Cell Lineage Analyses and Gene Function Studies Using Twin-spot MARCM
Summary
Here, we present a protocol for a mosaic labeling technique that permits the visualization of neurons derived from a common progenitor cell in two distinct colors. This facilitates neural lineage analysis with the capability of birth-dating individual neurons and studying gene function in the same neurons of different individuals.
Abstract
Mosaic analysis with a repressible cell marker (MARCM) is a positive mosaic labeling system that has been widely applied in Drosophila neurobiological studies to depict intricate morphologies and to manipulate the function of genes in subsets of neurons within otherwise unmarked and unperturbed organisms. Genetic mosaics generated in the MARCM system are mediated through site-specific recombination between homologous chromosomes within dividing precursor cells to produce both marked (MARCM clones) and unmarked daughter cells during mitosis. An extension of the MARCM method, called twin-spot MARCM (tsMARCM), labels both of the twin cells derived from a common progenitor with two distinct colors. This technique was developed to enable the retrieval of useful information from both hemi-lineages. By comprehensively analyzing different pairs of tsMARCM clones, the tsMARCM system permits high-resolution neural lineage mapping to reveal the exact birth-order of the labeled neurons produced from common progenitor cells. Furthermore, the tsMARCM system also extends gene function studies by permitting the phenotypic analysis of identical neurons of different animals. Here, we describe how to apply the tsMARCM system to facilitate studies of neural development in Drosophila.
Introduction
The brain, comprised of a vast number and diverse types of neurons, endows animals with the ability to perceive, process, and respond to challenges from the external world. Neurons of the adult Drosophila central brain are derived from a limited number of neural stem cells, called neuroblasts (NBs), during development1,2. Most of the NBs participating in brain neurogenesis in Drosophila undergo asymmetric division to generate self-renewing NBs and ganglion mother cells (GMCs), and the GMCs then go through another round of division to produce two daughter cells that differentiate into neurons3 (Figure 1A). Because of the intricacy of neuronal morphology and the challenges associated with identifying specific neurons, a positive mosaic labeling technology, mosaic analysis with a repressible cell marker (MARCM), was invented to enable the visualization of a single neuron or a small subset of neurons out of the population of surrounding, unlabeled neurons4.
MARCM utilizes the flippase (FLP)/FLP recognition target (FRT) system to mediate site-specific recombination between homologous chromosomes within a dividing precursor cell carrying a heterozygous allele of a repressor gene, whose expression normally inhibits the expression of a reporter gene4. After the mitotic division, the recombinant chromosomes are segregated into twin cells, such that one cell contains homozygous alleles of the repressor gene and the other cell has no repressor gene-the expression of the reporter in that cell (and its descendants) is no longer blocked4. Three clonal patterns are usually found in a MARCM experiment when FLP is stochastically induced in NBs or GMCs: single-cell and two-cell-GMC clones, which depict neuronal morphology at single-cell resolution, and multi-cellular-NB clones, which reveal entire morphological patterns of neurons derived from a common NB (Figure 1B). The MARCM technique has been widely applied in Drosophila neurobiological studies, including in neuronal type identification for reconstructing brain-wide wiring networks, neural lineage analyses for disclosing the developmental history of neurons, phenotypic characterization of gene functions involved in cell fate specification, and neuronal morphogenesis and differentiation studies5,6,7,8,9,10. Because conventional MARCM only labels one of the two daughter cells (and lineages) after the induced mitotic recombination event, potentially useful information from the unmarked side is lost. This limitation precludes the application of the basic MARCM system to high-resolution analyses of many neural lineages that switch cell fates in fast tempo or to precision analyses of gene functions in identical neurons of different animals11,12.
Twin-spot MARCM (tsMARCM) is an advanced system that labels neurons derived from a common progenitor with two distinct colors, which enables the recovery of useful information from both sides of the twin cells, thus overcoming the limitation of the original MARCM system11 (Figure 2A–2C). In the tsMARCM system, two RNA interference (RNAi)-based suppressors are situated at trans-sites of homologous chromosomes within a precursor cell, and the expression of those suppressors independently inhibit the expression of their respective reporters (Figure 2B). Following site-specific mitotic recombination mediated through the FLP/FRT system, the two RNAi-based suppressors become segregated into twin cells to permit the expression of distinct reporters (Figure 2B). Two clonal patterns, single-cell associated with single-cell clones and two-cell-GMC associated with multi-cellular-NB clones, are typically seen in a tsMARCM experiment (Figure 2C). Information derived from one side of the twin cells can be utilized as the reference for the other side, enabling high-resolution neural lineage analyses, such as birth-dating the labeled neurons, and phenotypic analyses of identical neurons in different animals for the precise investigation of neural gene function11,12. Here, we present a step-by-step protocol describing how to conduct a tsMARCM experiment, which can be used by other laboratories to broaden their studies of neural development (as well as the development of other tissues, if applicable) in Drosophila.
