A Guide to Production, Crystallization, and Structure Determination of Human IKK1/α
Summary
IκB Kinase 1/α (IKK1/α CHUK) is a Ser/Thr protein kinase that is involved in a myriad of cellular activities primarily through activation of NF-κB transcription factors. Here, we describe the main steps necessary for the production and crystal structure determination of this protein.
Abstract
A class of extracellular stimuli requires activation of IKK1/α to induce generation of an NF-κB subunit, p52, through processing of its precursor p100. p52 functions as a homodimer or heterodimer with another NF-κB subunit, RelB. These dimers in turn regulate the expression of hundreds of genes involved in inflammation, cell survival, and cell cycle. IKK1/α primarily remains associated with IKK2/β and NEMO as a ternary complex. However, a small pool of it is also observed as a low molecular weight complex(es). It is unknown if the p100 processing activity is triggered by activation of IKK1/α within the larger or the smaller complex pool. Constitutive activity of IKK1/α has been detected in several cancers and inflammatory diseases. To understand the mechanism of activation of IKK1/α, and enable its use as a drug target, we expressed recombinant IKK1/α in different host systems, such as E. coli, insect, and mammalian cells. We succeeded in expressing soluble IKK1/α in baculovirus infected insect cells, obtaining mg quantities of highly pure protein, crystallizing it in the presence of inhibitors, and determining its X-ray crystal structure. Here, we describe the detailed steps to produce the recombinant protein, its crystallization, and its X-ray crystal structure determination.
Introduction
Transcriptional activities of the NF-κB family of dimeric transcription factors are required for diverse cellular functions ranging from inflammation and immunity to survival and death. These activities are stringently controlled in cells and a loss of regulation leads to various pathological conditions, including autoimmune disorders, and cancer1,2,3. In the absence of a stimulus, the activities of NF-κB are kept inhibited by IκB (Inhibitor of -κB) proteins4. The phosphorylation of specific Ser residues on IκB proteins marks them for ubiquitination and subsequent proteasomal degradation or selective processing5. Two highly homologous Ser/Thr kinases, IKK2/β and IKK1/α, act as central regulators of NF-κB activities by carrying out these phosphorylation events6,7.
Interactions between a ligand and a receptor transduces a signal through a series of mediators leading to the activation of NF-κB factors. The NF-κB signalling process can broadly be classified into two distinct pathways – canonical and non-canonical (alternative)8. The activity of IKK2/β primarily regulates the NF-κB signalling of the canonical pathway that is essential for inflammatory and innate immune responses9. A distinct feature of this pathway is a rapid and short-lived activation of IKK2/β10 within a hitherto biochemically uncharacterized IKK complex — presumed to be composed of IKK1 and IKK2, as well as a regulatory component, NEMO (NF-κB Essential Modulator)11,12,13. Between the two catalytic IKK subunits of the IKK complex, IKK2 is primarily responsible14 for the phosphorylation of specific residues of prototypical IκBs (α, -β, & -γ) bound to NF-κB, and also an atypical IκB protein, NF-κB1/p105, which is a precursor of the NF-κB p50 subunit5. Phosphorylation induced ubiquitination and proteasomal degradation of IκB (or processing of p105) leads to the release and activation of a specific set of NF-κB dimers15. Aberrant NF-κB activity due to mis-regulated function of IKK2 has been observed in many cancers as well as in autoimmune disorders2,3,16.
In contrast to IKK2/β, activity of IKK1/α regulates NF-κB signalling of the non-canonical pathway, which is essential for development and immunity. IKK1 phosphorylates specific residues of NF-κB2/p100 on its C-terminal IκBδ segment, which leads to its processing and the generation of p52. The formation of transcriptionally active p52:RelB heterodimer initiates a slow and sustained response to developmental signals7,17,18,19,20. Interestingly, the generation of the central NF-κB factor p52 of this pathway is critically dependent on another factor, NF-κB Inducing Kinase (NIK)21,22, but not on IKK2 or NEMO. In resting cells, the level of NIK remains low due to its constant proteasome-dependent degradation23,24,25. Upon stimulation of cells by 'non-canonical' ligands, and in certain malignant cells, NIK becomes stabilized to recruit and activate IKK1/α. Kinase activities of both NIK and IKK1 are essential for efficient processing of p100 into p527. IKK1 and NIK phosphorylate three serines (Ser866, 870 and 872) of NF-κB2/p100 on its C-terminal IκBδ segment leading to its processing and the generation of p52. Aberrant activation of the non-canonical pathway has been implicated in many malignancies including multiple myeloma26,27,28.
Several highly efficient and specific inhibitors for IKK2/β are known, although none so far have turned out to be an effective drug. In contrast, IKK1/α-specific inhibitors are sparse. This may stem partly from our lack of structural and biochemical information on IKK1/α, which limits our understanding of the mechanistic basis of activation of NF-κB by IKK1 in cells, and rational drug design. The X-ray structures of IKK2/β provided us with insights into the activation mechanism of IKK2/β29; however, these structures could not reveal how different upstream stimuli trigger activation of IKK1/α or IKK2/β to regulate distinct sets of NF-κB activities 30,31. To understand the mechanistic basis underlying the distinct signalling function of IKK1/α, and to establish a platform for rational drug design, we focused on determining the structure of IKK1/α.
Protocol
1. Preparation of Recombinant Virus Suitable for Large Scale Expression of IKK1/α
- P1 virus preparation32
- Day 1: Plate Sf9 cells (~6 X 105) (passage number less than 10) in 2 mL of Sf900 III insect cell medium in each well of a 6-well plate and incubate at 27 °C. Passage cells when they reach a density of 2 to 3 X 106 cells per mL in suspension by diluting into fresh media at a density of ~6 X 105.
- Day 2: Dilute 8 µL of Sf9-transfection reagent in 100 µL of Sf900 III or Grace's Insect Medium without antibiotic and serum. Vortex briefly.
- Dilute 1 µg of kit-purified recombinant bacmid (details are provided in 'Representative Results')32 into another 100 µL of the same medium, in a separate tube.
- Combine diluted DNA and transfection reagent (total volume ~210-220 μL), and incubate at room temperature for 15-30 min.
- Remove the medium from the well. Add 1 mL of fresh medium without antibiotic and serum.
- Add the diluted DNA-transfection reagent mixture dropwise onto the cells. Adjust the drop size such that it does not dislodge adherent cells.
- After ~6 h, add 1.5 mL of fresh medium on top or remove the medium and add 2.5 mL of fresh medium. Warm fresh medium to 25-27 °C before addition.
- Incubate the cells at 27 °C. Check the wells periodically every ~12 h for signs of viral infection. Carry out all Sf9 maintenance and expression at 27 °C.
- Day 5: Take out the medium containing cells 60-72 h post infection. Centrifuge for 5 min at 500 x g and 4 °C. Save the supernatant; this is the P1-virus stock. Freeze small aliquots at -80 °C.
- Selecting the best expressing P1 virus clone.
- Plate cells similarly as described in step 1.1.
