Conditional Reprogramming of Pediatric Human Esophageal Epithelial Cells for Use in Tissue Engineering and Disease Investigation
Summary
Expansion of human pediatric esophageal epithelial cells utilizing conditional reprogramming provides investigators with a patient-specific population of cells that can be utilized for engineering esophageal constructs for autologous implantation to treat defects or injury and serve as a reservoir for therapeutic screening assays.
Abstract
Identifying and expanding patient-specific cells in culture for use in tissue engineering and disease investigation can be very challenging. Utilizing various types of stem cells to derive cell types of interest is often costly, time consuming and highly inefficient. Furthermore, undesired cell types must be removed prior to using this cell source, which requires another step in the process. In order to obtain enough esophageal epithelial cells to engineer the lumen of an esophageal construct or to screen therapeutic approaches for treating esophageal disease, native esophageal epithelial cells must be expanded without altering their gene expression or phenotype. Conditional reprogramming of esophageal epithelial tissue offers a promising approach to expanding patient-specific esophageal epithelial cells. Furthermore, these cells do not need to be sorted or purified and will return to a mature epithelial state after removing them from conditional reprogramming culture. This technique has been described in many cancer screening studies and allows for indefinite expansion of these cells over multiple passages. The ability to perform esophageal screening assays would help revolutionize the treatment of pediatric esophageal diseases like eosinophilic esophagitis by identifying the trigger mechanism causing the patient’s symptoms. For those patients who suffer from congenital defect, disease or injury of the esophagus, this cell source could be used as a means to seed a synthetic construct for implantation to repair or replace the affected region.
Introduction
Esophageal tissue engineering and eosinophilic esophagitis (EoE) have been the focus of research in many laboratories over the last decade. Congenital defects, such as esophageal atresia, are seen in approximately 1 in 4,000 live births, which results in the incomplete development of the esophagus leading to the inability to eat1. The incidence and prevalence of EoE have been on the rise ever since the identification of the disease entity in 1993. The incidence of EoE varied from 0.7 to 10/100,000 per person-year and the prevalence ranged from 0.2 to 43/100,0002. A new attractive surgical approach to treating long gap esophageal atresia consists in generating tissue constructs for implantation utilizing the patient's own cells. These cells in conjunction with synthetic scaffolding will generate an autologous construct that does not require immune suppression. Some groups have already begun to investigate the use of stem-like cells for esophageal tissue engineering 3 as well as the use of native esophageal epithelial cells to repopulate the mucosa4–7. Diseases that are present in the esophagus of pediatric patients are often hard to diagnose or study without intervention. Furthermore, utilizing animal models or in vitro immortalized cell line models for pediatric diseases like EoE do not encompass the exact disease pathogenesis or patient specific differences8. Therefore, the ability to study a patient's disease process in vitro in order to identify specific disease triggering antigens, evaluate underlying mechanisms and investigate drug treatments would be novel and provide clinicians with information that can aid in patient treatment.
There have been many autologous or patient-specific cell types that have been proposed for use in tissue engineering and studying human disease pathogenesis. However, some of these cell types are limited in their capability to generate enough cells of a specific phenotype to seed a large scaffold or perform high throughput in vitro studies. The use of pluripotent or multipotent stem cells has been the topic of much research discussion, however, limitations and shortcomings for using these cells have been well described9. The use of human embryonic stem cells is highly debated and presents many ethical issues. Most importantly, these cells form teratomas, which are similar to a tumor, if they are not differentiated from their pluripotent state prior to delivering them into a living host10. Furthermore, the use of embryonic stem cells would not be patient-specific and could elicit an allogenic response and the need for immune suppression10. Induced pluripotent stem cells (iPSCs) are pluripotent cells that can be derived from a patient's own cells. Somatic cells, such as skin cells, can be induced to a pluripotent state using a variety of integrative and non-integrative techniques. These cells then serve as a patient-specific cell sources for tissue engineering or disease investigation. The integration of unwanted genetic material into these cells is a concern many have described and even if sequences are completely removed iPSCs appear to conserve an epigenetic "memory" towards the cell type from which they were derived11. These cells also will form teratomas in vivo if not differentiated prior to transplantation11. Many differentiation protocols have been investigated focusing on epithelial lineages12,13,14, however, it is very important to note that the cell types resulting at the end of differentiation are not homogenous and only possess a fraction of the cell type of interest. This results in low yield and the need to purify the desired cell type. Although iPSCs are a potential patient-specific cell source, the process to obtain a cell type of interest for either tissue engineering or disease investigation is very inefficient.
