1 cells grown in suspension into 384-well dishes. (B) Addition of compounds of interest to microplates. (C) Simultaneous infection and addition of nucleic acid-binding dyes; incubation; and readout. Please click here to view a larger version of this figure.
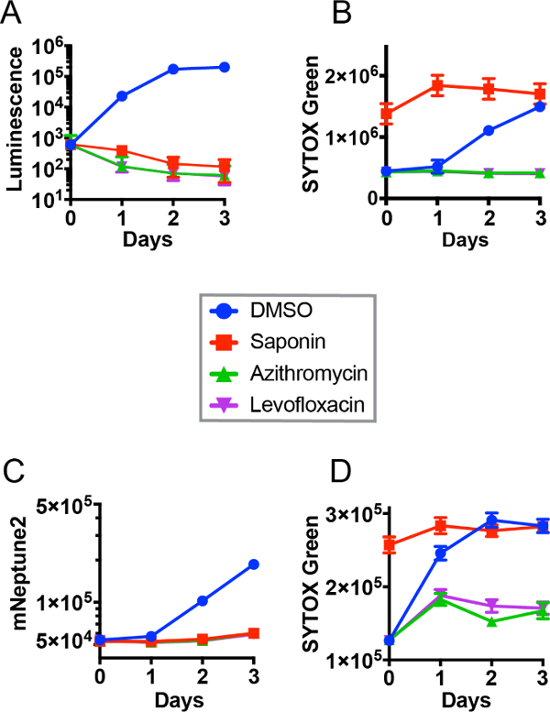
Figure 2: Dual, real-time readout of intracellular bacterial growth and eukaryotic cytotoxicity in high-throughput format. Corresponding luminescence (A) and fluorescent (B) signals during intracellular infection of J774A.1 cells with lux operon-expressing Legionella pneumophila. Corresponding fluorescent protein (C) and fluorescent (D) signals during intracellular infection of J774A.1 cells with mNeptune2-expressing Legionella pneumophila. Four treatments are shown: negative DMSO control ( ); positive cytotoxicity control, saponin (
); positive cytotoxicity control, saponin ( ); and positive bacterial growth inhibition controls, azithromycin (
); and positive bacterial growth inhibition controls, azithromycin ( ) and levofloxacin (
) and levofloxacin (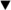 ). Data points represent mean and standard deviation of 96 replicates performed in duplicate, 384-well plates. Note, standard deviation error bars for some data points were minimal and therefore hidden within data point symbols. Panels A and B have been modified from Chiaraviglio and Kirby 2014.4 Please click here to view a larger version of this figure.
). Data points represent mean and standard deviation of 96 replicates performed in duplicate, 384-well plates. Note, standard deviation error bars for some data points were minimal and therefore hidden within data point symbols. Panels A and B have been modified from Chiaraviglio and Kirby 2014.4 Please click here to view a larger version of this figure.
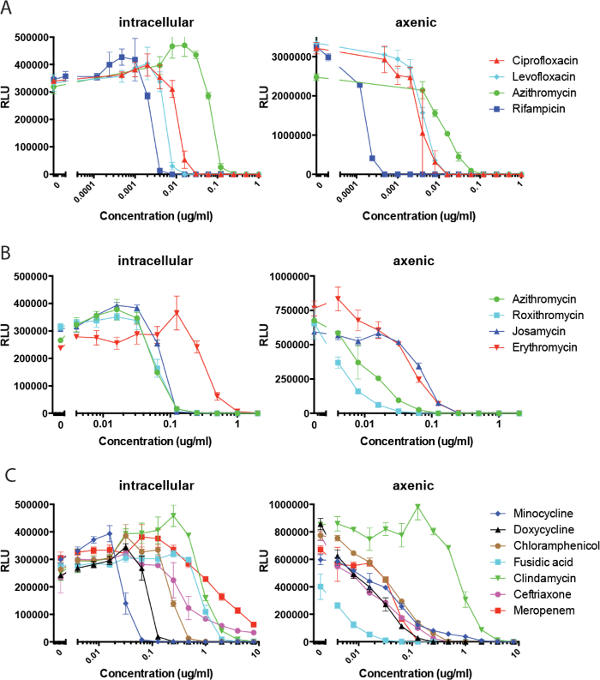
Figure 3: Representative examples of one-dimensional, dose-response assays. J774A.1 cells were infected with lux operon-expressing, L. pneumophila and treated with a two-fold dilution series of indicated antimicrobials (labeled as 'intracellular'). In parallel, lux operon-expressing, Legionella were diluted to the same final concentration in aces yeast extract medium (AYE) and treated with an identical, two-fold dilution series (labeled as 'axenic'). Luminescence was read two days post bacterial inoculation and plotted versus antimicrobial concentration. Panel A includes antimicrobials commonly used to treat Legionella infection. Panel B compares activity of different macrolide antibiotics. Panel C includes several different classes of antimicrobials with varying and sometimes contrasting intracellular and axenic potency. Data points represent the averages and standard deviations of three separate test wells per condition. This figure has been modified from Chiaraviglio and Kirby 2015.6 Please click here to view a larger version of this figure.