- Next day or ~6 h post plating, add 20 µL and 50 µL of different P1 virus clone stocks onto the cells in different wells.
- Dislodge the adherent cells by gentle pipetting and harvest the infected cells 60-72 h post infection by centrifuging for 5 min at 500 x g and 4 °C. Save the supernatant. This is the P2 virus stock. Store small aliquots of the stock at -80 °C.
- Add 100 µL of 2x Laemmli SDS buffer to the harvested cell pellet. Heat at >95 °C for 10 min. Centrifuge (typically > 10,000 x g) for 5 min.
- Run 10-15 µL of each sample on 8-10% SDS-PAGE at 120-160 V till the dye front reaches bottom of the gel.
- Electro-transfer the resolved proteins by standard semi-dry transfer protocol (20 V, 30-45 min) on a Nitrocellulose/PVDF membrane.
- Blot with anti-IKK1 antibody (dilution ~1:2,000, must be optimized) or anti-PentaHis antibody (dilution ~1:3,000, must be optimized) for expression.
- Choose the best P1 virus clone by comparing expression of recombinant IKK1.
- Preparation of large scale P3 virus stock.
- Plate cells similarly as described in step 1.1 in a 15 cm plate containing 25-30 mL of Sf900 III medium.
- ~6 h post plating, add 150-300 µL of the best-expressing P1 virus stock onto the cells.
- After 72 h post infection, aspirate the medium carefully from the plate in a suitable tube (sterile), and centrifuge the tube at 500-1000 X g for 10 min at 4 °C. Discard the cell pellet (if one is formed) and save the supernatant. This is P3 virus stock.
- Determining the optimum virus titre for large scale protein expression
- Plate cells in 2 mL of Sf900 III medium, as described in 1.1, in a 6-well plate.
- Add 20, 30, 40, and 50 µL of P3 virus stock in separate wells ~6 h post plating. Include an uninfected control.
- Harvest cells from each well 48-60 h post infection.
- Add 100 µL of 2X Laemmli SDS buffer. Heat at >95 °C for 10 min. Centrifuge (typically at 14,000 x g) for 5 min.
- Resolve 10-15 µL of each sample in an 8-10% SDS-PAGE gel.
- Transfer resolved protein to a Nitrocellulose/PVDF membrane as mentioned in 1.2.6.
- Blot with anti-IKK1 antibody (dilution ~1:2000, must be optimized) or anti-PentaHis antibody (dilution ~1:3000, must be optimized).
- Use the virus with the best expressing titre for large scale expressions.
2. Large Scale Expression of 6x-His Tagged Human IKK129,33
- Carry out large scale expression in a suspension culture. Split cells in 1 L of Sf900 III medium in a 2 L Erlenmeyer Flask with a vented cap. The cell density should be ~5×105 cells/mL.
- Grow these cells in suspension at ~100 rpm at 27 °C in a shaker for ~2 days until it reaches a density of 1-2×106/mL. Cell density must not exceed 2×106/mL.
- Add the desired amount of P3 virus, as assessed in 1.4.
- Grow the infected culture for 48-60 h at ~100 rpm at 27 °C in a shaker.
- Harvest cells by centrifugation at <1,500 x g at 4 °C.
- Save the cell pellet and discard the medium.
- Proceed to protein purification directly. Flash freeze the cell pellet in liquid nitrogen and store at -80 °C for future use.
3. Purification of His-IKK133
- On day 1, resuspend the cell pellet in 40 mL of Lysis Buffer (25 mM Tris pH 8.0, 0.2% NP-40, 200 mM NaCl, 10 mM imidazole, 10% Glycerol, 5 mM β-mercaptoethanol, and protease inhibitor cocktail).
NOTE: The wet cell pellet mass was not determined. Typically, ~2 L of culture was used for this step. - Lyse the cells by sonication at 60-70% duty cycles, 5-10 pulses of 30 s duration at an interval of >1 min, keeping the cell suspension chilled on ice.
NOTE: This step requires optimization with each model of sonicator. Do not let the sample warm up above 10 °C. - Clarify the lysate by centrifugation at ≥ 28,000 x g for 45 min at 4 °C.
- Equilibrate 4 mL of Ni-NTA Agarose resin with lysis buffer, and mix the clarified lysate (supernatant) following the manufacturer's protocol.
- Allow the protein to bind the resin by incubating for 2 h on a rotary mixer in a 4 °C room.
- Wash the resin thoroughly with the lysis buffer containing 30 mM imidazole. Check protein concentration in the wash fraction periodically using Bradford Assay. Wash until this protein concentration in the wash fraction reaches 0.1 mg/mL. Typically > 20 bed volume of wash buffer is required to reach this point.
- Elute His-tagged IKK1/α protein under gravity flow, using 20 mL of elution buffer (the lysis buffer containing 250 mM imidazole). Collect 1-1.5 mL fractions.
- Check the purity of eluted protein on an 8 or 10% SDS PAGE gel by loading 5-10 μL of sample from all fractions mixed with Laemmli buffer.
- Pool peak fractions (concentration ≥ 2mg/mL), and measure protein concentration using the Bradford assay.
- Digest with TEV (1:30-50 (TEV protease:IKK1)) protease overnight (≥12 h) at 4 °C.
NOTE: Optimize the amount of TEV protease required for complete (≥ 95%) digestion of the 6X-His tag a priori. TEV protease was purified in house. - On the morning of day 2, incubate the protein with 1 mM ATP in the presence of 20 mM MgCl2, 20 mM β-glycerophosphate, 10 mM NaF, and 1 mM sodium orthovanadate for 1 h at 27 °C.
- Filter the protein solution through a 0.45 or 0.22 μm filter.
- Load 6 mL of the sample onto a ~120 mL preparative size-exclusion column attached to an automated liquid chromatography system. Equilibrate the column with Buffer A (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM DTT, 5% Glycerol), prior to applying the protein sample.
NOTE: A smaller column could be used depending on the yield of protein after the Ni-NTA affinity purification step. Adjust/calculate load volume to be 5% of column volume). - Run the size-exclusion chromatography at a flow rate of 1 mL/min, and monitor elution at 280 and 254 nm. Collect fractions of 2 mL size.
- Check purity of the peak fractions (typically around ~60-70 mL) on an 8-10% SDS-PAGE gel.
- Pool the pure fractions (purity ≥95% with a concentration of ≥0.5 mg/mL) and concentrate in a 10 kDa or 30 kDa molecular weight cut-off centrifugal protein concentrator following the manufacturer's instructions.
- Periodically check protein concentration of the concentrate by the Bradford method, and concentrate the sample up to 8-12 mg/mL.
- Dispense 25 μL aliquots and flash freeze in liquid nitrogen. Store these flash frozen samples in the -80 °C freezer.
4. Screen for Crystallization Condition of IKK1
- Calculate the total amount (by volume) of protein required to screen for crystals following the methods described below.
- Try different inhibitors (generic kinase inhibitors: AMPPNP, and Staurosporine; IKK-specific inhibitors: MLN120B, Compound A, and IKK-inhibitor XII) to perform screening for crystallization. Make stock solutions of these compounds following the manufacturer's instructions. Ensure that the maximum concentration of the compound in its stock solution does not exceed 20 μM.