Human epithelial cells have been successfully isolated from a variety of both diseased and non-diseased tissues in the human body including: lung15, breast16, small intestine17, colon18, bladder19 and esophagus20. It is important to note that human primary cells have a finite number of passages in which the phenotype is maintained21,22. Unfortunately, this means that the number of cells needed for disease investigation or for seeding an engineered scaffold for implantation may not be achieved. Therefore, new techniques are needed to expand patient cells while still maintaining an epithelial phenotype. Conditional reprogramming of normal and cancerous epithelial cells utilizing feeder cells and ROCK inhibitor was described in 2012 by Liu et al.23. This technique was utilized to expand cancerous epithelial cells obtained from biopsies of prostate and breast cancer using irradiated feeder cells, ROCK inhibitor and conditional reprogramming medium. The goal was to generate enough cells for in vitro assays such as drug screening. This technique is capable of expanding epithelial cells indefinitely by "reprogramming" these cells to a stem or progenitor-like state, which is highly proliferative. It has been demonstrated that these cells are non-tumorigenic and do not possess the capability to form teratomas23,24. Furthermore, no chromosomal abnormalities or genetic manipulations were present after passaging these cells in culture using this technique23,24. Most importantly, these cells are only able to differentiate into the native cell type of interest. Therefore, this technique offers a large reservoir of patient-specific epithelial cells for disease investigation or tissue engineering without the need for immortalization.
Obtaining epithelial tissue from a specific organ in order to study disease processes is often limited and not always possible due to patient risk. For those patients suffering from esophageal disease or defects, endoscopic biopsy retrieval is a minimally invasive approach for obtaining epithelial tissue that can be dissociated and conditionally reprogrammed to provide an indefinite cell source that is specific to the mucosa of that patient's esophagus. This then allows for in vitro studies of the epithelial cells to evaluate disease processes and screen for potential therapeutics. One disease process that could greatly benefit from this approach is Eosinophilic Esophagitis, which has been described as allergic disease of the esophagus8. Allergy tests as well as therapeutic approaches could be evaluated in vitro using the patient's own epithelial cells and this data can then be passed onto the treating physician to develop individualized treatment plans. The technique of conditional reprogramming in conjunction with obtaining endoscopic biopsies from pediatric patients offers the ability to expand normal esophageal epithelial cells indefinitely from any patient. This cell source could therefore be teamed together with natural or synthetic scaffolding to provide a patient-specific surgical option for defects, disease or trauma. Having an indefinite cell number would help engineer esophageal constructs that possess a completely reseeded lumen with esophageal epithelial cells in order to help facilitate regeneration of the remaining cell types.
Protocol
Esophageal biopsies were obtained after informed consent was obtained from the parents/guardians of the pediatric patients and in accordance with institutional review board (IRB#13-094).
1. Sterilizing Instruments and Gelatin Solution
- Autoclave forceps, razor blades and scissors prior to handling tissue to prevent contamination.
- To make 200 mL of 0.1% gelatin solution, combine 200 mL of distilled water with 0.2 g of gelatin. Autoclave and cool prior to use.
2. Coating Tissue Culture Plates
- Approximately 2 h prior to cell isolation, add enough 0.1 % gelatin to cover a tissue culture dish (100 mm) and incubate at 37 °C in a cell culture incubator.
NOTE: These plates can also be made ahead of time and kept in the incubator before use.
3. Making Enzyme Solution for Dissociation
- Approximately 30 min prior to cell isolation, weigh out 10 units of dispase on a balance based on the units per milligram concentration on the vial. Place the powder in a 15 mL conical tube with 10 mL of Dulbecco's Modified Eagle Medium (DMEM) and place in a 37 °C water bath.
4. Making Cell Culture Medium
- To make 50 mL of conditional reprogramming medium, mix the following ingredients into a sterile conical tube: 35 mL of F12 (Ham's Medium), 11.5 mL of DMEM, 0.5 mL of L-glutamine, 0.5 mL of 100x penicillin/streptomycin, 2.5 mL of fetal bovine serum (FBS), 250 μg of human insulin, 500 ng of human epidermal growth factor (EGF).
- To make 500 mL of 3T3 medium, mix 50 mL of FBS, 5 mL of 100x penicillin/streptomycin and 5 mL of L-glutamine with 500 mL of DMEM.