Protocol
1. Build tsMARCM-ready Flies Using the Required Transgenes11,13
- Generate the original version of tsMARCM-ready flies from transgenes that are carried by individual flies11 (see Table 1). Conduct standard fly genetic crossing schemes, which have been described previously, by putting multiple transgenes in the same fly stocks13.
- In one parental line, assemble the transgenes of FRT40A, UAS-mCD8::GFP (the first reporter, mCD8::GFP; expression of the transgene is under the control of the upstream activation sequence promoter, which produces a membrane-tethered reporter of mouse cluster of differentiation 8 protein fused with green fluorescent protein), and UAS-rCD2RNAi (the suppressor that inhibits the expression of the second reporter; the RNAi against the expression of rat cluster of differentiation 2, rcd2) on the left arm of the 2nd chromosome. Place tissue-specific-GAL4 driver on the X or 3rd chromosome, if possible.
- In the other parental line, assemble the transgenes of FRT40A, UAS-rCD2::RFP (the second reporter, rCD2::RFP; the other membrane-tethered reporter consisting of rCD2 protein fused with red fluorescent protein), and UAS-GFPRNAi (the suppressor that inhibits the expression of mCD8::GFP; the RNAi against the expression of gfp) on the left arm of the 2nd chromosome. Place heat-shock (hs)-FLP or tissue-specific-FLP on the X or 3rd chromosome, if possible.
- Generate the new version of tsMARCM-ready flies from transgenes that are carried by individual flies14 (see Table 1). Conduct standard fly genetic crossing schemes, which have been described previously, by putting multiple transgenes in the same fly stocks13.
- In one parental line, assemble the transgenes of an FRT site and UAS-mCD8.GFP.UAS-rCD2i (a combined transgene of mCD8::GFP and the suppressor that inhibits the expression of rCD2::RFP) together in the same arm of the 2nd or 3rd chromosome (appropriate pairs of transgenes of FRT and UAS-mCD8.GFP.UAS-rCD2i are indicated in Table 1). Place tissue-specific-GAL4 driver on chromosomes other than the chromosome of the chosen FRT site, if possible.
- In the other parental line, assemble the transgenes of an FRT site and UAS-rCD2.RFP.UAS-GFPi (the other combined transgene of rCD2::RFP and the suppressor that inhibits the expression of mCD8::GFP) in the same arm of the 2nd or 3rd chromosome (appropriate pairs of transgenes of FRT and UAS-rCD2.RFP.UAS-GFPi are indicated in Table 1). Place hs-FLP or tissue-specific-FLP on chromosomes other than the chromosome of the chosen FRT site, if possible.
2. Cross tsMARCM-ready Flies to Generate tsMARCM Clones in Their Progeny
- Cross tsMARCM-ready flies together (e.g., put 10-15 male flies from step 1.1.1 or 1.2.1 and 20-30 virgin female flies from step 1.1.2 or 1.2.2 together) in a fly-food vial with fresh-made yeast paste on the wall of the vial.
- Maintain the crossed tsMARCM-ready flies at 25 °C for 2 days to enhance the chance of mating before collecting fertilized eggs.
- Frequently transfer the crossed tsMARCM-ready flies into freshly yeasted vials to avoid crowding (tap down the flies or use carbon dioxide to anesthetize the flies to facilitate the transfer; keep no more than 80-100 fertilized eggs in a 10 mL fly-food vial to avoid too many hatched larvae).
- Culture the tsMARCM animals (i.e., fertilized eggs; an example of the genotype of tsMARCM animals, as used in Figure 3, is hs-FLP[22],w/w; UAS-mCD8::GFP,UAS-rCD2RNAi,FRT40A,GAL4-GH146/UAS-rCD2::RFP,UAS-GFPRNAi,FRT40A; +; +) that have been laid in the serially transferred vials at 25 °C until they develop to the desired stages (e.g., embryos, larvae, or pupae).
- Place the transferred vials that contain tsMARCM animals at the same developmental stage to a 37 °C water bath for 10, 20, 30, 40, and 50 min to determine the optimal time of heat-shock that induces the expression of FLP to generate the tsMARCM clones of interest. Skip this step if tissue-specific-FLP is being used in the tsMARCM experiment (hs-FLP is preferred for neural lineage analyses; the reasons for this preference have been described previously15).
NOTE: Repeat step 2.5 using the optimal time of heat-shock in future tsMARCM experiments if more tsMARCM clones of interest are wanted; generally, more FLP expression is induced in hs-FLP[22] compared to hs-FLP[1] with the same heat-shock time. - Culture the heat-shocked tsMARCM animals at 25 °C until they have reached the appropriate developmental stage, and then proceed to step 3.
