In Vivo Single-Molecule Tracking at the Drosophila Presynaptic Motor Nerve Terminal
Summary
Here we illustrate how single molecule photo-activated localization microscopy can be carried out on the motor nerve terminal of a live Drosophila melanogaster third instar larva.
Abstract
An increasing number of super-resolution microscopy techniques are helping to uncover the mechanisms that govern the nanoscale cellular world. Single-molecule imaging is gaining momentum as it provides exceptional access to the visualization of individual molecules in living cells. Here, we describe a technique that we developed to perform single-particle tracking photo-activated localization microscopy (sptPALM) in Drosophila larvae. Synaptic communication relies on key presynaptic proteins that act by docking, priming, and promoting the fusion of neurotransmitter-containing vesicles with the plasma membrane. A range of protein-protein and protein-lipid interactions tightly regulates these processes and the presynaptic proteins therefore exhibit changes in mobility associated with each of these key events. Investigating how mobility of these proteins correlates with their physiological function in an intact live animal is essential to understanding their precise mechanism of action. Extracting protein mobility with high resolution in vivo requires overcoming limitations such as optical transparency, accessibility, and penetration depth. We describe how photoconvertible fluorescent proteins tagged to the presynaptic protein Syntaxin-1A can be visualized via slight oblique illumination and tracked at the motor nerve terminal or along the motor neuron axon of the third instar Drosophila larva.
Introduction
Neuronal communication relies on neurotransmitter release, which occurs through the regulated fusion of neurotransmitter-containing synaptic vesicles with the plasma membrane1. This process called exocytosis2,3,4,5,6 is highly dynamic and can be up- or down-regulated according to fluctuations in the rate of stimulation7. The proteins involved in these processes are subject to Brownian motion, and therefore display nanoscopic organization capable of sustaining a range of binding actions that underpin these physiological functions. Plasma membrane proteins are highly dynamic, allowing lateral trapping of molecules into nanoclusters on the plasma membrane7,8,9,10,11,12. As such, investigating the mobility of these proteins helps to elucidate their mode of action8. Synaptic proteins such as the soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs), for example Syntaxin-1A, are now known to exhibit lateral fast mobility, as well as lateral trapping within nanoclusters that may serve as molecular depots and sites for vesicle fusion9,13,14,15.
The recent development of photoconvertible fluorescent proteins, such as monomeric (m)Eos16,17 and Kaede18, has allowed the nanoscopic resolution of neuronal plasma membrane proteins through the use of approaches such as photo-activated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM)14,15,19. These developments have enabled investigations into the mobility and nanoclustering of biological proteins in cultured live cells and neurons. More recently, studies have been carried out to elucidate (at similar high resolutions) the dynamics and organization of these membrane proteins in the neurons of live intact organisms such Mus musculus, Caenorhabditis elegans, and Drosophila melanogaster14,20,21,22.
Investigating the dynamics and organization of a protein in vivo requires not only use of the recently developed super-resolution imaging tools (such as PALM, STORM, and photoconvertible fluorophores), but also the ability to overcome hindrances such as accessibility of the protein and the penetration depth of the imaging laser. Imaging intracellular proteins in an intact animal is inherently challenging as is imaging proteins on structures embedded deep within living tissues of the intact animal, such as the hippocampus of an intact mouse. Hence, proteins on or close to the surface of the tissue are more easily imaged. Another difficulty with imaging in vivo is in ensuring reproducibility across animals. As the anatomy might differ from one sample to the other, selecting a structure with ease of replication in different intact animal samples could also prove challenging. The use of stereotypical structures, such as synapses, help with overcoming some of these difficulties.
Recent studies have imaged protein mobility in animals with an optically transparent epidermis such as Caenorhabditis elegans22, while other investigations have imaged protein organization on the surface of a tissue/organ, for example actin imaging in cortical neurons through the skull of a live mouse21. In some cases, anesthetics have been used to keep the animal immobile; however, specific anesthetics may adversely affect the protein of interest. Alternative means to keep animals immobile during in vivo imaging should therefore be explored to minimize error.