- To make 500 mL of keratinocyte serum-free medium, thaw EGF and bovine pituitary extract (BPE) supplements (provided with the media). Add the supplements to the 500 mL bottle of base medium and add 5 mL of 100x penicillin/streptomycin
5. Culturing and Irradiating 3T3 Cells as a Feeder Source
- Prepare 3T3 medium, comprising DMEM, 10% FBS, 1x penicillin/streptomycin and 2 mM L-glutamine.
- Culture 3T3 cells in tissue-treated T-75 flasks at least 3 days before the arrival of patient samples.
- Just prior to the arrival of patient samples, trypsinize 3T3 cells using 5 mL of 0.25% Trypsin-EDTA per flask and incubate at 37 °C for 5 min or until most cells are floating
- Add 5 mL of 3T3 medium to stop the reaction. Aspirate the cell suspension and transfer to a 15 mL conical tube. Spin for 5 min at 300 x g.
- Aspirate the medium and resuspend the cells in a defined volume of Trypan blue for cell counting. Combine 10 µL of cells with 10 µL of Trypan blue to exclude dead cells. Load 10 µL of this solution on a hemocytometer for cell counting.
NOTE: For this protocol, seeding density for irradiated 3T3 cells is 10,900 cells per cm2, which is equal to 600,000 cells per 100 mm plate. - Aliquot the number of cells necessary to plate a 100 mm plate per patient sample and resuspend in 25 mL of 3T3 medium in a 50 mL conical tube and irradiate with 3000 rads.
- Following irradiation, spin cells down for 5 min at 300 x g and resuspend in conditional reprogramming medium at 5 mL per 100 mm plate.
- Set the tube aside until the plating of patient cells takes place later in the protocol (see step 7.6).
6. Obtaining, Preserving and Transporting Pediatric Esophageal Biopsies
- Add 5 mL of complete keratinocyte serum-free medium with 100 µg/mL primocin to 15 mL conical tubes and bring it to the medical facility on ice.
NOTE: These tubes of medium can be stored at 4 °C for up to 2 months. - Obtain approximately 8 esophageal biopsies at the time of endoscopy and separate as follows:
- Place six biopsies in the 15 mL conical containing complete keratinocyte serum free medium with 100 µg/mL primocin and place on wet ice.
- Place one biopsy in 5 mL of buffered formalin and place on wet ice.
- Place one biopsy in a small biopsy cryomold with optimal cutting temperature compound.
- Immediately drop this mold into a beaker of isopentane placed in liquid nitrogen. Once frozen, place the cryomold in a specimen bag and store on dry ice in a polystyrene foam container.
- Place the tubes containing biopsies in medium or formalin inside a sealable bag and then into a shipping box containing wet ice. Seal and label the box with a biohazard label and transport it to the laboratory.
NOTE: The container that contains the dry ice and frozen biopsy is also sealed and labeled with a biohazard label.
7. Processing Patient Tissue for Downstream Culture and Analysis
- Upon arrival at the laboratory, transfer the frozen biopsy on dry ice to -80 °C freezer for downstream sectioning or RNA extraction. Place the sample in formalin. Store in a 4 °C refrigerator and allow to fix overnight.
- Spin the remaining biopsies down at 300 x g, and aspirate the medium. Add 10 mL of dispase solution to the conical tube, seal and allow it to incubate at 37 °C in a water bath for 15 min.
- Following the incubation, spin the biopsies down at 300 x g. Aspirate the medium and resuspend biopsies in 5 mL of 0.05% Trypsin-EDTA.
- Transfer the 5 mL of trypsin-EDTA containing the biopsies to a 100 mm sterile plate and mince the biopsies using 2 sterilized razor blades as fine as possible.
- Transfer the 5 mL of trypsin-EDTA containing minced biopsies to a new 15 mL conical. Rinse the plate twice with an additional 5 mL of 0.05% Trypsin-EDTA and add to the conical tube. Incubate the tube for 10 min in a 37 °C water bath.
- Following the incubation, spin the biopsies down at 300 x g (1.4 RPM) for 5 min and aspirate the medium. Add 5 mL of conditional reprogramming medium to the conical tube as well as the 5 mL of conditional reprogramming medium containing the irradiated feeder cells (see step 5.7).
- Add ROCK inhibitor (1 μL/mL) to the 10 mL of cell suspension and plate after aspirating the gelatin from the tissue culture dish. Incubate at 37 °C.