Figure 4: Example of a dual-readout, dose-response experiment for determining IC50, CC50 and selectivity. Effects of doxycycline on intracellular growth and J774A.1 cell cytotoxicity were determined on days 1 and 2 post infection. Data points represent the averages and standard deviations of three separate test wells per condition. Please click here to view a larger version of this figure.
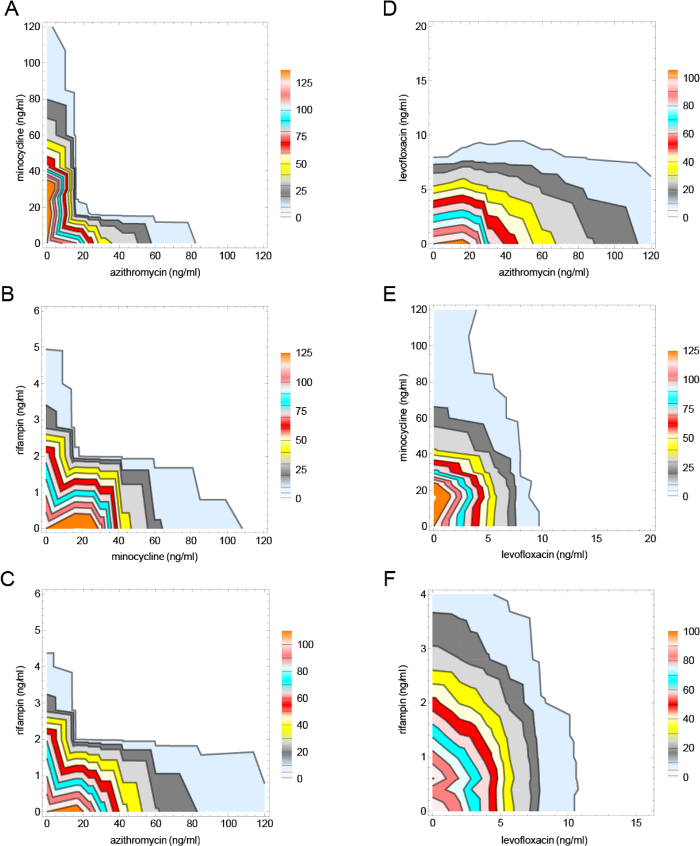
Figure 5: Example of two-dimensional synergy assays. Combinatorial serial dilutions of pairs of antimicrobials were tested for synergy against intracellular growth of Legionella in J774A.1 macrophages. Isocontours lines connect points of similar percent inhibition. The rightmost isobologram connects combinations with complete intracellular growth inhibition (>99% luminescence reduction compared to untreated controls) and corresponds to the standard isobologram plot. Shading indicates percent inhibition as delineated by the color-coded key in each graph. Isocontours from left to right correspond to 100, 90, 80, 70, 60, 50, 40, 30, 20, 10, 5, and 1% growth relative to untreated controls. This figure has been modified from Chiaraviglio and Kirby 2015.6 Please click here to view a larger version of this figure.