Another caveat to super-resolution imaging in vivo is that the lasers used to illuminate the proteins of interest could adversely affect the surrounding tissue, and hence keeping the excitation intensity as low as possible is recommended. Illumination of the surrounding tissue by the laser also tends to increase the background signal. The use of low excitation intensity helps reduce the background, the caveat being it could also lead to a decrease in fluorescent protein photon yield. Background subtraction during analysis could be used as an alternative means to reduce the background signal prior to image analysis.
Drosophila presents an ideal organism for in vivo imaging as it provides well-established genetic tools for the investigation of specific protein function. The upstream activating sequence (UAS)-Gal4 system enables temporal and spatial expression of proteins and can therefore be used to manipulate neurotransmission23, such that thermo-genetic stimulation using Drosophila transient receptor potential sub-family A1 (dTRPA1)24 or opto-genetic stimulation using far-red-shifted CsChrimson25 can be combined with imaging14. We have overcome many of the complications of performing in vivo single-molecule imaging in this system and now describe how single-particle tracking can be carried out in Drosophila melanogaster third instar larvae.
Protocol
1. Design of Transgenic Drosophila Expressing mEos2-Tagged Proteins
- Select the protein to be tagged with the mEos2 protein. To increase the imaging success rate, select proteins with a transmembrane domain in the neuronal plasma membrane14 (e.g., Syntaxin-1A), proteins on synaptic vesicles or autophagosomes26,27 (e.g., Synaptobrevin-2), or cytosolic proteins with close interaction with neuronal plasma membrane proteins (e.g., Munc18-1).
- Construct transgenes encoding the mEos2-tagged protein (Figure 1). Design the forward and reverse primers for the protein and mEos2.
NOTE: The mEos2 coding sequence can be amplified from the pmEos2-N1 plasmid, which is designed to fuse to the C-terminus of the protein of interest. Cloning can for example be performed by in vivo recombination in Saccharomyces cerevisiae using the pFL44S-attB-MCS-w+ vector14,28, alternately other cloning methods could be used such as Gibson assembly protocol29.- Linearize the vector with BamHI and XbaI and co-transform into ura3- yeast cells together with the PCR fragments encoding mEos2 and the protein of interest. Inject Drosophila embryos with the construct to generate a transgenic fly line30.
- Rear the transgenic fly cultures on standard yeast and molasses medium or some other suitable substitute contained in plastic vials. To ensure healthy and fairly large synaptic boutons31, avoid overcrowding flies in the vials, and flip flies to new vials after each day of egg-laying; this ensures the third instar larvae are well fed and reach proper size by the time they commence wandering on the medium. Raise the transgenic fly at room temperature (25 °C).
NOTE: Larger synaptic boutons tend to yield larger quantity of proteins visualized during imaging.
2. Dissection of Third Instar Larva
- Make a cylindrical or semicylindrical shaped polydimethylsiloxane (PDMS) base ≤20 mm in diameter and ≤20 mm in height using an appropriate silicone elastomer kit (Figure 2A). Mix silicone elastomer with a silicone hardening agent at a ratio of 10:1.
- Stir the mixture thoroughly for 5 min. Pour the mixture into a mold with the dimsensions listed above. Let it harden in an oven at 100 °C for 45 min. The PDMS base will serve as the foundation on which to carry out dissection. Ensure the PDMS base fits into the well of a glass-bottomed culture dish.
- Select a wandering third instar larva from the vial wall. Use an optical stereo-microscope at 4.5 magnification to visualize the larva placed on the PDMS cylindrical base with the dorsal side up.
- Use curved and angled tweezers to stick minutien pins on the head over the mouth hooks, between the two extending anterior spiracles, and on the tail over the anus, between the two posterior spiracles. Add 40 µL of Schneider's insect medium at room temperature onto the immobilized larva.
NOTE: Hemolyph-like solutions (HL3 or HL6) may also be used in the place of Schneider's solution32,33. - Provide access through the dorsal side of the abdominal body wall of the larva by use of a minutien pin to incise into the midline of the wall.