3. Prepare, Stain, and Mount Fly Brains Containing tsMARCM Clones
- Prepare forceps-protection dishes. Mix part A (30 g) and part B (3 g) of the Silicone Elastomer Kit with activated charcoal (0.25 g). Pour the mixture into the appropriate dishes.
- Dissect larval, pupal, or adult brains out of the tsMARCM animals in 1x phosphate-buffered saline (PBS) using forceps on a forceps-protection dish viewed through a dissection microscope. NOTE: A protocol for fly brain dissection has been described previously16.
- Fix the fly brains in a glass spot plate with 1x PBS containing 4% formaldehyde at room temperature for 20 min. Process the fly brains in this glass spot plate for the remainder of the steps.
- Rinse the fly brains three times with 1% PBT (1x PBS containing 1% Triton X-100) and wash them three times for 30 min each in 1% PBT (rinse: remove PBT and add new PBT quickly; wash: remove PBT, add new PBT, and incubate the fly brains in the new PBT using an orbital shaker).
- Incubate the fly brains with the mixture of primary antibodies overnight at 4 °C or for 4 h at room temperature. An example of the mixture of primary antibodies is rat anti-CD8 antibody (1:100), rabbit anti-DsRed antibody (1:800), and mouse anti-Bruchpilot (Brp) antibody (1:50) in 1% PBT containing 5% normal goat serum (NGS).
- Rinse the fly brains three times with 1% PBT, and then wash them three times for 30 min each with 1% PBT.
- Incubate the fly brains with the mixture of secondary antibodies overnight at 4 °C or for 4 h at room temperature. An example of the mixture of secondary antibodies is goat anti-rat IgG antibody conjugated with green-fluorescent dye (1:800), goat anti-rabbit IgG antibody conjugated with yellow-fluorescent dye (1:800), and goat anti-mouse IgG antibody conjugated with far-red-fluorescent dye (1:800) in 1% PBT containing 5% NGS.
- Rinse the fly brains three times with 1% PBT, and then wash them three times for 30 min each with 1% PBT. Mount them on a micro slide with an anti-quenching reagent. Put a micro cover glass on the micro slide to cover and protect the fly brains. Seal the edges of the micro cover glass and micro slide using clear nail polish. Store these mounted and sealed fly brain specimens at 4 °C.
4. Take, Process, and Analyze Fluorescent Images of tsMARCM Clones
- Capture fluorescent images of tsMARCM clones from the samples created in step 3.8 using the confocal microscopy system of choice.
- Project stacks of tsMARCM confocal fluorescent images into two-dimensional, flattened images using the image processing software supplied with the confocal microscopy system of choice (see Figure 3B–3E, 3G–3J, 3L–3O, and 3Q–3T for examples of flattened confocal fluorescent images).
- Count the cell number of the multi-cellular-NB sides of the tsMARCM clones using an appropriate image processing software (e.g., the "Cell Counter" function in ImageJ17; see Figure 3E, 3J, 3O, and 3T for examples of the cell bodies of neurons).
Representative Results
The tsMARCM system has been used to facilitate neural lineage analyses and gene function studies by retrieving important information on neurons derived from common NBs. The system has been used to identify most (if not all) neuronal types, determine the cell number of each neuronal type, and ascertain the birth-order of these neurons11,12,18 (see the Discussion Section for details on using tsMARCM to conduct neural lineage analyses or gene function studies). Here, we used four tsMARCM clones of anterodorsal olfactory projection neurons (adPNs in the ALad1 neural lineage of the Drosophila olfactory system) labeled with GAL4-GH146 as examples to illustrate how to conduct a tsMARCM experiment1,2. After FLP expression was induced at the embryonic stage, a single, green VL2p adPN associated with 45 magenta adPNs was detected in a tsMARCM clone (Figure 3A–3E). In contrast, when FLP was induced later, at different larval stages, single, green adPNs (DL1 adPN or DC3 adPN) or no-GH146+ adPNs associated with different numbers of magenta adPNs (31, 21, or 15) were observed in three other tsMARCM clones (Figure 3F–3T). The birth of VL2p adPNs prior to the birth of DL1-, DC3- and no-GH146+-adPNs was determined by counting the cell numbers of adPNs in their associated multi-cellular-NB sides of the tsMARCM clones (45, 31, 21, and 15 in Figure 3E, 3J, 3O, and 3T, respectively). In addition to counting the cell numbers, the complexity of the axonal and dendritic patterns in the multi-cellular-NB sides of the tsMARCM clones can be used to determine the relative birth-order of their associated two-cell-GMC sides of the tsMARCM clones. Mosaic clones induced at early developmental stages normally contain more neurons and thus have more complex axonal and dendritic patterns, whereas mosaic clones induced at late developmental stages contain fewer neurons and thus have less complex axonal and dendritic patterns (Figure 3C–3D, 3H–3I, 3M–3N, and 3R–3S).