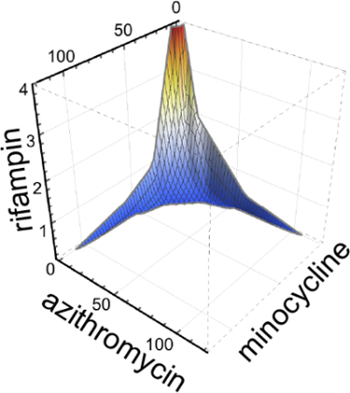
Figure 6: Example of a three-dimensional synergy assay. The lowest combinatorial concentrations of azithromycin, minocycline and rifampicin that resulted in >99% growth inhibition were connected to form an isobologram surface plot. This figure has been modified from Chiaraviglio and Kirby 2015.6 Please click here to view a larger version of this figure.
Discussion
We describe real-time assays for simultaneous detection of intracellular bacterial growth and host cell cytotoxicity. There are several critical steps in the protocol. First, for robust assay performance, there must be sufficient spectral separation between bacterial and cytotoxicity readouts. Such separation is intrinsic for combinations of luciferase operon reporters and fluorescent DNA-binding dyes. However, based on our experience (Table 1-3, Figure 2), use of dual, fluorescent readout requires non-overlapping fluorescent signals (e.g., use of a far-red fluorescent protein reporter and green nucleic acid stain) to obtain robust, solution-based data. Furthermore, from prior experience, potentially related to available reader optics or solution fluorescence properties, only DNA stains with green or orange emission gave usable cytotoxicity data, i.e., Z' greater than >0.5, a measure indicative of strong assay performance.4 Therefore, solution-based, dual-readout assays are somewhat constrained in terms of what reporters can be used.
Another critical step is the ratio of infecting bacteria to macrophages. Bacterial infection induces host cell death. Therefore, too high an inoculum may lead to early host cell death, relatively low levels of bacterial replication, and potential obfuscation of direct cytotoxic effects of antimicrobials under study. The appropriate infecting ratio can be determined empirically and may depend on virulence of the particular bacterial strain under study. A low ratio of approximately 1:1 is a good starting point. Experiments such as those shown in Figure 2 and Tables 1–3 performed with several infection ratios will allow assessment and optimization of conditions for use in future assays. Of note, for Legionella in particular, use of a strain background with natively lower host cell cytotoxicity (a flagellin mutant)8 contributed significantly to the high assay robustness obtained. Furthermore, translation of the dual-readout technique to other intracellular, pathogen-host systems may also require additional adjustments to the infection protocol. Legionella does not grow in tissue culture medium, allowing us to attribute increased reporter signal to intracellular growth alone. However, many intracellular pathogens (e.g., Brucella) can grow to varying extents both intracellularly and in tissue culture medium. Therefore, additional steps such as gentamicin treatment and washing of host cells after initial infection may be necessary to reduce extracellular replication and allow selective examination of intracellular replication capacity.22
Assay scale-up for use in high-throughput screening and synergy testing is associated with its own set of challenges. Most particularly, we found that the large numbers of J774A.1 cells needed could best be cultured in bacterial rather than traditional tissue culture flasks (see Figure 1). However, to maintain culture sterility, sponge foam or other filter caps for these flasks were found preferable to standard bacterial flask caps. Placement of a small orbital shaker platform inside a standard tissue culture incubator permitted us to perform scale-up culture with equipment on hand. Lastly, use of automated dispensing systems (Figures 1A, B) greatly facilitated assay setup and testing of combinatorial dilution series.
Notably, adequate detection of bacterial replication relies on the ability to sufficiently express either a lux operon or fluorescent protein in the bacterial strain of interest. To this end, we have created a set of fluorescent and lux operon reporter transposons driven by a strong constitutive promoter that should allow facile marking of diverse types of bacteria (Kang and Kirby, data not shown). Alternatively, for pathogens whose intracellular replication proves cytotoxic for the host cell, real-time cytotoxicity measurements alone can be used as a surrogate for intracellular replication. However, in this case, cytotoxic and inactive compounds cannot be reliably distinguished in a single assay.