- Use spring scissors to cut the body wall open from the incision point. Cut along the midline in the anterior-posterior direction34.
- Use fine forceps and spring scissors to disembowel the larva: remove the internal organs including the salivary glands, trachea and gut. Wash the larva twice with Schneider's insect medium.
- With fine forceps, hold the two body wall flaps, gently stretch them out, and stick two minutien pins on each side. This should produce a hexagon-shaped preparation (Figure 2A).
- Detach the brain from the ventral nerve cord using spring scissors to decrease the possibility of muscle movement during imaging; this also serves to hold the preparation flat against the imaging dish. Use the curved tweezers to push all six minutien pins into the PDMS base.
NOTE: Only a small portion of the pins should be embedded within the abdominal wall. - To confirm that the larva has survived the dissection procedure, slightly poke the ventral nerve cord, this should elicit a peristaltic contraction of the larva abdominal wall.
3. Slightly Oblique Illumination Single-Particle Tracking Microscopy
- Invert the dissected larva-PDMS base onto a glass-bottomed culture dish. Fill the imaging dish with 2 mL of room temperature Schneider's insect medium (Figure 2B).
- Locate motor neurons in the second and third segments with synapses on abdominal muscles 6 and 735 using the 63x/1.2 NA water-immersion objective on a fluorescent total internal reflection fluorescence (TIRF) microscope.
- Use the eyepieces of the microscope to locate the dissected larva; identify muscle 6 by use of bright field illumination.
- Use the microscope acquisition software (see Table of Materials), in TIRF configuration; slide the collimator camera angle to 1° to increase the penetration depth, otherwise move out of TIRF critical angle to achieve oblique illumination.
- Switch the 488 nm laser on to visualize mEos2 in green. Use a low laser power (less than 5% of 22.8 mW) to avoid bleaching the green fluorescence. Locate the neuromuscular junctions (which look like beads on a string) and acquire a low resolution TIRF image.
- Simultaneously switch on the 405 nm laser (photoconverts mEos2 from green to red species) and the 561 nm laser (image the photoconverted red mEos2) to stochastically localize single mEos2 fluorescent proteins.
NOTE: Use of low 405 nm laser power is recommended (0.005 to 0.9% of 4.78 mW) as this allows for a relatively constant photoconversion rate during movie acquisition. Use between 50 and 75% of the 561 nm laser (20.1 mW), as this is sufficient to acquire fluorescence from the red mEos2 species without adversely affecting the morphology of the muscle tissue surrounding the neuromuscular junction. - For each neuromuscular junction chain, acquire a time series composed of 15,000 frames or more at 33 Hz. Save as.czi format in order to preserve the accompanying meta-data for possible reference.
- Use ImageJ to convert '.czi' movie files to '.tiff' files.
- Analyze the acquired movies with suitable single-molecule tracking analytical software11,14,36 such as TrackMate on FIJI/ImageJ37 (see Table of Materials). Locates the 'x' and 'y' coordinates of each localization by the Gaussian fit of the intensity point spread function (PSF).
Note: Troubleshooting: If green fluorescence is not observed or is too dim at the neuromuscular junction upon exposure to the 488 nm laser, briefly switch on the photoconverting and imaging lasers (405 nm and 561 nm), as this helps to produce more of the red species of mEos2. Then switch back to the 488 nm laser and take a TIRF image.
4. Single-Molecule Localization in Fixed Larvae
- To measure the protein nanocluster size via PALM, replace the Schneider's insect medium after dissection with a fixative – 4% paraformaldehyde diluted in phosphate buffered saline (PBS).
- Incubate the larva for 30 min at room temperature.
- Remove the dissection pins and place the fixed larva in a 1.7 mL clear snap-lock microcentrifuge tube.
- Dissect and fix as many other larvae as needed, then wash them three times with PBS.
- Replace the PBS with block buffer (a solution of PBS, 0.1% Triton X-100, and 3% normal goat serum).
- Wash the larvae with PBS three times, then incubate with the necessary primary antibody (diluted in the block buffer solution).
- Incubate overnight at 4 oC.