- Prepare the IKK:inhibitor complex for the crystallization screen by mixing 100 μM of IKK1 with 200 μM inhibitor, and incubate at 18-20 °C for 30-40 min. Centrifuge this mixture at >15,000 x g for 2 min at room temperature. Save the supernatant for crystallization screen.
- Use the commercially available crystallization screens (see the Table of Materials).
- Pipette 80-100 μL of each crystallization reagent (reservoir solutions), using a multichannel pipette or a robot, into the reservoir of a 96-well plate. The plate is now ready to set up drops. Ensure that the plate accommodates 2 to 3 crystallization drops so that 2 to 3 inhibitors can be screened in a single plate.
- Using a crystallization robot, dispense and mix 0.2–0.25 μL of IKK1:inhibitor complex with the same volume of reservoir solution.
NOTE: Screening can be performed manually using larger drop volumes (0.5-1.0 μL of IKK1:inhibitor complex with the same volume of reservoir solution). However, use of a robot would help save valuable reagents. - Seal the plates immediately after setting the drops with optically clear films to avoid evaporation. Seal properly using appropriate applicator. Make replica plates for each inhibitor set. Incubate one plate at 18 °C, and the other in the cold room (temperature range ~4–6 °C). Make sure that incubators are not affected by vibration.
- Use a stereomicroscope with a polarizer to check for appearance of crystals in each drop every day for the first seven days, and then at longer intervals. Systematically note and score these observations.
NOTE: Use a stereomicroscope with a polarizer so that the growth defects in crystals can be visually assessed. Here, crystals appeared in a condition where the reservoir solution contained 3% w/v Dextran sulfate sodium salt Mr 5,000, 0.1 M BICINE pH 8.5, 15 % w/v Polyethylene glycol 20,000. Crystals of IKK1 grew only in the presence of IKK-inhibitor XII.
5. Crystal Growth Optimization29,33
- Preparation of stock solutions.
- Prepare 30% w/v solutions of Dextran sulfate of different molecular weight (Avg. MW 5,000, 8,000, 15,000, 40,000 Da) in deionized H2O. Filter solution using 0.22 μm filters.
- Prepare 1 M or 0.5 M stocks of different buffers in the pH range of 6.5 to 9. Filter solutions using 0.22 μm filters.
- Prepare 25% or 50% (w/v) PEG solutions of different molecular weight (Avg. MW:1000, 1,500, 3,350, 4,000, 6,000, 8,000, 10,000, 12,000, 20,000 Da). Filter solutions using 0.22 μm filters.
- Perform a grid screen by systematically varying pH, buffer type, concentration, and type of both PEG and Dextran Sulfate.
- Optimize screening both at 18 °C, and in the cold room (temperature range ~4-6 °C).
- Mix an equal amount of the IKK1:Inhibitor complex with well solution and incubate over the well solution in a sealed environment. This step can be performed manually or using a dispensing robot.
NOTE: Here, the largest and well-defined crystals (as assessed by uniform color under the polarizer which indicated uniform growth) were obtained under the following conditions: 100 mM BisTris pH 7.0, 2.8-3.5% of Dextran Sulfate (average MW 15 kDa), 8.5-10% PEG 20 kDa in the cold room.
6. Growing Crystals in Large Numbers
- Prepare the following stock solutions and filter them through 0.22 μm filter.
- Prepare 0.5M BisTris pH 7.0, and BisTris propane pH 7.0 and 7.5.
- Prepare 30% (w/v) Dextran Sulfate (Average MW ~15 kDa). Store at 4 °C.
- Prepare 50% PEG (Average MW ~12 kDa).
- Use 24 well hanging drop plates.
- Apply sealant grease on the raised rings of each well.
- Prepare the well solution as follows (final concentration): 100 mM BisTris pH 7.0–7.5, 2.8–3.5% Dextran Sulfate ~15 kDa, 8.5–10% PEG ~12 kDa. Prepare 1 mL well solution in 1.5 mL tubes.
NOTE: A minor variation in crystallization condition was observed depending upon the batch of protein, and/or Dextran Sulfate, thus a trial of a narrow range is recommended. Both BisTris and BisTris propane buffers of the same pH range yield single crystals. - Keep the well solutions and greased plates in the cold room for at least 1 h.
- Prepare IKK1:Inhibitor complex as described in 4.3. Transfer the well solutions from 1.5 mL tubes to the individual wells of the 24 well plate.
- Clean siliconized (commercial or in-house (siliconization/silanization)) glass cover slips using lint-free wipes and apply high pressure pinpointed air jets using commercially available aerosol duster.
- Place 1–1.5 μL of well solution on a clean cover slip. Pipette equal volume of IKK1:inhibitor complex, mix gently by pipetting up and down 3-4 times. Turn the glass cover slip and place on the respective well with forceps and seal the well by pressing on the grease.
- After setting up all the drops of a 24-well plate, incubate the plate in the cold room in the dark. Occasionally check the drops under a microscope for appearance and growth of crystals.
NOTE: It has not been tested whether incubating crystal plates under the light would change the outcome. It is critical, however, that the plates are incubated in an environment with minimum/no vibration.
7. Cryo Protection of Crystals
- Preparation of cryo solutions.
- Prepare Cryo A (~2% Dextran Sulfate, ~10% PEG 12000, 60 mM BisTris propane (of the same pH as the intended well solution), 5 mM Tris pH 7.5, 40-50 mM NaCl, 2.5% Glycerol).
- Prepare Cryo B (~0.3% Dextran Sulfate, ~10% PEG 12000, 60 mM BisTris propane (of the same pH as the intended well solution), 5 mM Tris pH 7.5, 40-50 mM NaCl, 20 or 25% Glycerol or 20 or 25% Ethylene Glycol).
NOTE: Test different cryo-protectants in addition to Glycerol and Ethylene Glycol (e.g., PEG 200, PEG 400, PEG MME 550, PEG 1000).
- Gently remove the glass cover slip containing the crystal and place it onto a solid surface with the crystal drop facing upwards. Gently add 10 μL of Cryo A solution on top of the drop, and gently mix by pipetting so that the crystal is not touched.
NOTE: This cleans the crystal surface of debris and help equilibrate the crystal with the initial cryo buffer. Avoid mechanical and thermal stress to the crystal. Perform all the subsequent steps under the microscope to avoid any damage to the crystal. - Slowly remove some liquid from the crystal and keep about 5-8 μL of the solution. Avoid dehydration of the crystal. Take extreme care to avoid mechanical stress and dehydration of the crystal in all the subsequent steps.
- Gently add 2.5–4 μL of the Cryo B solution onto the drop, mix very gently by pipetting. Cover with a small Petri dish to avoid direct air flow over the crystal. Wait for 5 min. Always be careful the crystal does not suffer rapid changes of osmotic pressure or dehydration.
- Repeat step 7.4 four times to slowly acclimatize the crystal into the cryo solution.