The availability of real-time readout out provides particular advantage for high-throughput screening assays. Specifically, avoidance of endpoint assessment of bacterial growth (colony forming unit determination)7 and cytotoxicity (addition of cell viability reagent at termination of assay)23,24 eliminates several experimental steps and associated labor. Moreover, time courses can easily be followed in the same assay wells with minimal increase in experimental effort. Importantly, the availability of two distinct types of bacterial replication reporters (bacterial lux operon and fluorescent protein) has particular significance for high-throughput screening assay development. Specifically, false positive antimicrobial readout resulting from spurious interference with lux operon or fluorescent reporter output can be addressed through use of both reporters sequentially in complementary orthogonal assays in primary and secondary screening, respectively25. Notably, we found that the bacterial lux operon was the more sensitive reporter of bacterial growth with a roughly 20-fold to 100-fold lower detection limit than our fluorescent protein reporters. However, lux output is likely subject to more false positives based on the several gene products and steps required for light output.26 More specifically, in contrast to eukaryotic single gene luciferase reporters, the bacterial lux operon is a five-gene cluster that encodes both a two-component luciferase enzyme and genes for endogenous synthesis of luciferase substrate. It would make sense therefore to use the lux operator reporter in a primary screen to ensure sufficient sensitivity, and the fluorescent protein reporters in secondary assays to ensure specificity.
Taken together, the methodology described provides real-time, non-destructive alternatives to endpoint assays for measuring bacterial replication and eukaryotic cytotoxicity and therefore represents a methodological simple and versatile method for assessing antimicrobial effects on intracellular pathogens.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Research reported in this manuscript was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI099122 to J.E.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank Jennifer Smith, David Wrobel, Su Chiang, Doug Flood, Sean Johnston, Jennifer Nale, Stewart Rudnicki, Paul Yan, Richard Siu, and Rachel Warden from the ICCB-Longwood Screening Facility and/or the National Screening Laboratory for the New England Regional Centers of Excellence in Biodefense and Emerging Infectious Diseases (supported by U54AI057159) for their assistance in development and performance of high-throughput screening assays. We also would like to thank Kenneth P. Smith for helpful comments on the manuscript.
Materials
| J774A.1 cells | American Type Culture Collection | TIB-67 | Host cell |
| ACES | Sigma-Aldrich | A9758 | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| Yeast extract, ultrafiltered | Becton-Dickinson/Difco | 210929 | For making buffered charcoal yeast extract agar and buffered yeast extract medium; lower grades may cause impaired growth and/or alter sensitivity of Legionella to growth inhibitors |
| Alpha-ketoglutaric acid, monopotassium salt | Sigma-Aldrich | K2000 | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| Sodium pyruvate | Sigma-Aldrich | P5280 | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| Potassium phosphate, dibasic | Thermo Fisher Scientific | P288-500 | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| L-cysteine | Sigma-Aldrich | C-7755 | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| Ammonium iron(III) citrate | Sigma-Aldrich | F5879 | For making buffered charcoal yeast extract agar and buffered yeast extract medium; ferric pyrophosphate may be used instead but is more difficult to weigh accurately |
| Potassium hydroxide solution, concentrated | Thermo Fisher Scientific | SP236-500 | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| Deonized water | N/A | N/A | For making buffered charcoal yeast extract agar and buffered yeast extract medium |
| Thymidine (tissue culture grade) | Sigma-Aldrich | T1895 | For supplementing both RPMI 1640 and buffered yeast extract agar/medium — lower grade thymidine may be used for the latter, but may cause impaired cell growth and/or cell death in RPMI 1640 |
| RPMI 1640, standard formulation | Corning via Thermo Fisher Scientific | 10-040-CV | For growing J774A.1 cells prior to plating; includes 2 mM L-glutamine |
| RPMI 1640 lacking phenol red | Corning via Thermo Fisher Scientific | 17-105-CV | For plating J774A.1 cells in 384 well dishes (not suitable for growth prior to plating); also lacks L-glutamine — supplement to 2 mM before use |
| L-glutamine, 200 mM in 0.85% NaCl (tissue culture grade) | HyClone via Thermo Fisher Scientific | SH30034.02 | For supplementing RPMI 1640 lacking L-glutamine, to 2 mM final concentration |
| Iron-supplemented calf serum | Gemini Bioproducts | 100-510 | For supplementing RPMI 1640, to 9.1% final concentration |