- Wash the larvae three times with PBS, then incubate with the necessary secondary antibody (diluted in the block buffer solution).
- Wash the larvae three times with PBS, attach the larva back onto the PDMS base, and image as previously described for single-molecule tracking.
Representative Results
Using the protocol listed above, Syntaxin-1A-mEos2 transgenic Drosophilamelanogaster were generated (Figure 1). Transgenic third instar Drosophila larvae were dissected on the PDMS cylindrical base (Figure 2A-F). To visualize single molecules of Syntaxin-1A, we used PALM on a TIRF microscope with slightly oblique laser illumination14.
Single molecules of Syntaxin-1A were tracked on the presynaptic motor nerve terminals on muscle 6 of the second abdominal segment. The green fluorescence of mEos2 could be detected as shown by the low-resolution image of Syntaxin-1A-mEos2 on type 1b boutons when excited using the 488 nm laser (Figure 3A). In the imaging experiments, the 405 nm and 561 nm excitation laser imaging depths were 133.5 nm and 165.6 nm, respectively. The green image was acquired as an average of 16 individual frames. This green image was used to create the region of interest used to limit analysis of localizations in the photoconverted movie to the presynaptic motor nerve terminals alone.
The analysis of the acquired photoconverted sptPALM movies generated a super-resolved intensity, diffusion coefficient, and trajectory maps (Figure 3A). Syntaxin-1A-mEos2 mobility was further quantified by analysis of the mean square displacement (MSD) of the individual trajectories (Figure 3B). Statistical comparisons can be done using the area under the MSD curve (Figure 3B). Further analysis of mobility yields the diffusion coefficient distribution of Syntaxin-1A-mEos2, and this can be statistically evaluated by comparison of the mobile to the immobile populations (Figure 3C).
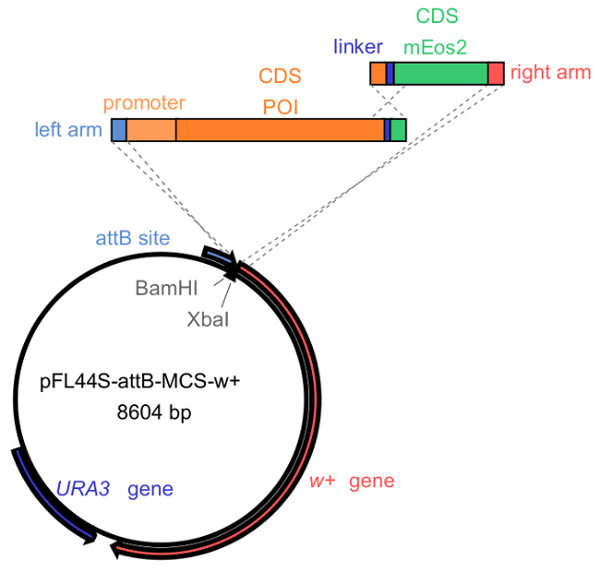
Figure 1: Generating mEos2-tagged constructs. The pFL44S-attB-MCS-w+ vector is linearized with BamHI and XbaI. Overlapping polymerase chain reaction (PCR) fragments encoding the protein of interest (POI) and mEos2, respectively, can be cloned, for example, by in vivo recombination in ura3- yeast cells or by Gibson assembly. Note that to generate the Syntaxin-1A-mEos2 transgene, we cloned the construct flanked by the endogenous Syntaxin-1A promoter and terminator sequences. The resulting construct can subsequently be injected into Drosophila embryos to create a transgenic fly line expressing the protein of interest (POI) fused to mEos2. Please click here to view a larger version of this figure.