8. Culturing and Expanding Cells
- After approximately 5 days of culture, aspirate the initial medium containing the minced-biopsy pieces and rinse the dish 2-3 times with 5 mL sterile PBS. Add new medium to the plate including ROCK inhibitor at 10 μM.
- Upon reaching 70% confluency, expand the cells. Add 5 mL of 0.05% Trypsin-EDTA to the plate and incubate in a 37 °C incubator no more than 2.5 min to remove the feeder cells. Following that incubation, aspirate the trypsin-EDTA and rinse the plate with 5 mL of PBS.
- Aspirate the PBS and add 5 mL of 0.25% Trypsin-EDTA to the plate and incubate for 5-10 min or until the cells are loose from the plate. Add equal amounts of medium to the plate to stop the reaction and transfer all of the fluid to a 50 mL conical tube and spin down at 300 x g for 5 min.
- Resuspend cells in a defined volume of conditional reprogramming medium and mix 10 μL of the suspension with 10 μL of trypan blue. Count on a hemocytometer to determine total cell number.
- To expand cells, add 1 million human esophageal cells to a 150 mm plate along with 1.2 million irradiated 3T3 cells in a total volume of 15 mL. Add fresh ROCK inhibitor at 10μM and add the cell suspension to a gelatin-coated 150 mm dish.
9. Freeze and Store Human Esophageal Epithelial Cells
- To make freezing medium, add 10% DMSO to conditional reprogramming medium. After trypsinizing and counting, aliquot the cells to freeze into a new 50 mL conical tube and spin down at 300 x g for 5 min.
- Aspirate the medium and add freezing medium at a volume of 2 mL per cryovial. Once the cell suspension is homogenous, aliquot 106 cells in pre-labeled cyrovials. Place in a freezing container at -80 °C for at least 24 h prior to moving the vials to long-term liquid nitrogen storage.
Representative Results
A summary of the key steps in isolating esophageal epithelial cells from patient biopsies is summarized in Figure 1. Colonies of epithelial cells will form in approximately 4-5 days and will be surrounded by fibroblast feeder cells (Figure 2A). As these colonies expand they will merge with other colonies to form larger colonies (Figure 2B). Once the cultures have become 70% confluent, they need to be expanded (Figure 2C). To ensure that fibroblasts are removed from the plate prior to removing the epithelial cells, 0.05% trypsin-EDTA is used for up to 2.5 min to remove the feeders. The absence of feeders should look similar to Figure 2D. The adherent epithelial cells are then trypsinized with 0.25% Trypsin-EDTA and replated on new irradiated feeder cells (Figure 2E).
Phenotypic characterization of these cells via qRT-PCR demonstrates upregulation of P63, a marker of progenitor cells at the end of passage 1, while still maintaining E-cadherin and TJP1 expression as compared to the initial biopsy sample. This indicates that cells have been "reprogrammed" to a more progenitor/stem-like state that are more proliferative compared to non-reprogrammed cells. When these cells are taken from the conditional reprogramming conditions and plated on normal tissue culture plates, the expression of P63 decreases and TJP1 and E-Cadherin (CDH1) increases demonstrating a shift in phenotype from progenitor cell to a more differentiated epithelial cell (Figure 3A). These cells were also analyzed at passage 1 for epithelial markers via flow cytometry and greater than 90% of the cells present from three patients were positive for EpCAM (Figure 3B). Cells were also analyzed via immunofluorescence at passages 1 and 3 to ensure the phenotype over those 3 passages was not changing. The staining demonstrates the persistence of epithelial marker E-cadherin (Figure 4A-D), progenitor marker P63 (Figure 4E-H), proliferation marker KI-67 (Figure 4I-L) and epithelial tight junction protein TJP1 (Figure 4M-P).
Lastly, a growth curve was plotted using a cell line from a non-diseased patient to evaluate the doubling time of these cells in conditional reprogramming culture. Cells were plated at 100,000 cells at day 0 and were found to be just slightly over 500,000 cells at day 4 (Figure 5). The doubling time was calculated to be approximately 36-40 h. It is expected that diseased cells will have a slower doubling time, however, every patient should be assessed separately to account for individual differences.