- Keep 10–20 μL of the cryo solution at this stage (i.e., allow the crystal to be bathed in about 20–25% cryo-protectant containing solution). Soak for different periods of time (5 min to 1 h) in the final cryo-solution.
- Freeze multiple crystals at different time-points.
- Carefully pick a single crystal from the drop in an appropriate cryo-loop mounted on a proper base and plunge-freeze in liquid N2. Perform this step smoothly and quickly to avoid ice formation on the crystal.
NOTE: Pick the crystal using a loop diameter slightly larger than the longest axis of the crystal so that it can be mounted in the robotic goniometer used in Advanced Photon Source, Argonne National Laboratory. - Store the crystal containing cryo-loops in pucks immersed in liquid N2. Store these pucks in liquid N2 Dewar flask until they are ready for shipment for X-ray diffraction at the synchrotron. Proceed to X-ray diffraction data collection and processing (sections 8 and 9).
NOTE: Diffraction data of IKK1 crystals from home X-ray sources were not of high enough resolution for structure determination trials, although it indicated the quality of crystals to be screened at the synchrotron. All diffraction data were collected at different beamlines (primarily 19ID) of the Advance Photon Source, Argonne National Laboratory, USA.
8. X-ray Data Collection
- Collect X-ray diffraction data at the ID19 beamline of the APS synchrotron source, using remote data collection software34.
NOTE: Upwards of 100 crystals were tested for their diffraction properties. The crystals routinely diffracted to a resolution of 7-11 Å, only rarely crystals that diffracted to beyond 5 Å were observed, and only one large crystal diffracted beyond 4.5 Å. Since the crystals decayed rapidly during the data collection, datasets at different parts of the same crystal were collected. This was possible since the crystals were large (larger than 400 µm), and the beam diameter used was 100 µm.
9. X-ray Data Processing
- Merge all datasets collected on different parts of the same crystal34.
NOTE: seven datasets were merged from the best crystal and the resultant data was 93% complete. The overall I/σ was ~6.8) and Rmerge was 89% in the highest resolution bin (5.4-4.5 Å). - Process datasets with HKL200034 to obtain space group, unit cell dimensions, solvent content, and plausible composition of the asymmetric unit.
10. Structure Solution
- Preparation of search models
- Based on available structural models of hIKK2 (pdb id 4E3C and 4KIK35), generate a series of dimers with different orientations of the N-terminal KD (that renders an opening of 30 and 62 Å, between P578 of two KD in the dimer). None of these models fetched a molecular replacement search solution after extensive trials.
- Generate a map of IKK1 from single-particle cryo-EM data33.
- Generate a model of human IKK1 from the highest resolution structure of IKK2 (pdb id 4KIK).
- Dock the individual domains of the human IKK1 model into the cryo EM density map of human IKK1 using COOT36. This rotated the KD orientation about 24 degrees and modified the N-terminal opening to ~58 Å.
- Apply the KD orientation of the cryo-EM fitted model (the monomer) to all dimers generated in 10.1.1.
- Molecular replacement
- Use dimers from 10.1.1 and 10.1.5 as search models in programs PHASER37, MOLREP38 and CNS39,40.
NOTE: Here, using one of the dimers from 10.1.5 (52 Å opening) as a search model, six dimers (two hexamers) were located in the asymmetric unit by using MOLREP. The 52 Å opening in the search model is similar to the 49-50 Å opening of the dimers (all six dimers) in the refined structure.
- Use dimers from 10.1.1 and 10.1.5 as search models in programs PHASER37, MOLREP38 and CNS39,40.
11. Structure Refinement
- Structure refinement procedure
- Fix the orientation and position of this initial model by rigid body refinement in CNS (cns_solve_1.3)39,40.
- Refine the structure further using minimization and simulated annealing with a maximum likelihood target function and a flat bulk-solvent correction using the CNS system39,40.
- Build the model based on 2FO-FC and Fo-Fc maps using Xtalview41.
NOTE: Apply DEN-assisted refinement42 and NCS restrain during the refinements for better accuracy of the model. Indeed, DEN refinement dramatically improved geometries and R-factors of the structure, and the R factor was 22.2% and free R factor was 27.8% for the final model. The dramatic improvement of the R free must be due to the high accuracy model of IKK1 domains that were generated from the IKK2 model (4KIK).
Representative Results
Cloning and expression of different constructs of IKK1/α
Full length human IKK1/α was cloned into the baculovirus expression vector pFastBacHTa within its EcoRI and NotI restriction sites to obtain an N-terminal hexa-Histidine tagged IKK1. The tag could be removed by TEV protease digestion. Since full length IKK1/α contains flexible regions on both ends, and flexible regions usually render a protein difficult to crystallize, we cloned various truncated fragments of IKK1/α within the abovementioned sites of pFastBacHTa vector. Various truncation mutants of IKK1/α were generated with both wild-type (wt) and S176E,180E (EE) backbones. IKK1/α EE refers to the mimetic of the constitutively active form of the phosphorylated kinase. Recombinant baculovirus production and protein expression were performed using a previously published protocol with minor modifications43 (Figure 1).
Difficulty in IKK1/α crystallization
Since human IKK2/β and IKK1 are homologous, and we could crystallize different versions of human IKK2/β under several different conditions, we expected IKK1/α to crystallize under similar conditions using similar strategies. However, after extensive trials with several different IKK1 variants, we obtained crystals with only one truncated construct (IKK1 10-667 EE) (Figure 2), and that also only in the presence of the IKK inhibitor XII44 that displayed suitable X-ray diffraction properties.
Structure solution
IKK1/α crystals suffered from numerous growth problems, and the data often displayed very high mosaicity. Weak crystal packing associated with large solvent content and the dynamic nature of the IKK1 monomer/dimer are the likely reason behind this. To circumvent these problems, painstaking efforts were taken to obtain the best possible crystals, and the cryo-preservation procedure was performed with utmost care under various conditions and with various cryo-preservative buffers. The data was also collected with ultimate precision to minimize beam damage. Since the crystals were large (largest dimension often exceeding 400 microns), different areas of the same crystals were exposed to the X-ray beam to collect multiple datasets. This enabled different datasets to be scaled, and to obtain a reasonably complete dataset with higher redundancy and minimized error. Soaking with heavy atoms, or heavy atom clusters (e.g., TaBr and TAMM), caused the crystals to diffract inadequately. Although we could locate the position of TaBr clusters in derivative data, the poor quality of the data in addition to the non-isomorphocity provided little, if any, phase information.
We processed datasets with HKL2000 using various available parameters. The diffraction pattern revealed that the crystal belongs to the space group P21 with unit cell dimensions, a = 174.51, b = 186.94, c = 275.83 Å and β = 98.84 degrees. We used mainly I/σ to estimate the cut-off and tested various cut-offs for the datasets. We obtained a merged data set of 4.5 Å resolution from 4 of 7 sets of diffraction data collected on one crystal. We decided to keep data until 4.5 Å from visual inspection of the final density map. It may be worthwhile to use CC1/2, since we can analytically estimate CC of the merged dataset against the true (usually unmeasurable) intensities using CC*45. Because of the large unit cell combined with the low symmetry of the space group, we anticipated the asymmetric unit to contain 12 to 24 molecules based on a solvent content of 70% to 40%.