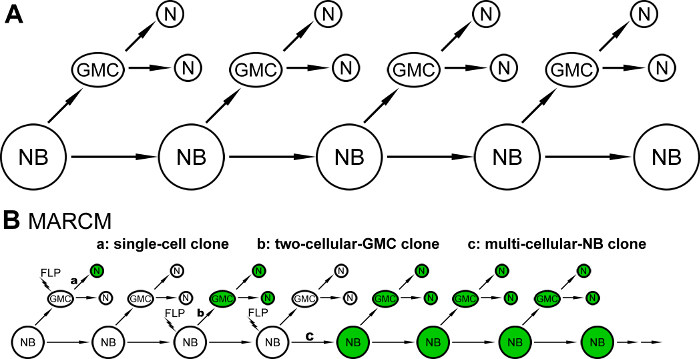
Figure 1: Schematic illustrations of Drosophila neurogenesis and the three types of clonal patterns found in a MARCM experiment. (A) A schematic diagram illustrating neurogenesis in most neural lineages of the Drosophila central brain: neuroblasts (NBs) undergo an asymmetric division to generate self-renewing NBs and ganglion mother cells (GMCs), and GMCs divide one more time to produce two daughter neurons (Ns). (B) Three clonal patterns are observed in a MARCM experiment: single-cell clones (a), when flippase (FLP) is induced in GMCs, and two-cell-GMC clones (b) and multi-cellular-NB clones (c), when FLP is induced in NBs. Please click here to view a larger version of this figure.
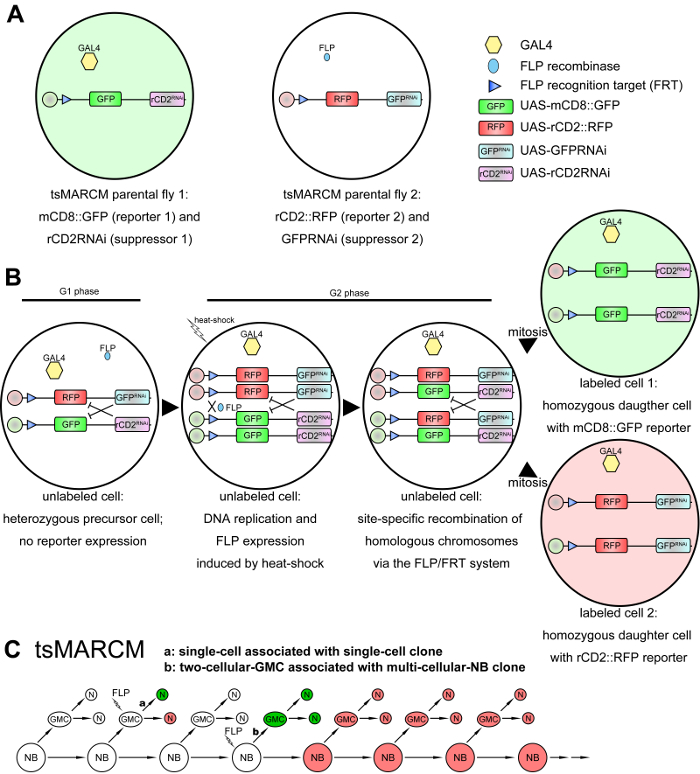
Figure 2: Schematic diagrams of the tsMARCM system. (A) tsMARCM-ready flies: one parental fly contains transgenes of an FRT site and UAS-mCD8::GFP (the first reporter), UAS-rCD2RNAi (the first suppressor that inhibits the expression of the second reporter), and a tissue-specific-GAL4; the other parental fly contains an FRT site, UAS-rCD2::RFP (the second reporter), UAS-GFPRNAi (the second suppressor that inhibits the expression of the first reporter), and hs-FLP. (B) The presence of the two RNAi-based suppressors in the progeny of the crossed tsMARCM-ready flies inhibits the expression of reporters (i.e., mCD8::GFP and rCD2::RFP in the G1 and G2 phases of panel B). The expression of FLP is induced by heat-shock, and FLP then mediates the site-specific recombination of homologous chromosomes within a dividing precursor cell upon binding to FLP recognition targets (FRTs). This results in the segregation of the RNAi-based suppressors in the twin cells after mitotic division to permit the expression of distinct reporters. (C) Two clonal patterns are observed in a tsMARCM experiment: single-cell, associated with single-cell clones (a) when FLP is induced in GMCs, and two-cell-GMC, associated with multi-cellular-NB clones (b) when FLP is induced in NBs. Please click here to view a larger version of this figure.