| Trypan Blue solution | Sigma-Aldrich | T8154 | For staining for J774A.1 cell death determination while counting cell density |
| SYTOX Green, 5 mM solution in DMSO | Thermo Fisher Scientific | S7020 | For staining for J774A.1 cell death determination by fluorescence reading or epifluorescence microscopy (in conjunction with orange-red or far red fluorescent bacteria). Use at 125 nM final concentration. |
| GelRed, 10000X solution in water | Biotium | 41003 | For staining for J774A.1 cell death determination by epifluorescence microscopy (in conjunction with green fluorescent bacteria). Use Gel Red at 1X final concentration. |
| Cell culture incubator | Thermo Fisher Scientific | 13-255-26 | For incubation of J774A.1 cells (both before and after infection); can also be used for incubation of bacteria, or a standard atmosphere incubator can be used instead) |
| Orbital shaker | BellCo Glass | 7744-01010 | For shaking incubation of J774A.1 cells before infection; fits inside cell culture incubator; includes shaker base 7744-01000 and tray 7740-01010 (these are also available separately) |
| Shaker flasks (250 ml) | ChemGlass Life Sciences | CLS-2038-04 | For shaking incubation of J774A.1 cells before infection |
| Shaker clamps for flasks (250 ml) | BellCo Glass | 7744-16250 | For shaking incubation of J774A.1 cells before infection |
| Shaker flasks (1000 ml) | ChemGlass Life Sciences | CLS-2038-07 | For shaking incubation of J774A.1 cells before infection |
| Shaker clamps for flasks (1000 ml) | BellCo Glass | 7744-16100 | For shaking incubation of J774A.1 cells before infection |
| Sponge foam caps for flasks (250 ml – 1000 ml) | ChemGlass Life Sciences | CLS-1490-038 | For shaking incubation of J774A.1 cells before infection; reduces risk of contamination relative to standard metal caps |
| MultiDrop Combi programmable multichannel peristaltic pump | Thermo Fisher Scientific | 5840300 | For dispensing J774A.1 cells, medium, and bacterial suspension containing fluorophores to large numbers of 384 well dishes |
| Combi standard bore manifold | Thermo Fisher Scientific | 24072670 | Default predispense volume of 20 ml is insufficient to compensate for settling — increase to 80 ml |
| White 384 well dishes treated for tissue culture | Corning | 3570 | For reading luminescence and fluorescence; Greiner catalog # 781080 also tested successfully |
| DMSO (tissue culture grade, in sealed ampoules) | Sigma-Aldrich | D2650 | For dissolving positive control and test compounds |
| Azithromycin | Sigma-Aldrich | PHR1088 | Antibiotic positive control |
| Saponin (from Quillaja bark) | Sigma-Aldrich | S-4521 | Cytoxicity positive control |
| Multichannel pipettor | Thermo Fisher Scientific | Finnpipette | For transfer of fixed amounts of positive control compounds; pipettor must have digital dispensing with detents to enable repetitive fixed volume dispensing |
| Epson pin transfer robot | Epson/ICCB-L | (Custom equipment) | For transfer of fixed amounts of test compounds from library arrays |
| D300 digital dispensing system | Hewlett-Packard via Tecan | D300 | For transfer of variable amounts of test compounds ranging from 11 picoliters to 10 microliters |
| T8+ cartridges for D300 digital dispensing system | Hewlett-Packard via Tecan | T8+ | For dispensing test compounds |
| EnVision multi-mode plate reader | Perkin-Elmer | (Contact manufacturer) | For optimal detection of SYTOX Green fluorescence, use excitation filter 485/14, emission filter 535/25, and dichroic mirror 505 nm, with selection of minimum gain and transmittance, and “high concentration mode. For luminescence detection, use the "USLUM" protocol for high-sensitivity detection. For mNeptune2 detection, use excitation filter 600/8, emission filter 665/7.5, and dichroic mirror 658 nm, with selection of gain and transmittance to achieve the highest maximum signal possible without saturating the photomultiplier. |
| Epifluorescence microscope with computer-connected digital camera | Nikon | Ti | For live cell imaging; any standard fluorescent microscope can substitute, with phase contrast or DIC optics, capable of imaging green (fluorescein), orange-red to red (Texas Red), and far-red (Cy5) fluorescence, with 100X oil objective for highest resolution |
| Glass-bottom tissue culture dishes | MatTek Corporation | P35G-1.5-20-C | For live cell imaging. Dishes such as the MatTek allow microscopic visualization at 600X or 1000X magnification through use of an inverted epifluorescent or confocal microscope. These specific dishes are 3.5 cm nominal diameter, 3.3 cm inside diameter, with 20 mm diameter #1.5 thickness cover slips inserted into the bottoms. |
| Photoshop CS6 | Adobe | Adobe photoshop or similar programs can be used to pseudocolor and merge light microscopic and fluorescent images. | |
| Mathematica 10 | Wolfam | For generation of two-dimensioonal isocontour isobolograms and three-dimensional surface isobolograms. |
Riferimenti
- Garzoni, C., Kelley, W. L. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 17 (2), 59-65 (2009).