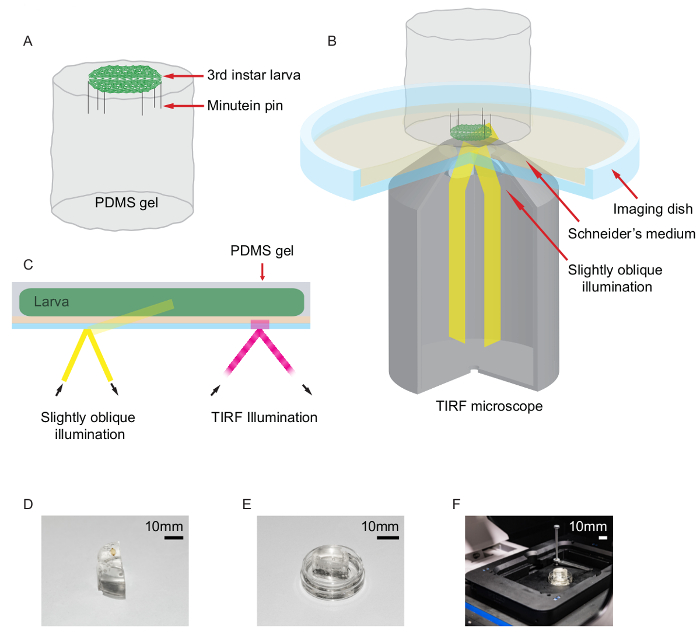
Figure 2: Drosophila larva dissection and oblique illumination. (A) Schema showing Drosophila larva dissection carried out on half-cylindrical PDMS base. A filleted larva was held onto the PDMS gel by use of minutien pins. (B-C) The larva-PDMS prep was placed inside a glass-bottomed culture dish containing Schneider's insect medium. A TIRF microscope in a slightly oblique illumination configuration was used to visualize Syntaxin-1A-mEos2 in the motor nerve terminals. (D-F) A half-cylindrical PDMS base with dissected Drosophila larva. The larva-PDMS preparation was placed in an imaging dish filled with Schneider's medium, and the larva was imaged on the super-resolution PALM microscope. Scale bar, 10 mm. (This figure has been modified from14). Please click here to view a larger version of this figure.
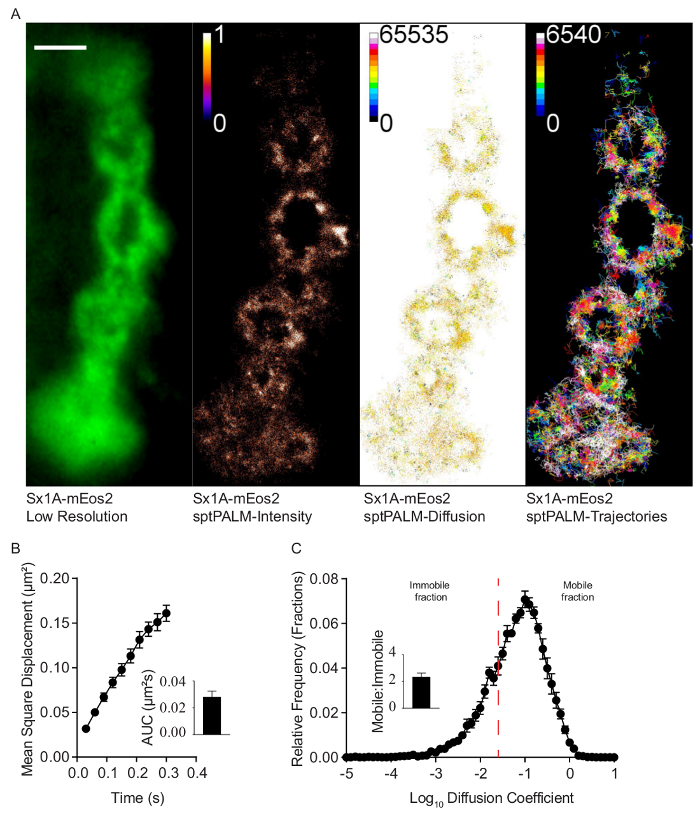
Figure 3: In vivo tracking of Syntaxin-1A-mEos2 at the presynaptic motor nerve terminal. (A) A low-resolution TIRF image of Syntaxin-1A-mEos2 on a type 1b motor nerve terminal. Analysis of photoconversion movies imaged at 33 Hz generated sptPALM super-resolved intensity, diffusion coefficient, and trajectory maps. 5,309 individual trajectories were imaged over 16,000 frames movie acquisition. Scale bar, 3 μm. (B-C) Mean square displacement (MSD) of Syntaxin-1A-mEos2 was plotted from 6 different motor nerve terminals; area under the MSD curve (AUC) was used to quantify the level of mobility. The diffusion coefficient distribution reveals mobile and immobile Syntaxin-1A populations that can be quantified as ratios of each other; an average of ~2,400 trajectories were obtained per motor nerve terminal (n = 3 larvae). Data presented as mean standard error of mean. Please click here to view a larger version of this figure.