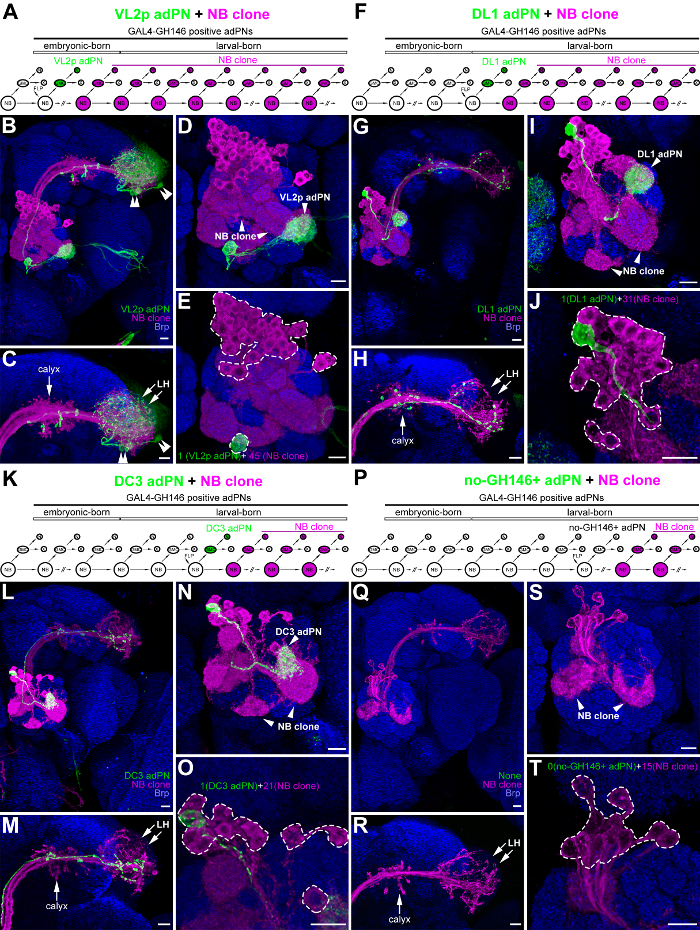
Figure 3: Using four tsMARCM clones of adPNs as examples of how to conduct a tsMARCM experiment in a neural lineage analysis. Four tsMARCM clones of adPNs are illustrated, with diagrams of their neurogenesis patterns (A, F, K, and P); confocal images of the overall brain (B, G, L, and Q), showing the lateral horn (LH; C, H, M, and R), the antennal lobe (AL; D, I, N, and S) and the soma (E, J, O, and T); and the patterns of their neuronal morphologies. Single adPNs (VL2p, DL1, DC3, and no-GH146+ adPNs; green) associated with multi-cellular-NB adPNs (magenta) of the tsMARCM clones were derived by the induction of FLP to mediate mitotic recombination in the adPN NB at the embryonic (A–E) and larval (F–T) stages. Note that adPNs are one of the hemi-lineages in the ALad1 neural lineage, which results in tsMARCM clones of a single, green adPN associated with magenta adPNs in Figure 3 (for unknown reasons, neurons from the other hemi-lineage of the ALad1 neural lineage die). The birth-order of VL2p, DL1, DC3, and no-GH146+ adPNs was determined by counting the cell numbers of adPNs in the magenta multi-cellular-NB sides of the tsMARCM clones (45, 31, 21, and 15 within the dashed lines of panels E, J, O, and T). The neurite patterns are usually more complex in the magenta multi-cellular-NB sides when the tsMARCM clones are induced at early developmental stages (e.g., the dendritic pattern is more complex in panel E compared to panels I, N, and S). tsMARCM analyses are sometimes confusing when neurons of other neural lineages are also labeled by the same GAL4 driver (e.g., green LH neurons, shown by double-arrows, obstruct the visualization of the axonal elaboration pattern of the VL2p adPNs in panels B and C). Brain neuropiles (shown in blue) were stained with antibody against Bruchpilot (Brp). Heat-shock time: 12 min (panels A–E), 15 min (panels F–J), and 35 min (panels K–T). Scale bar in panels B–E, G–J, L–O, and Q–S = 10 µm. Please click here to view a larger version of this figure.