- Rosen, D. A., et al. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect Immun. 76 (7), 3337-3345 (2008).
- Blango, M. G., Mulvey, M. A. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother. 54 (5), 1855-1863 (2010).
- Chiaraviglio, L., Kirby, J. E. Evaluation of impermeant, DNA-binding dye fluorescence as a real-time readout of eukaryotic cell toxicity in a high throughput screening format. Assay Drug Dev Technol. 12 (4), 219-228 (2014).
- Edelstein, P. H. Antimicrobial chemotherapy for legionnaires’ disease: a review. Clin Infect Dis. 21 Suppl 3, S265-S276 (1995).
- Chiaraviglio, L., Kirby, J. E. High-Throughput Intracellular Antimicrobial Susceptibility Testing of Legionella pneumophila. Antimicrob Agents Chemother. 59 (12), 7517-7529 (2015).
- Pedro-Botet, L., Yu, V. L. Legionella: macrolides or quinolones?. Clin Microbiol Infect. 12 Suppl 3, 25-30 (2006).
- Coers, J., Vance, R. E., Fontana, M. F., Dietrich, W. F. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 9 (10), 2344-2357 (2007).
- Chu, J., et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 11 (5), 572-578 (2014).
- Eliopoulos, G. M., Moellering, R. C., Lorian, V. Ch. 9. Antimicrobial combinations. In: Antibiotics in Laboratory Medicin. , 330-396 (1996).
- Jones, R. E., Zheng, W., McKew, J. C., Chen, C. Z. An alternative direct compound dispensing method using the HP D300 digital dispenser. J Lab Autom. 18 (5), 367-374 (2013).
- Odds, F. C. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 52 (1), (2003).
- Shaner, N. C., et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 22 (12), 1567-1572 (2004).
- Lam, A. J., et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 9 (10), 1005-1012 (2012).
- Berger, K. H., Merriam, J. J., Isberg, R. R. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol. 14 (4), 809-822 (1994).
- Berger, K. H., Isberg, R. R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 7 (1), 7-19 (1993).
- Vogel, J. P., Isberg, R. R. Cell biology of Legionella pneumophila. Curr Opin Microbiol. 2 (1), 30-34 (1999).
- Kirby, J. E., Isberg, R. R. Legionnaires’ disease: the pore macrophage and the legion of terror within. Trends Microbiol. 6 (7), 256-258 (1998).
- Kirby, J. E., Vogel, J. P., Andrews, H. L., Isberg, R. R. Evidence for pore-forming ability by Legionella pneumophila. Mol Microbiol. 27 (2), 323-336 (1998).
- Vogel, J. P., Andrews, H. L., Wong, S. K., Isberg, R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 279 (5352), 873-876 (1998).
- Song, H., Fares, M., Maguire, K. R., Sidén, A., Potácová, Z. Cytotoxic Effects of Tetracycline Analogues (Doxycycline, Minocycline and COL-3) in Acute Myeloid Leukemia HL-60 Cells. PLoS ONE. 9 (12), e114457 (2014).
- Isberg, R. R., Falkow, S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 317 (6034), 262-264 (1985).
- Niles, A. L., Moravec, R. A., Riss, T. L. Update on in vitro cytotoxicity assays for drug development. Expert Opin Drug Discov. 3 (6), 655-669 (2008).
- Niles, A. L., Moravec, R. A., Riss, T. L. In vitro viability and cytotoxicity testing and same-well multi-parametric combinations for high throughput screening. Curr Chem Genomics. 3, 33-41 (2009).
- Mayr, L. M., Bojanic, D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 9 (5), 580-588 (2009).
- Dunlap, P., Thouand, G., Marks, R. Biochemistry and genetics of bacterial bioluminescence. Bioluminescence: Fundamentals and Applications in Biotechnology – Volume 1. 1, 37-64 (2014).