The combined effect of low resolution X-ray data with weak intensities (i.e., significant errors in data) (Figure 3), large number (12) of IKK1 molecules in the asymmetric unit, and conformational variation of IKK1 monomeric/dimeric model compared to known IKK2 models, initially prevented us from obtaining a molecular replacement solution IKK1, and thus determining its structure.
IKK1/α and IKK2/β both contain a kinase domain (KD), a ubiquitin-like domain (ULD), and an a-helical scaffold dimerization domain (SDD). The structure of IKK1 revealed that it forms dimers similarly to IKK2 utilizing a nearly identical inter-subunit interface of the distal region of SDD. However, IKK1 dimer displayed a significantly different relative positioning of the N-terminal KD relative to SDD+ULD compared to that in known IKK2 dimers. Different IKK2 structures earlier indicated different intermonomer orientation within its dimers (Figure 4, panel A), so that the distances between the alpha carbons of P578 in the two KD in four different dimer models varied between 39 and 61 Å. Since the sequences of these two kinases are very similar and residues at the dimer interface are identical, we anticipated IKK1/α would form a similar dimer; however, since there are significant differences in residues of the KD-SDD interface (e.g., W424 and F111 in IKK2 are V and P respectively), we anticipated perhaps a yet different KD orientation in IKK1. Indeed, IKK1/α structure indicated that the related orientations of KD to SDD is unique, and it deviates from all known models of IKK2/β. In excess of a hundred dimer models were used as search models in programs PHASER, MOLREP, and CNS without any success, indicating the need for a rather accurate search model for the success of molecular replacement trials, especially with our low-resolution data. A monomer model has too little mass to pick up any solution in the weak diffraction data of the large asymmetric unit containing 12 monomers. Also, the inclusion or omission of water in the search model made no difference in molecular replacement searches, especially because of the weak diffraction data. The CC1/2 and CC* indicates that data up to 4.5 Å is quite precise. We used a conservative resolution cut-off of 4.5 Å based on I/σ of 1.5. The datasets with different resolution cut-offs did not show any stark difference during molecular replacement search operations, and final maps refined against these datasets did not show any distinct improvement in map features upon extending resolution cut-offs. The final build of the model is quite accurate, more than what could be built from the density alone, likely since the initial molecular replacement models were built from models derived with a high-resolution data, and we utilized the cryo-EM map/density to cross-check the features of the map, especially in regions where maps were unclear.
In hindsight, the obtainment of a useful search model was possible only because of the availability of the low resolution cryo-EM map, and a rather high accuracy model of IKK1 domains that could be generated based on high resolution IKK2 structure (Figure 4, panel B). We could dock the IKK1 domains in the cryo-EM map of IKK1 to obtain a reasonably close dimer model. The initial model indicated an orientation of KD rotated about 24 degrees relative to that of a IKK2 monomer, and N-terminal opening of 58 Å (between P578 of two KDs in a dimer). Our prior knowledge of different IKK2/β dimer structures (and their variation) enabled us to fine-tune the search model by changing the openings between 30-62 Å. Using one of the dimers (52 Å opening) as a search model, we located six dimers in the asymmetric unit using program MOLREP46. These six dimers are organized into two hexamers in the asymmetric unit, and the calculated solvent has a volume of 68%.
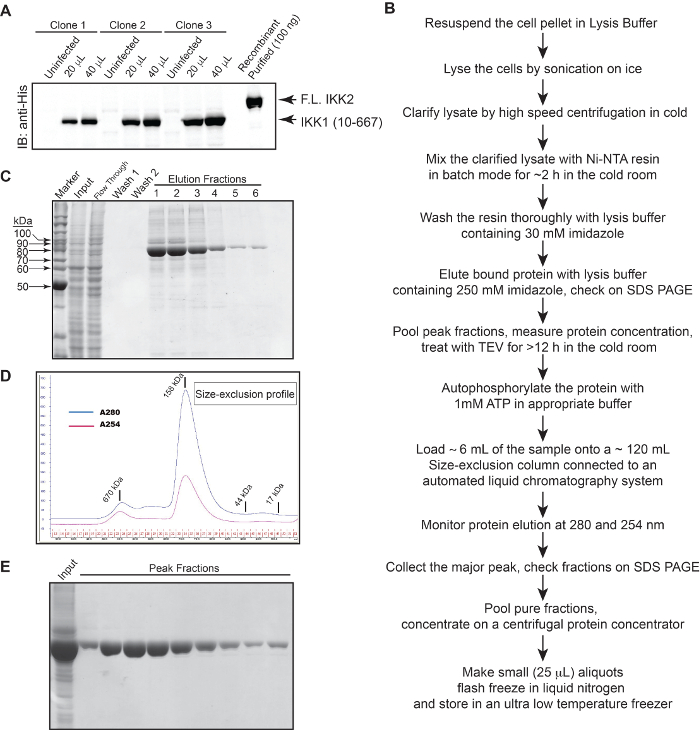
Figure 1: Purification of IKK1. (A) Expression of IKK1 (10-667) in insect cells. (B) A flow chart for purification of IKK1; (C) An SDS-PAGE gel showing purity of IKK1 after the Ni-NTA affinity chromatography step. (D) Size-exclusion profile indicating dimer of IKK1. (E) SDS-PAGE gel showing purity of IKK1 from the peak size-exclusion fractions. Please click here to view a larger version of this figure.
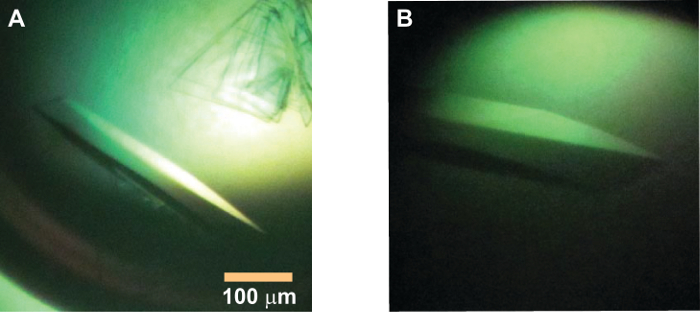
Figure 2: Morphology and size of IKK1 crystals. (A) A crystal of IKK1 construct 10-667; the crystallization drop also shows crystals of another morphology (thin plates) which do not diffract well. (B) Zoomed in view of the crystal that diffracted to beyond 5 Å. Please click here to view a larger version of this figure.