Discussion
Critical Steps within the Protocol
Steps 1.1.1, 1.2.1, 2.3, 2.5, and 3.2 are critical for obtaining good tsMARCM results. Tissue-specific-GAL4 drivers that do not express GAL4 in neural progenitors are preferred for steps 1.1.1 and 1.2.1. Avoid over-crowding the animals grown in the fly-food vials in step 2.3. Because the induction latency, the expression level of FLP upon heat-shock, and the average dividing time of neural progenitors (i.e., NBs and GMCs) are not known, it is best to vary the heat-shock time (10, 20, 30, 40, and 50 min) on different samples of the same genotype and developmental stage in step 2.5. This will allow for the determination of the optimal heat-shock time in order to obtain the best ratio of tsMARCM clones of interest. In addition, be careful during fly brain dissection in step 3.2 in order to obtain intact brain samples; this is crucial for accurately counting neuronal cell numbers. For researchers who are interested in using tsMARCM for studying the function of genes on other chromosome arms (e.g., 2R, 3L, and 3R), tsMARCM-ready flies should be assembled as in step 1.2 by choosing appropriate pairs of transgenes of FRT, UAS-mCD8.GFP.UAS-rCD2i,and UAS-rCD2.RFP.UAS-GFPi, which are indicated in Table 1. We also recommend that the female tsMARCM-ready flies carry hs-FLP on the X chromosome (e.g., hs-FLP[22],w; UAS-rCD2::RFP,UAS-GFPRNAi,FRT40A; +; +), which facilitates repeated tsMARCM experiments.
Modifications, Troubleshooting, and Limitations of the Technique
The efficiency and perdurance of RNAi suppressors and reporters are the major determinants for whether neural lineage analyses and gene function studies can be successfully performed in tsMARCM experiments. Generally, the combination of low perdurance of RNAi suppressors and reporters derived from neural progenitors, together with the high expression of RNAi suppressors and reporters in post-mitotic neurons, permits better tsMARCM results. Because the expression of RNAi suppressors and reporters is controlled by GAL4 drivers, researchers should select strong GAL4 drivers that express GAL4 in differentiated neurons but not in neural progenitors. For example, GAL4-OK107 is a strong GAL4 driver that expresses GAL4 in mushroom body (MB) progenitors and their derivatives, MB γ, α'/β', and α/β neurons11,19. Unfaithful labeling of MB γ neurons has been observed in two clonal patterns (i.e., single-cell associated with single-cell clones and two-cell-GMC associated with multi-cellular-NB clones) when GAL4-OK107 was used in tsMARCM experiments11. Intriguingly, the above issue can be solved by using MB247-GAL4, a different MB GAL4 driver that expresses GAL4 in differentiated MB neurons11.
Not all GAL4 drivers can be used in the tsMARCM system or in other mosaic labeling systems (e.g., MARCM, Q-MARCM, and twin-spot generator)4,20,21 if the driver is not expressed in the neurons of interest. Therefore, prior to using tsMARCM for neural lineage analyses, GAL4 drivers should be tested for their coverage of the neurons of interest in the dual-expression-control MARCM with a pan-cell driver (e.g., tubulin promoter-LexA::GAD)22. We note that tsMARCM clones sometimes cannot be analyzed unambiguously, especially when a GAL4 driver with a complex neural expression pattern is used (e.g., green lateral horn (LH) neurons prevent clear visualization of the axonal elaboration pattern of the VL2p adPN in Figure 3B and 3C). However, the above issue of ambiguous patterns also occurs in other mosaic labeling systems (e.g., MARCM, Q-MARCM, and twin-spot generator)4,20,21 because the limitation originates with the GAL4 driver. We also note that, to conduct rescue experiments for the authentication of gene functions, the rescue transgenes should be engineered with the target sequences of the RNAi suppressors in 5' or 3' untranslated regions of the transgenic rescue constructs, or an alternative twin-spot labeling system (e.g., coupled MARCM, a combination of Q-MARCM and conventional MARCM)21 should be adopted.
Significance of the Technique with Respect to Existing/Alternative Methods
Besides tsMARCM, twin-spot generator and coupled MARCM would also, at least in theory, allow for the visualization of neurons derived from common neural progenitors in two distinct colors (a comparison of these three mosaic labeling technologies has been presented previously15). The minimum number of transgenes required for assembling twin-spot generator, the original version of tsMARCM (or the new version of tsMARCM), and coupled MARCM is 4, 8 (or 6), and 9, respectively. However, tsMARCM is the only system that has been used to conduct comprehensive neural lineage analyses12,15,18,23. Site-specific recombination does occur in non-mitotic cells in the twin-spot generator system, and this leads to the co-expression of the two reporters in the same cell, making it challenging to use this system to study the development of the central nervous system15. In contrast, mosaic clones obtained in the tsMARCM and coupled MARCM systems are derived from mitotic cells. However, because the coupled MARCM system requires two independent drivers to control the expression of the different reporters in the Q-MARCM and conventional MARCM systems, this raises concerns about the labeling of neurons derived from common neural progenitors if the two independent drivers do not faithfully label the same neurons in the neural lineages of interest15.