Discussion
This protocol describes single-molecule tracking of mEos2-tagged proteins at the neuromuscular junction of the Drosophila third instar larva. To avoid over-expression of transgenic protein, Syntaxin-1A-mEos2 expression was driven by endogenous Syntaxin-1A promoter. Studies of synaptic protein mobility have mostly been limited to in vitro investigations of cultured cells and cortical neurons9,11,12,13. Whether these proteins exhibit similar mobility signatures in a live animal is largely unknown. To visualize single-protein mobility in vivo, limitations such as optical transparency, accessibility, and penetration depth have to be overcome. By dissecting the larval preparation and visualizing the neuromuscular junctions embedded on the surface of the muscle, we avoid the issue of optical transparency and accessibility. Attempts to image single-molecule mobility in normal TIRF configuration proved unsuccessful. However, use of slightly oblique illumination allows for increased excitation laser penetrating depth. Furthermore, the minutien pins used to immobilize the larva helped to avoid any possible adverse effect of anesthetic use on protein mobility. It is possible to use higher magnification objectives, such as the 100x oil immersion objectives, to visualize single molecules in vivo. However, increased magnification may increase the occurance of drifts out of focus. In addition, we have used a lower intensity (~30%) 561 nm laser successfully to image single molecules at motor nerve terminals. Additional testing may be required to use lower laser power.
This protocol is suitable to visualize the mobility of synaptic proteins within the proximity of the neuronal plasma membrane. Using this approach, the mobility of the SNARE protein – Syntaxin-1A and the autophagosome bound protein Atg18a – have been successfully imaged in vivo14,26. The mobility of proteins on motor neuron axons can also be investigated27. However, investigation of the mobility of proteins on the brain or ventral nerve cord may prove difficult to assess due to the high background signal produced by the underlying tissue; in addition, visualizing protein mobility on the same brain structure will require fluorescently tagging the brain structure (to ensure replicability), and use of dual-camera imaging would be needed.
Current methods available for super-resolution microscopy in Drosophila are mostly limited to imaging embryos, rather than the more developed third instar larva38,39. The main issue with these protocols is that they yield a low number of trajectories in the area imaged, and hence prevent in-depth analysis of mobility states22,40. Our in vivo single molecule tracking protocol has the capacity to generate a high number of trajectories, and could therefore be used in other animal models such as Caenorhabiditis elegans and zebrafish. Indeed, every NMJ chain generated ~2,400 trajectories making it suitable for analyzing the different mobile states and transitions between these states14,27. Additionally, many in vivo single molecule imaging strategies rely on the use of anesthetics to immobilize the animals20,22, which might alter the physiologic function under which the imaging is done. Here we optimized an 'anesthesia-free' protocol to immobilize the larvae using minutien pins.
A potential use of this protocol may be for visualizing the mobility of proteins in three dimensions. Recent advances in super-resolution microscopy allow protein mobility to be visualized in vitro in 'x', 'y', and 'z' dimensions41,42. Our current method does not account for the mobility of proteins in the 'z' axis. Yet the Drosophila motor nerve terminal is a spherical structure and a valuable future approach will be to quantify protein mobility in a three-dimensional volume43. Imaging in the z-dimension is feasible using an astigmatic lens. Alternatively, in TIRF mode44,45, the z-resolution is also increased. However, in our experiments, we have used slightly oblique illumination to visualize single molecules of syntaxin1A-mEos2 without the astigmatic lens.