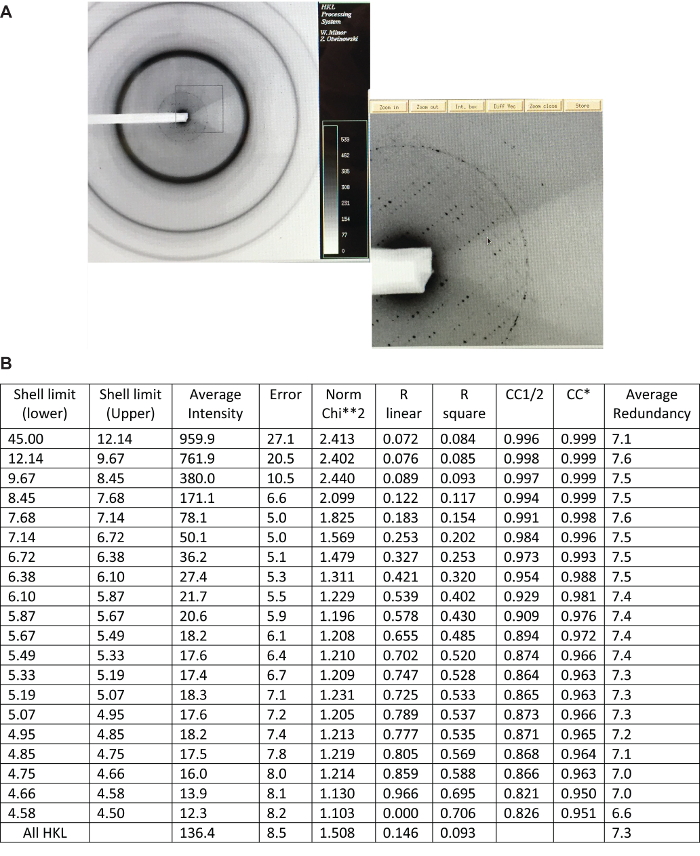
Figure 3:. Crystallographic data obtained from IKK1 crystals. (A) An X-ray diffraction profile of an IKK1 crystal indicating poor diffraction properties of typical IKK1 crystals. (B) HKL2000 scaling statistics of the merged dataset used for IKK1 structure determination, and redundancy of the data. R linear = SUM (ABS(I – <I>)) / SUM (I), R square = SUM ( (I – <I>) **2) / SUM (I **2), Chi**2 = SUM ( (I – <I>) ** 2) / (Error ** 2 * N / (N-1) ) ), CC1/2 = Correlation coefficient, CC* = Correlation coefficient of merged dataset against true intensities. Please click here to view a larger version of this figure.
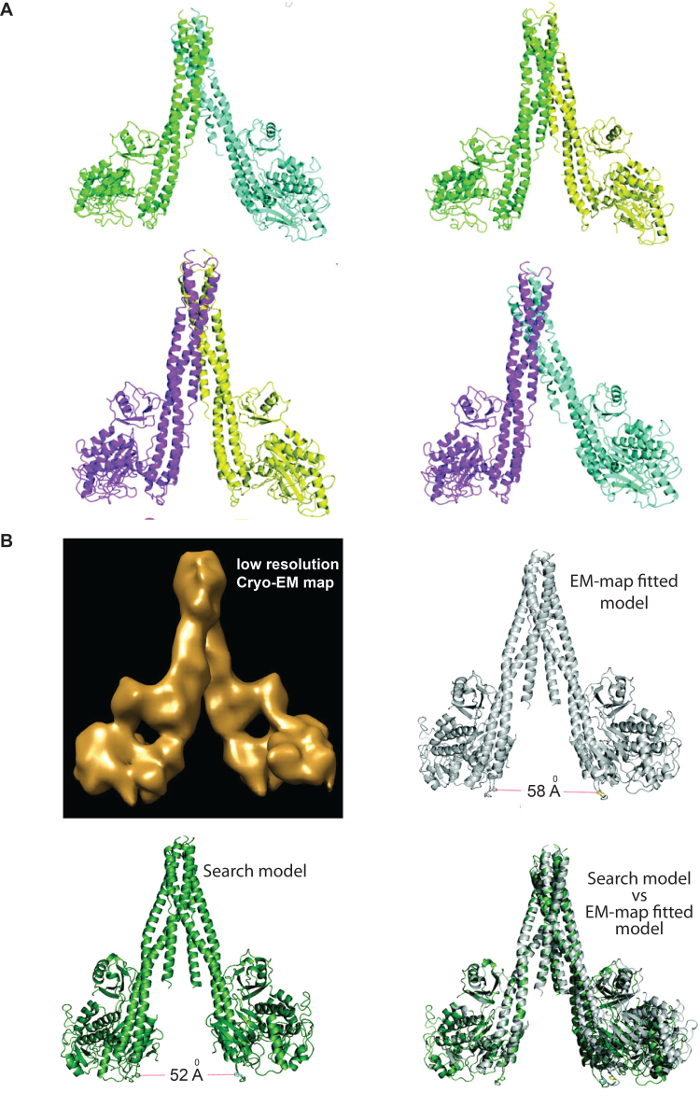
Figure 4: Preparation of search models. (A) Different models of IKK2 dimer indicating different orientations of the KD relative to SDD (giving rise to different separation between two KD); these models were tested initially to find a molecular replacement solution without any success. (B) Generation of IKK1 search models – Cryo-EM map of IKK1 dimer at 11 Å resolution; EM-map fitted initial IKK1 search model, individual domains are based on IKK2 model (4KIK); An IKK1 search model further fine-tweaked (appropriate KD orientation) based on various possibilities as judged from IKK2 models describe above in 4A; Superposition of EM-map fitted model to the search model that yielded the molecular replacement solution. Please click here to view a larger version of this figure.
Discussion
Production, crystallization and structure solution of two related IKK proteins
We set out to determine the X-ray crystal structure of IKK1/α with the notion that it would be a relatively straightforward exercise given our experience with IKK2/β protein production, crystallization, and structure determination. However, we were highly surprised that these two related proteins behaved very differently regarding the ease of crystallization. Despite efforts from several high-profile laboratories, the determination of IKK1 structure took nearly two decades. This largely stemmed from a few key bottlenecks.
The first difficulty was to obtain a highly pure and soluble protein form. The insect cell expression system enabled us to obtain mg quantities of pure protein that was reasonably soluble. In regard to the level of expression, all IKK1/α constructs expressed at much lower levels than IKK2/β constructs in insect cell expression systems. It should be noted that neither protein is soluble and functional when expressed in E. coli. In an insect cell expression system, IKK2/β fragments expressed at high levels ranged from 10 to 30 mg/L of culture depending on the constructs. In contrast, IKK1/α constructs could be generated only at 0.5 to 2.0 mg/L of culture. The reason for this difference is still unknown to us. We should also try expression of IKK1 in native host cells (i.e., mammalian overexpression system in case of a mammalian IKK protein), especially since the post-translational modifications such as auto- or trans-phosphorylation commonly occurring in IKK will occur most efficiently and appropriately in its native environment. Purification of IKK complexes directly from large scale culture of mammalian cells is also currently amenable.
While working with IKK2/β, we noticed that it can be homogeneously auto-phosphorylated by treating with ATP, and this makes the protein amenable to crystallization. Similarly, the incubation of IKK1 protein with ATP, and the resultant auto-phosphorylation, yielded an activated IKK1 which was soluble, and suitable for both biochemical characterization and crystal structure determination.