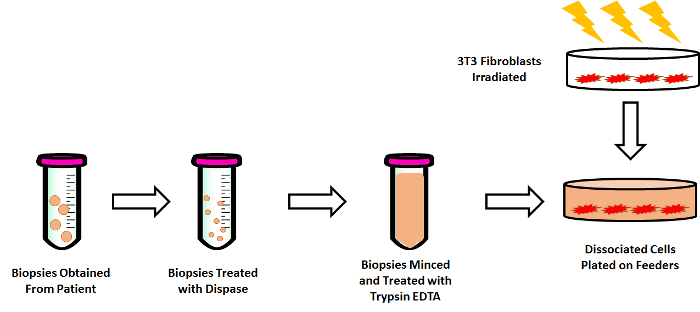
Figure 1: Overview of Isolation and Culturing Human Esophageal Epithelial Cells from Pediatric Biopsies. Biopsies are obtained from pediatric patients after informed consent and transported in keratinocyte serum-free medium to the lab. These biopsies are then treated with 1 U/ mL of dispase, followed by mincing the tissue with sterile razor blades and finally treating with 0.05% Trypsin-EDTA. The cell suspension is then added to irradiated feeder cells in conditional reprogramming medium. Please click here to view a larger version of this figure.
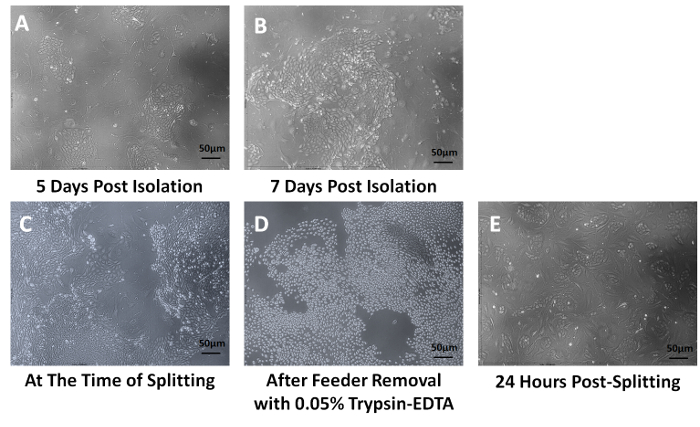
Figure 2: Culturing and Expanding Human Esophageal Epithelial Cells. After 5 days in conditional reprogramming medium, esophageal epithelial cells begin to form small cobblestone colonies (A), which eventually converge to form larger colonies (B). Once the tissue culture dish reaches 70% confluence (C), the plates are treated for a very brief time with 0.05% Trypsin-EDTA to remove the feeder cells, while still maintaining the adherence of the epithelial cells (D). The epithelial cells are then removed with 0.25% Trypsin-EDTA and replated with new feeder cells. Just 24 h after splitting these cells they will form new colonies and continue to proliferate (E). Magnification is 100X. Please click here to view a larger version of this figure.
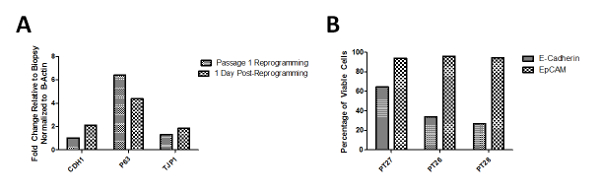
Figure 3: Flow Cytometry and qRT-PCR Analysis of Cells Expanded in Culture. (A) Representative gene expression of cells in culture at passage 1 of conditional reprogramming demonstrated a 6 fold increase in P63 expression, a marker of progenitor cells as well as a decrease in epithelial markers CDH1 and TJP1 as compared to the biopsy gene expression. This indicates the cell population is in a more progenitor-like state and therefore is more highly proliferative. Gene expression 24 h post-reprogramming (after the removal of feeder cells and ROCK Inhibitor) demonstrates increased CDH1 and TJP1 expression but a decrease in P63 expression. (B) Flow cytometry analysis of cells from multiple patients obtained at passage 1 demonstrate greater than 90% of the cells expressing EpCam. Patient 27 represents non-EoE patient cells, while Patients 26 and 28 are EoE patient cells. Please click here to view a larger version of this figure.