In conclusion, availability of both a transgenic Drosophila fly stock expressing proteins tagged with a photoconvertible protein, a PDMS base to dissect the transgenic larva, as well as a TIRF microscope equipped with the possibility of changing the angle of illumination are the most important tools needed to visualize single proteins as described in this protocol.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank Jean-Baptiste Sibarita and Daniel Choquet (IINS, CNRS/University of Bordeaux) for their kind support with the single-molecule analysis. We especially thank Nick Valmas and Jessica Mcgaw (QBI, University of Queensland) for their help with the video recording, voice-over as well as figure graphic design. This work was supported by the Australian Research Council (AR) discovery project (DP170100125 to F.A.M.), the Australian Research Council (LIEF Grant LE0882864 to F.A.M.), Future Fellowship (FT100100725 to B.V.S.), Australian Research Council (LIEF Grant LE130100078 to F.A.M.) and NHMRC Project grant (APP1103923 to B.V.S.). F.A.M. is a NHMRC Senior Research Fellow (ONT1060075).
Materials
| PDMS (Sylgard 184 silicone elastomer kit) | Dow Corning Corporation | 000000000001064291 | |
| Glass-bottomed Culture Dish | In Vitro Scientific | D29-20-1.5-N | |
| Optical Stereo-microscope | Olympus | SZ51 | |
| Curved Tweezers (Style 5/45) | Electron Microscopy Sciences | 0208-5AC-PO | |
| Minutien Pins (0.1mm) | Fine Science Tools | 26002-10 | |
| Schneider's Insect Medium | Sigma, Life Sciences | S0146 | |
| Vannas Spring Scissors | Fine Science Tools | 15000-00 | |
| Fine Forceps (Dumont #5) | Fine Science Tools | 11254-20 | |
| ELYRA PS.1 Microscope | Zeiss | PS.1 | |
| Phosphate Buffered Saline (PBS) | Lonza | 17-515Q | |
| Paraformaldehyde (16%) | Electron Microscopy Sciences | 15710 | |
| Triton X-100 | Sigma-Aldrich | T-9284 | |
| Normal Goat Serum | Sigma-Aldrich | G9023 | |
| Maxyclear snaplock microcentrifuge Tube (1.7 mL) | Axygen | MCT-175-C | |
| Zen Black acquisition software | Carl Zeiss, 2012 | ||
| Metamorph software | Molecular Devices | 7.7.8 | PALM Tracer Plugin |
| Fly strain | w1118; HA-Sx1A-mEos2 | ||
Riferimenti
- Sudhof, T. C. The molecular machinery of neurotransmitter release (Nobel lecture). Angew Chem Int Ed Engl. 53 (47), 12696-12717 (2014).
- Papadopulos, A., et al. Activity-driven relaxation of the cortical actomyosin II network synchronizes Munc18-1-dependent neurosecretory vesicle docking. Nat Commun. 6, 6297 (2015).
- Papadopulos, A., Tomatis, V. M., Kasula, R., Meunier, F. A. The cortical acto-Myosin network: from diffusion barrier to functional gateway in the transport of neurosecretory vesicles to the plasma membrane. Front Endocrinol (Lausanne). 4, 153 (2013).
- Tomatis, V. M., et al. Myosin VI small insert isoform maintains exocytosis by tethering secretory granules to the cortical actin. J Cell Biol. 200 (3), 301-320 (2013).
- Wen, P. J., et al. Phosphatidylinositol(4,5)bisphosphate coordinates actin-mediated mobilization and translocation of secretory vesicles to the plasma membrane of chromaffin cells. Nature Communications. 2, (2011).
- Malintan, N. T., et al. Abrogating Munc18-1-SNARE complex interaction has limited impact on exocytosis in PC12 cells. J Biol Chem. 284 (32), 21637-21646 (2009).
- Choquet, D., Triller, A. The dynamic synapse. Neuron. 80 (3), 691-703 (2013).
- Triller, A., Choquet, D. New concepts in synaptic biology derived from single-molecule imaging. Neuron. 59 (3), 359-374 (2008).
- Gandasi, N. R., Barg, S. Contact-induced clustering of syntaxin and munc18 docks secretory granules at the exocytosis site. Nat Commun. 5, 3914 (2014).