A critical step to obtain well-diffracting crystals was to fine tune the crystallization conditions, in particular the addition of the appropriate type and concentration of Dextran sulfate molecules. The density indicates bound Dextran sulfate molecules, and its resultant effect on IKK1 was likely critical for crystallization.
The inherent flexibility of IKK1, which is often observed in kinases, is the primary deterrent for crystallization and is often linked to engagement of the kinase domain with the activation loop. IKK2 with both native (S177 and S181) and phosphomimetic (EE) forms of activation loop serines could be crystallized, but we only obtained crystals with truncated IKK1/α of the SS to EE mutant version and that too only in presence of IKK inhibitor XII. These crystals diffracted well enough for structure determination only when grown in the cold room (~4-6 °C). If we encounter a similarly obstinate behavior with any IKK1 homolog (or an IKK ortholog), we can try various backbone modifications (especially of the active site loop serine residues which are commonly phosphorylated). We can also try co-crystallization in presence of interacting partner proteins. Many kinases, including those of the IKK family, interact with partner proteins and reside within high-order complexes.
Several inhibitors helped produce IKK2 crystals in various conditions (e.g., with both high salt, PEG as a precipitant), both at 18 °C, and in the cold room. However, under similar conditions and with a variety of inhibitors, IKK1/α did not produce any crystal. Thus, the use of a larger repertoire of inhibitors will certainly increase the chances of finding one that enable crystallization.
Another critical step in structure determination was the careful cryo-preservation and data collection procedure. The weak data and rapid decay of the crystals in the X-ray beam warranted the need for larger crystals and collection of multiple data sets from the same crystals. Without a reasonable complete dataset of high accuracy, we could not have succeeded in obtaining a molecular replacement solution.
Finally, we failed to determine the structure of IKK1/α by molecular replacement using more than 100 models created from known IKK2/β structural models available in the public database. Thus, obtaining an appropriate model for a molecular replacement procedure was a must. The cryo-EM map, high accuracy structural models of individual domains, along with comparative studies of previously determined IKK2 structures led us to obtain a search model that fetched us the molecular replacement solution.
The crystallization of a protein is inherently random, so despite following the above structured guidance, we might face problems in crystallizing another IKK1 homolog or another full-length or truncated construct of the human IKK1. Regardless, knowledge of these critical steps will enable a researcher to explore rational conditions that will increase chances of obtaining a well-diffracting IKK crystal.
IKK1/α and IKK2/β display similar domain organization but different interdomain organization
Not surprisingly, both IKK1/α and IKK2/β adopt a highly similar domain architecture stemming from their high sequence similarity33,43. As indicated earlier, both IKK1/α and IKK2/β fold into three distinct domains: N-terminal kinase domain, ubiquitin like domain (ULD), and scaffold dimerization domain (SDD). Both IKK1/α and IKK2/β form stable dimers in which the C-terminal half of the SDD participate in dimerization. Residues involved in the dimer formation are identical. However, the inter-domain interactions between SDD/ULD and KD are somewhat different since these involve different residues. This also likely caused the relative orientation of the kinase domain to the SDD in IKK1/α to differ from that in IKK2/β. This was not entirely unexpected since we also observe differences among available IKK2/β dimer models. Interestingly, IKK2/β dimers can organize into tetramers in crystal lattice even though this tetramerization propensity is weak in solution. However, IKK1/α dimers formed unique hexamers. These hexamers are also observed in solution, although only a small pool of IKK1/α dimers exist as hexamers. These suggest that hexamerization is likely to have a specific and critical function.
Differentiating functional differences from structural differences
IKK1/α and IKK2/β perform distinct functions in cells. However, the relationship between IKK1/α function and structure has remained elusive. IKK1/α clearly exists in different forms: as free dimer or hexamer, and within a hitherto uncharacterized hetero-trimer complex along with IKK2/β and NEMO. Since IKK2/β activation by canonical signalling does not require IKK1/α, the functional role of IKK1/α in the heterotrimer (canonical IKK-complex) remains unclear. It is also unclear if the activation of IKK1/α induced by non-canonical signalling relies on free dimeric IKK1/α and/or hexameric IKK1/α. Since mutational experiments reveal that disruption of the hexamer interface strongly affects non-canonical signalling, hexamerization is likely to be essential at some stage during non-canonical signalling.
Designing small molecule inhibitors using IKK1/α structure
Knowing the structure of IKK1/α allows us to investigate surface patches on a monomer or pockets at inter-domain or inter-monomer interfaces, which can be targeted by small molecules. IKKs have long been considered important drug targets. However, the essentiality of these kinases in a variety of functions including cellular homeostasis, organismal viability, and immunity makes it extremely difficult to target them. So far, no IKK-targeted molecule has been found that could be used safely in patients.
IKK1/α and IKK2/β are often targeted simultaneously by their inhibitors since their kinase domains are highly similar in sequence and in structure. Considerable research effort has been spent into finding specific inhibitors targeting only one of these two kinases. Consequently, several IKK2/β-specific inhibitors have been discovered, some of which do not affect IKK1/α function. However, to our knowledge no such compound exists that effectively inhibits IKK1/α without perturbing IKK2/β function. This absence could be attributed to the lack of structural information on IKK1/α. Our structural and cellular studies indicate that, although domain arrangement and global subunit structures are highly similar in these two proteins, they display unique high-order arrangements employing specific surface patches33. These differences that enable different interactions with associated proteins must translate into their functional differences. We are hopeful that these distinctions revealed between the IKK1/α and IKK2/β inter- and intra-subunit interactions can be exploited to make subunit-specific allosteric inhibitors.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank the staff at the beamlines 19ID, 24ID, and 13ID at Advanced Photon Source, Lemont, IL, for support during data collection on various crystals. We are grateful to Dmitry Lyumkis, Salk Institute for fetching us the low resolution cryo-EM map at early stages of EM map/model building, which was used to build the initial IKK1 molecular replacement search model. The research leading to these results has received funding from NIH grants AI064326, CA141722, and GM071862 to GG. SP is currently a Wellcome Trust DBT India Intermediate Fellow.