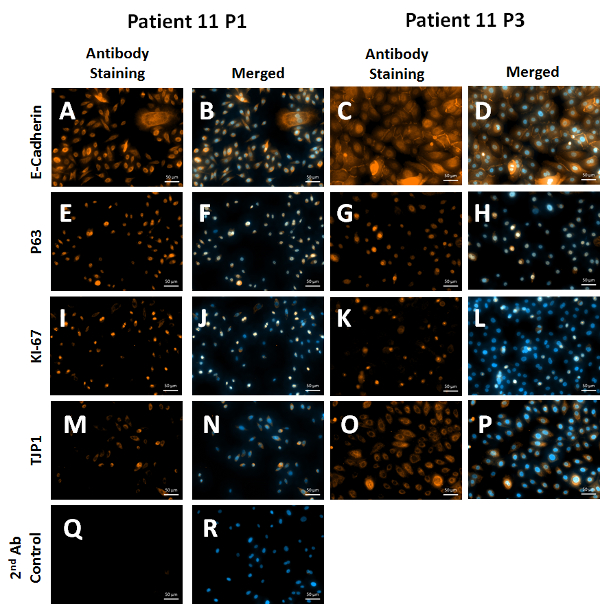
Figure 4: Immunofluorescence Staining of Cells at Passages 1 and 3. Cells grown in conditional reprogramming medium with irradiated feeder cells express E-Cadherin (A–D) as well as progenitor marker P63 (E–H). These cells are still proliferative at passage 3 (I–L). Lastly, these cells express tight junction protein 1 (TJP1), which is also consistent with an epithelial phenotype (M–P). Nuclei are counterstained with DAPI (Blue) and antibodies are detected using an Alexa Fluor 546 Antibody (Orange). 2nd Antibody (Ab) control represents non-specific binding of the secondary antibody to the tissue (Q, R). Scale bars = 50 µm. Magnification is 200X. Please click here to view a larger version of this figure.
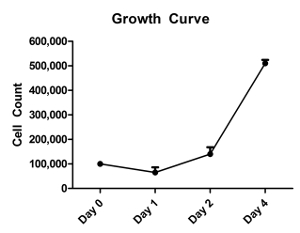
Figure 5: Growth Curve of Cells in Conditional Reprogramming. Cells in conditional reprogramming medium from a non-diseased patient were seeded at a defined density and replicates of three were counted each day for 4 days. The growth curve was generated using these cell counts and doubling time was calculated based on the numbers obtained at Day 4 and Day 0. The doubling time of these cells is approximately 36-40 h. Error bars indicate standard error. Please click here to view a larger version of this figure.
Discussion
The most important steps in order to isolate and expand esophageal epithelial cells from patient biopsies are: 1) adequately dissociating biopsy tissue with minimal cell death; 2) ensure ROCK inhibitor is added to the cell culture medium at every medium change; 3) Do not use more feeder cells than recommended; 4) maintain a clean aseptic culture; and 5) passage cells just prior to reaching confluence.
Due to the patient-related differences in biopsy samples obtained for conditional reprogramming, it is reasonable to expect differences in growth kinetics, phenotype and ability to expand to multiple passages. Typically, 4 esophageal biopsies are obtained from each patient for cell isolation. Once dissociated, we net approximately 300,000-600,000 cells at day 0. This expands to over 10 million cells by passage 3 on average. Morphology of colonies that form should look similar to cobblestone morphology described for epithelial cell lines25 (Figure 2). A sample of cells in culture should be analyzed via qRT-PCR, flow cytometry and immunofluorescence for epithelial markers to ensure the populations expanding are indeed epithelial in origin (Figure 3). Should the cells not express epithelial markers (E-Cadherin, EpCAM, TJP1), the cell line should be terminated and discarded. It is expected that greater than 90% of the cells present are EpCAM positive if they are epithelial in origin. In order to evaluate if the cultures are changing phenotype or losing epithelial expression, immunofluorescence or flow cytometry should be utilized to evaluate epithelial markers (E-cadherin), progenitor expression (P63), and epithelial tight junction proteins (TJP1). As the cells are passaged it is expected that phenotype should not change, proliferation should not decrease and these cells should display epithelial expression as well as progenitor expression (Figure 4). Furthermore, normal esophageal tissue that is dissociated and plated in conditional reprogramming culture appears to have a population doubling time of approximately 36-40 h (Figure 5). Cells that are found not to double for more than 5 days are likely to be senescent and should be discarded.
It has been described that cells isolated from patients with inflammatory processes present, such as EoE, possess epithelial cells that have a decreased proliferative rate, decreased E-cadherin expression (Figure 3, Patients 26 and 28) and a high rate of apoptosis26. Therefore, it is not unreasonable to have difficulty culturing cells from a severely ill patient using this method. It is possible that cells will only withstand a few passages or even just initial passage. This should still provide the investigator with a greater number of cells than originally isolated. However, this number may not be adequate for all proposed studies. Since these cells are being used to ultimately study esophageal disease or construct an esophageal replacement, it is important to remember these cells are only responsible for mucosal regeneration. Other cell types such as fibroblasts, neurons, smooth muscle cells and blood vessels are also just as important to the study of esophageal disease and for tissue engineering applications.