Future Applications or Directions after Mastering the Technique
Using tsMARCM to conduct high-resolution neural lineage analyses
By applying the approach used in Figure 3 to systematically analyze tsMARCM clones of adPNs, 40 types of adPNs, with a total of 82 neurons, were identified in a previous study12. Among these 40 types of adPNs, 35 were uni-glomerular adPNs (i.e., innervating a single glomerulus in the antennal lobe (AL)), which relay olfactory information to other regions of the Drosophila brain12 (see Figures 2-4 in Yu et al., 2010). The other five types of adPNs were poly-glomerular adPNs (i.e., innervating multiple glomeruli in the AL), which could integrate olfactory information from the AL and provide an additional level of functional complexity to the olfactory system12 (see Figures 3E-3H and 4Z in Yu et al., 2010). Embryonic-born adPNs have only one neuron per type, whereas larval-born adPNs are represented by multiple neurons per type, suggesting that there is interplay between specificity and redundancy in the olfactory system12 (see Figures 2-4 in Yu et al., 2010). Interestingly, neurons produced within certain developmental windows are destined to die, and many types of adPNs are found with a fixed number of neurons in the ALad1 neural lineage, suggesting that the number and types of adPNs for assembling the olfactory system are tightly regulated12 (see Figures 2H, 4V, and 5 in Yu et al., 2010; also see Figure 3P–3T).
Using tsMARCM to accurately study gene functions
Besides birth-dating adPNs, tsMARCM has also been applied to determine the birth-order of six central complex neurons labeled by GAL4-OK107. These six neurons make up a small subset of neurons in the LALv1 neural lineage that contribute to the circuitry of the Drosophila central complex to control intricate motor patterns2,11 (see Figure 4a and 4b in Yu et al., 2009). Interestingly, each of these six central complex neurons possesses a distinct neuronal morphology11 (see Figure 4d-4h and 4o in Yu et al., 2009). All of them carry distinct axonal patterns in the lateral accessory lobe (LAL). The first four neurons distribute their dendrites to sub-compartments of the noduli, a sub-structure of the central complex, whereas the remaining two neurons project their dendrites to the ellipsoid body, another sub-structure of the central complex. Specifically, the 3rd-born central complex neuron projects its dendrites to the ventral noduli, with a compact axonal pattern in the LAL, whereas the 4th-born central complex neuron distributes its dendrites to the dorsal noduli, with a broad axonal pattern in the LAL11 (see Figure 4f and 4g in Yu et al., 2009).
Beyond comprehensively identifying neurons, the power of tsMARCM would be better exemplified by studying gene functions in neurons of interest, since the tsMARCM system permits better phenotypic analyses by enabling the examination of identical neurons in different animals. For example, tsMARCM detects a single temporal fate change that requires chronologically inappropriate morphogenesis (chinmo, a BTB-zinc finger nuclear protein) in the 3rd-born central complex neurons11 (see Figure 4i, 4p, and 4w in Yu et al., 2009). If the chinmo mutation had been characterized in the original MARCM system (i.e., single green-labeled neurons, without the labeling of neurons derived from the associated sides of the twin cells as the reference), the chinmo-deficient neuron shown in Figure 4i from Yu et al., 2009 would have been misidentified as the 4th-born, rather than the 3rd-born, central complex neuron. This would have resulted in the misinterpretation of chinmo function as controlling axonal guidance rather than specifying temporal fate11 (double-arrow in Figure 4i from Yu et al., 2009). However, by using the magenta multi-cellular-NB sides of tsMARCM clones as the reference (via counting the cell numbers in Figure 4m, 4n, and 4p from Yu et al., 2009), the chinmo-deficient neuron shown in Figure 4i from Yu et al., 2009) was proven to be the 3rd-born central complex neuron, which therefore led to the correct conclusion that chinmo functions in temporal fate specification for central complex neurons11. Taken together, the above results highlight the important advantages of the tsMARCM system for high-resolution phenotypic analyses, thereby accurately disclosing gene functions in neurons (or other types of cells, if applicable) for future neural developmental studies (or the developmental studies of other tissues).
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Ministry of Science and Technology (MOST 104-2311-B-001-034) and the Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan.