Materials
| Cellfectin/Cellfectin II | Thermo Fisher Scientific | 10362100 | Cellfectin is now discontinued, replaced by Cellfectin II |
| Sf900 III Insect cell medium | Thermo Fisher Scientific | 12658-027 | |
| SF9 cells | Thermo Fisher Scientific | 12659017 | |
| anti-IKK1 antibody | Novus Biologicals | NB100-56704 | Previously sold by Imgenex |
| anti-PentaHis antibody | Qiagen | 34460 | |
| PVDF membrane | Millipore | IPVH00010 | Nitrocellulose can also be used |
| Ni-NTA agarose | Qiagen | 30210 | |
| Bradford assay reagent | BioRad | 500001 | |
| Superdex 200 column | GE Healthcare | 28989335 | |
| Amicon concentrator | Millipore | UFC801008, UFC803008, UFC201024, UFC203024 | |
| Compound A | Bayer | ||
| Calbiochem IKK-inhibitor XII | Calbiochem | 401491 | |
| Staurosporine | SIGMA | S4400 | |
| MLN120B | Millenium | Gift item | |
| AMPPNP | SIGMA | A2647 | |
| Dextran sulfate | SIGMA | 51227, 42867, 31404, | |
| Dextran sulfate | Alfa Aesar | J62101 | |
| PEG | SIGMA | 93593, 81210, 88276, 95904, 81255, 89510, 92897, 81285, 95172 | Some of them are new Cat # on SIGMA catalogue. What we had was originally from Fluka that had different Cat #. |
| Crystallization Screens | |||
| Crystal Screen I and II (Crystal Screen HT) | Hampton Research | HR2-130 | |
| Index HT | Hampton Research | HR2-134 | |
| PEG/Ion and PEG/Ion2 (PEG/Ion HT) | Hampton Research | HR2-139 | |
| PEGRX 1 and PEGRx 2 (PEGRx HT) | Hampton Research | HR2-086 | |
| SaltRx 1 and SaltRx 2 (SaltRx HT) | Hampton Research | HR2-136 | |
| Crystal mounts | Hampton Research | HR8-188, 190, 192, 194 | |
| Synchrotron | The Advanced Photon Source (APS) at the U.S. Department of Energy’s Argonne National Laboratory | Beamline 19 ID | The Advanced Photon Source (APS) at the U.S. Department of Energy’s Argonne National Laboratory provides ultra-bright, high-energy storage ring-generated X-ray beams for research in almost all scientific disciplines. |
Riferimenti
- Xia, Y., Shen, S., Verma, I. M. NF-kappaB, an active player in human cancers. Cancer immunology research. 2 (9), 823-830 (2014).
- Grivennikov, S. I., Greten, F. R., Karin, M. Immunity, inflammation, and cancer. Cell. 140 (6), 883-899 (2010).
- Ben-Neriah, Y., Karin, M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nature immunology. 12 (8), 715-723 (2011).
- Hinz, M., Scheidereit, C. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO reports. 15 (1), 46-61 (2014).
- Karin, M., Ben-Neriah, Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 18, 621-663 (2000).
- Ghosh, S., Karin, M. Missing pieces in the NF-kappaB puzzle. Cell. 109, S81-S96 (2002).
- Sun, S. C. The noncanonical NF-kappaB pathway. Immunol Rev. 246 (1), 125-140 (2012).
- Bonizzi, G., Karin, M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25 (6), 280-288 (2004).
- DiDonato, J. A., Hayakawa, M., Rothwarf, D. M., Zandi, E., Karin, M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 388 (6642), 548-554 (1997).
- Werner, S. L., Barken, D., Hoffmann, A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 309 (5742), 1857-1861 (2005).
- Zandi, E., Rothwarf, D. M., Delhase, M., Hayakawa, M., Karin, M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 91 (2), 243-252 (1997).
- Rothwarf, D. M., Zandi, E., Natoli, G., Karin, M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 395 (6699), 297-300 (1998).
- Yamaoka, S., et al. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 93 (7), 1231-1240 (1998).
- Li, Z. W., et al. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med. 189 (11), 1839-1845 (1999).
- Hayden, M. S., Ghosh, S. Shared principles in NF-kappaB signaling. Cell. 132 (3), 344-362 (2008).
- Sun, S. C., Chang, J. H., Jin, J. Regulation of nuclear factor-kappaB in autoimmunity. Trends Immunol. 34 (6), 282-289 (2013).
- Claudio, E., Brown, K., Park, S., Wang, H., Siebenlist, U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 3 (10), 958-965 (2002).
- Senftleben, U., et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 293 (5534), 1495-1499 (2001).
- Coope, H. J., et al. CD40 regulates the processing of NF-kappaB2 p100 to p52. EMBO J. 21 (20), 5375-5385 (2002).
- Dejardin, E., et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 17 (4), 525-535 (2002).
- Xiao, G., Fong, A., Sun, S. C. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. The Journal of biological chemistry. 279 (29), 30099-30105 (2004).
- Xiao, G., Harhaj, E. W., Sun, S. C. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Molecular cell. 7 (2), 401-409 (2001).
- Qing, G., Qu, Z., Xiao, G. Stabilization of basally translated NF-kappaB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-kappaB2 p100. J Biol Chem. 280 (49), 40578-40582 (2005).
- Zarnegar, B. J., et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 9 (12), 1371-1378 (2008).
- Vallabhapurapu, S., et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 9 (12), 1364-1370 (2008).
- Annunziata, C. M., et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer cell. 12 (2), 115-130 (2007).
- Keats, J. J., et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer cell. 12 (2), 131-144 (2007).
- Staudt, L. M. Oncogenic activation of NF-kappaB. Cold Spring Harbor perspectives in biology. 2 (6), a000109 (2010).
- Polley, S., et al. A structural basis for IkappaB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS Biol. 11 (6), e1001581 (2013).
- Hinz, M., Scheidereit, C. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Rep. 15 (1), 46-61 (2014).
- Scheidereit, C. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene. 25 (51), 6685-6705 (2006).
- Luckow, V. A., Lee, S. C., Barry, G. F., Olins, P. O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 67 (8), 4566-4579 (1993).
- Polley, S., et al. Structural Basis for the Activation of IKK1/alpha. Cell reports. 17 (8), 1907-1914 (2016).
- Otwinowski, Z. a. M., W, Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 276 (Macromolecular Crystallography, part A), 307-326 (1997).
- Liu, S., et al. Crystal structure of a human IkappaB kinase beta asymmetric dimer. J Biol Chem. 288 (31), 22758-22767 (2013).
- Emsley, P., Lohkamp, B., Scott, W. G., Cowtan, K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 66 (Pt 4), 486-501 (2010).
- McCoy, A. J., et al. Phaser crystallographic software. Journal of applied crystallography. 40 (Pt 4), 658-674 (2007).
- Vagin, A. T., Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022-1025 (1997).
- Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature protocols. 2 (11), 2728-2733 (2007).
- Brunger, A. T., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica. Section D, Biological. 54 (Pt 5), 905-921 (1998).
- McRee, D. E. XtalView: a visual protein crystallographic software system for X11/Xview. J. Mol Graph. 10, 44-47 (1992).
- Schroder, G. F., Levitt, M., Brunger, A. T. Super-resolution biomolecular crystallography with low-resolution data. Nature. 464 (7292), 1218-1222 (2010).
- Polley, S., et al. A structural basis for IkappaB kinase 2 activation via oligomerization-dependent trans auto-phosphorylation. PLoS biology. 11 (6), e1001581 (2013).
- Christopher, J. A., et al. The discovery of 2-amino-3,5-diarylbenzamide inhibitors of IKK-alpha and IKK-beta kinases. Bioorganic & medicinal chemistry letters. 17 (14), 3972-3977 (2007).
- Karplus, P. A., Diederichs, K. Linking crystallographic model and data quality. Science. 336 (6084), 1030-1033 (2012).
- Chen, V. B., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 66 (Pt 1), 12-21 (2010).