This technique offers a patient-specific cell source that is an invaluable tool for studying disease pathogenesis as well as serving as a potential reservoir of patient-specific cells for esophageal tissue engineering applications. Cells utilized for seeding biomaterials for surgical implantation must be free of genetic abnormalities, viral sequences and not form teratomas. The use of stem cells (multipotent or pluripotent) generates too many unwanted cell types following differentiation, some of which can form teratomas if they did not undergo differentiation. Moreover, this cell source phenotype is consistent and does not require purification or removal of unwanted cells. The ideal cell source will be cells that are found in the same anatomical location that is being engineered. In this approach these esophageal epithelial cells are being used to repopulate the mucosa of an esophageal scaffold. Additional cell types are needed for esophageal regeneration; however, the focus of this technique was to ensure the mucosa was reseeded and not incomplete as this can lead to leakage, stricture and perforation.
In addition to utilizing these cells for engineering new surgical options for the esophagus, these cells can also be used to study esophageal disease processes as well as screen for new therapeutic approaches to treat these conditions. Eosinophilic esophagitis (EoE) is described as an allergic disease of the esophagus, with the exact mechanism and pathogenesis not yet fully described. Current treatment regiments include elimination diets, oral steroids and multiple endoscopies. No assay exists to evaluate what triggers the inflammatory process for each patient, which leads doctors to use a "guess and check" approach which is long and often frustrating for the patient. Since this method of patient-specific cell expansion does not utilize lentiviral transduction or genetic modification, the possibility of permanent changes to the genotype of the cells from the reprogramming process is minimal. This means the cells theoretically remain unchanged from the in vivo environment. The ability to test patient cells in vitro to identify the triggering agent would be novel and could ultimately lead to development of new diagnostics or therapeutics for these pediatric patients.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We would like to acknowledge Connecticut Children’s Medical Center Strategic Research Funding for supporting this work.
Materials
| Primocin | InVivogen | ant-pm2 | |
| Isopentane | Sigma Aldrich | 277258-1L | |
| Gelatin From Porcine Skin | Sigma Aldrich | G1890-100G | |
| DMEM | Thermofisher Scientific | 11965092 | |
| Cryomold | TissueTek | 4565 | |
| Cryomatrix OCT | Thermofisher Scientific | 6769006 | |
| 15ml Conical Tubes | Denville Scientific | C1017-p | |
| Complete Keratinocyte Serum Free Medium | Thermofisher Scientific | 10724011 | |
| Penicillin Streptomycin | Thermofisher Scientific | 15140122 | |
| Glutamax | Thermofisher Scientific | 35050061 | |
| Insulin Solution | Sigma Aldrich | I9278-5ml | |
| Human Epidermal Growth Factor (EGF) | Peprotech | AF-100-15 | |
| ROCK Inhibitor (Y-27632) | Fisher Scientific | 125410 | |
| F-12 Medium | Thermofisher Scientific | 11765054 | |
| Fetal Bovine Serum | Denville Scientific | FB5001 | |
| Dispase | Thermofisher Scientific | 17105041 | |
| 0.05% Trypsin-EDTA | Thermofisher Scientific | 25300062 | |
| 0.25% Trypsin-EDTA | Thermofisher Scientific | 25200072 | |
| 100mm Dishes | Denville Scientific | T1110-20 | |
| 150mm Dishes | Denville Scientific | T1115 | |
| 50ml Conicals | Denville Scientific | C1062-9 | |
| Phosphate Buffered Saline Tablets | Fisher Scientific | BP2944-100 | |
| 5ml Pipettes | Fisher Scientific | 1367811D | |
| 10ml Pipettes | Fisher Scientific | 1367811E | |
| 25ml Pipettes | Fisher Scientific | 1367811 | |
| 9" Pasteur Pipettes | Fisher Scientific | 13-678-20D | |
| NIH 3T3 Cells | ATCC | CRL1658 |
Riferimenti
- Clark, D. C. Esophageal atresia and tracheoesophageal fistula. Am Fam Physician. 59 (4), 910-920 (1999).