Materials
| Carbon dioxide (CO2) | Local vendor | n.a. | Anesthetize flies |
| CO2 pad | Local vendor | n.a. | Sort, cross and transfer flies |
| 10x PBS pH 7.4 | Uniregion Bio-Tech (or other vendors) | PBS001 | Dissect, rinse, wash and immunostain fly brains |
| Formaldehyde | Sigma-Aldrich | 252549 | Fix fly brains |
| Triton X-100 | Sigma-Aldrich | T8787 | Rinse, wash and immunostain fly brains |
| Normal goat | Jackson ImmunoResearch | 005-000-121 | Immunostain fly brains |
| serum | |||
| Rat anti-mouse CD8 antibody | Invitrogen | MCD0800 | Immunostain fly brains |
| Rabbit anti-DsRed antibody | Clonetech | 632496 | Immunostain fly brains |
| Mouse anti-Brp antibody | Developmental Studies Hybridoma Bank | nc82 | Immunostain fly brains |
| Goat anti-rat IgG antibody conjugated with Alexa Fluor 488 | Invitrogen | A11006 | Immunostain fly brains |
| Goat anti-rabbit IgG antibody conjugated with Alexa Fluor 546 | Invitrogen | A11035 | Immunostain fly brains |
| Goat anti-mouse IgG antibody conjugated with Alexa Fluor 647 | Invitrogen | A21236 | Immunostain fly brains |
| SlowFade gold antifade reagent | Molecular Probes | S36936 | Mount fly brains and protect quenching of fluorescence |
| PYREX 9 depression glass spot plate | Corning | 7220-85 | Dissect, rinse, wash and immunostain fly brains |
| Sylgard 184 silicone elastomer kit | World Precision Instruments | SYLG184 | Make black Sylgard dishes to protect forceps during brain dissection |
| Activated charcoal | Sigma-Aldrich | 242276-250G | Make black Sylgard dishes |
| Dumont #5 forceps | Fine Science Tools | 11252-30 | Dissect and mount fly brains |
| Micro slide | Corning | 2948-75×25 | Mount fly brains |
| Micro cover glass No.1.5 | VWR International | 48366-205 | Mount fly brains |
| Nail polish | Local vendor | not available | Seal micro cover glass on micro slides |
| Incubator | Kansin Instruments | LTI603 | culture flies at 25°C |
| (or other vendors) | |||
| Water bath | Kansin Instruments | WB212-B2 | Induce heat-shock in flies at 37°C |
| (or other vendors) | |||
| Orbital shaker | Kansin Instruments | OS701 | Wash and immunostain fly brains |
| (or other vendors) | |||
| Dissection microscope | Leica | EZ4 | Sort, cross and transfer flies; Dissect, rinse, wash and immunostain fly brains |
| Confocal microscope | Zeiss (or other vendors) | LSM 700 (or other models) | Image tsMARCM clones |
| image-processing software 1 (e.g., Zeiss LSM image browser) |
Zeiss | not available | Project stacks of confocal images |
| image-processing software 2 (e.g., Image-J) |
not available | not available | Count cell number of tsMARCM clones |
Riferimenti
- Ito, M., Masuda, N., Shinomiya, K., Endo, K., Ito, K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr Biol. 23 (8), 644-655 (2013).
- Yu, H. H., et al. Clonal development and organization of the adult Drosophila central brain. Curr Biol. 23 (8), 633-643 (2013).
- Goodman, C. S., Doe, C. Q., Bate, M., Arias, A. M. . The Development of Drosophila melanogaster. , (1993).
- Lee, T., Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22 (3), 451-461 (1999).
- Lee, T., Lee, A., Luo, L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 126 (18), 4065-4076 (1999).
- Lee, T., Marticke, S., Sung, C., Robinow, S., Luo, L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 28 (3), 807-818 (2000).
- Chiang, A. S., et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 21 (1), 1-11 (2011).
- Jefferis, G. S., Marin, E. C., Stocker, R. F., Luo, L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 414 (6860), 204-208 (2001).
- Hong, W., Luo, L. Genetic control of wiring specificity in the fly olfactory system. Genetica. 196 (1), 17-29 (2014).
- Zhu, S., et al. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 127 (2), 409-422 (2006).
- Yu, H. H., Chen, C. H., Shi, L., Huang, Y., Lee, T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 12 (7), 947-953 (2009).
- Yu, H. H., et al. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biol. 8 (8), (2010).
- Greenspan, R. J. . Fly pushing : the theory and practice of Drosophila genetics. , (1997).
- Awasaki, T., et al. Making Drosophila lineage-restricted drivers via patterned recombination in neuroblasts. Nat Neurosci. 17 (4), 631-637 (2014).
- Lee, T. Generating mosaics for lineage analysis in flies. Wiley Interdiscip Rev Dev Biol. 3 (1), 69-81 (2014).
- Wu, J. S., Luo, L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 1 (4), 2110-2115 (2006).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9 (7), 671-675 (2012).
- Lin, S., Kao, C. F., Yu, H. H., Huang, Y., Lee, T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biol. 10 (11), e1001425 (2012).
- Zhan, X. L., et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 43 (5), 673-686 (2004).
- Griffin, R., et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods. 6 (8), 600-602 (2009).
- Potter, C. J., Tasic, B., Russler, E. V., Liang, L., Luo, L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 141 (3), 536-548 (2010).
- Lai, S. L., Lee, T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 9 (5), 703-709 (2006).
- Kao, C. F., Yu, H. H., He, Y., Kao, J. C., Lee, T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 73 (4), 677-684 (2012).