- Soon, I. S., Butzner, J. D., Kaplan, G. G., deBruyn, J. C. Incidence and prevalence of eosinophilic esophagitis in children. J Pediatr Gastroenterol Nutr. 57 (1), 72-80 (2013).
- Sjöqvist, S., et al. Experimental orthotopic transplantation of a tissue-engineered oesophagus in rats. Nat Commun. 5, 3562 (2014).
- Saxena, A. K. Esophagus tissue engineering: designing and crafting the components for the ‘hybrid construct’ approach. Eur J Pediatr Surg. 24 (3), 246-262 (2014).
- Saxena, A. K., Ainoedhofer, H., Höllwarth, M. E. Culture of ovine esophageal epithelial cells and in vitro esophagus tissue engineering. Tissue Eng Part C Methods. 16 (1), 109-114 (2010).
- Saxena, A. K., Kofler, K., Ainodhofer, H., Hollwarth, M. E. Esophagus tissue engineering: hybrid approach with esophageal epithelium and unidirectional smooth muscle tissue component generation in vitro. J Gastrointest Surg. 13 (6), 1037-1043 (2009).
- Macheiner, T., Kuess, A., Dye, J., Saxena, A. K. A novel method for isolation of epithelial cells from ovine esophagus for tissue engineering. Biomed Mater Eng. 24 (2), 1457-1468 (2014).
- Mishra, A. Significance of Mouse Models in Dissecting the Mechanism of Human Eosinophilic Gastrointestinal Diseases (EGID). J Gastroenterol Hepatol Res. 2 (11), 845-853 (2013).
- Medvedev, S. P., Shevchenko, A. I., Zakian, S. M. Induced Pluripotent Stem Cells: Problems and Advantages when Applying them in Regenerative Medicine. Acta Naturae. 2 (2), 18-28 (2010).
- Metallo, C. M., Azarin, S. M., Ji, L., de Pablo, J. J., Palecek, S. P. Engineering tissue from human embryonic stem cells. J Cell Mol Med. 12 (3), 709-729 (2008).
- Lee, H., Park, J., Forget, B. G., Gaines, P. Induced pluripotent stem cells in regenerative medicine: an argument for continued research on human embryonic stem cells. Regen Med. 4 (5), 759-769 (2009).
- Ghaedi, M., et al. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J Clin Invest. 123 (11), 4950-4962 (2013).
- Huang, S. X., et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. , (2013).
- Ogaki, S., Morooka, M., Otera, K., Kume, S. A cost-effective system for differentiation of intestinal epithelium from human induced pluripotent stem cells. Sci Rep. 5, 17297 (2015).
- Ehrhardt, C., Kim, K. J., Lehr, C. M. Isolation and culture of human alveolar epithelial cells. Methods Mol Med. 107, 207-216 (2005).
- Zubeldia-Plazaola, A., et al. Comparison of methods for the isolation of human breast epithelial and myoepithelial cells. Front Cell Dev Biol. 3, 32 (2015).
- Spurrier, R. G., Speer, A. L., Hou, X., El-Nachef, W. N., Grikscheit, T. C. Murine and human tissue-engineered esophagus form from sufficient stem/progenitor cells and do not require microdesigned biomaterials. Tissue Eng Part A. 21 (5-6), 906-915 (2015).
- Roche, J. K. Isolation of a purified epithelial cell population from human colon. Methods Mol Med. 50, 15-20 (2001).
- Southgate, J., Hutton, K. A., Thomas, D. F., Trejdosiewicz, L. K. Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest. 71 (4), 583-594 (1994).
- Kalabis, J., et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc. 7 (2), 235-246 (2012).
- Geraghty, R. J., et al. Guidelines for the use of cell lines in biomedical research. Br J Cancer. 111 (6), 1021-1046 (2014).
- Lodish, H., Berk, A., Zipursky, S. L., et al. . Molecular Cell Biology. , (2000).
- Liu, X., et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 180 (2), 599-607 (2012).
- Suprynowicz, F. A., et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc Natl Acad Sci U S A. 109 (49), 20035-20040 (2012).
- Davis, A. A., et al. A human retinal pigment epithelial cell line that retains epithelial characteristics after prolonged culture. Invest Ophthalmol Vis Sci. 36 (5), 955-964 (1995).
- Carro, O. M., Evans, S. A., Leone, C. W. Effect of inflammation on the proliferation of human gingival epithelial cells in vitro. J Periodontol. 68 (11), 1070-1075 (1997).