- Milovanovic, D., Jahn, R. Organization and dynamics of SNARE proteins in the presynaptic membrane. Front Physiol. 6, 89 (2015).
- Kasula, R., et al. The Munc18-1 domain 3a hinge-loop controls syntaxin-1A nanodomain assembly and engagement with the SNARE complex during secretory vesicle priming. J Cell Biol. 214 (7), 847-858 (2016).
- Constals, A., et al. Glutamate-induced AMPA receptor desensitization increases their mobility and modulates short-term plasticity through unbinding from Stargazin. Neuron. 85 (4), 787-803 (2015).
- Ribrault, C., et al. Syntaxin1A lateral diffusion reveals transient and local SNARE interactions. Journal of Neuroscience. 31 (48), 17590-17602 (2011).
- Bademosi, A. T., et al. In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat Commun. 8, 13660 (2017).
- Bar-On, D., et al. Super-resolution imaging reveals the internal architecture of nano-sized syntaxin clusters. J Biol Chem. 287 (32), 27158-27167 (2012).
- McKinney, S. A., Murphy, C. S., Hazelwood, K. L., Davidson, M. W., Looger, L. L. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 6 (2), 131-133 (2009).
- Zhang, M. S., et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nature Methods. 9 (7), 727-U297 (2012).
- Ando, R., Hama, H., Yamamoto-Hino, M., Mizuno, H., Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 99 (20), 12651-12656 (2002).
- Schneider, R., et al. Mobility of calcium channels in the presynaptic membrane. Neuron. 86 (3), 672-679 (2015).
- Berning, S., Willig, K. I., Steffens, H., Dibaj, P., Hell, S. W. Nanoscopy in a living mouse brain. Science. 335 (6068), 551-551 (2012).
- Willig, K. I., et al. Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys J. 106 (1), L01-L03 (2014).
- Zhan, H., et al. In vivo single-molecule imaging identifies altered dynamics of calcium channels in dystrophin-mutant C. elegans. Nat Commun. 5, 4974 (2014).
- Duffy, J. B. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 34 (1-2), 1-15 (2002).
- Hamada, F. N., et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 454 (7201), 217-220 (2008).
- Klapoetke, N. C., et al. Independent optical excitation of distinct neural populations. Nat Methods. 11 (3), 338-346 (2014).
- Vanhauwaert, R., et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. , (2017).
- Joensuu, M., et al. Subdiffractional tracking of internalized molecules reveals heterogeneous motion states of synaptic vesicles. J Cell Biol. 215 (2), 277-292 (2016).
- Merhi, A., Gerard, N., Lauwers, E., Prevost, M., Andre, B. Systematic mutational analysis of the intracellular regions of yeast Gap1 permease. PLoS One. 6 (4), e18457 (2011).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6 (5), 343-345 (2009).
- Bischof, J., Maeda, R. K., Hediger, M., Karch, F., Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 104 (9), 3312-3317 (2007).
- Stewart, B. A., McLean, J. R. Population density regulates Drosophila synaptic morphology in a Fasciclin-II-dependent manner. J Neurobiol. 61 (3), 392-399 (2004).
- Stewart, B. A., Atwood, H. L., Renger, J. J., Wang, J., Wu, C. F. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 175 (2), 179-191 (1994).
- Macleod, G. T., Hegstrom-Wojtowicz, M., Charlton, M. P., Atwood, H. L. Fast calcium signals in Drosophila motor neuron terminals. J Neurophysiol. 88 (5), 2659-2663 (2002).
- Halachmi, N., Nachman, A., Salzberg, A. Visualization of proprioceptors in Drosophila larvae and pupae. J Vis Exp. (64), e3846 (2012).
- Menon, K. P., Carrillo, R. A., Zinn, K. Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip Rev Dev Biol. 2 (5), 647-670 (2013).
- Nair, D., et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci. 33 (32), 13204-13224 (2013).
- Tinevez, J. Y., et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2017).
- Ferguson, J. P., et al. Deciphering dynamics of clathrin-mediated endocytosis in a living organism. J Cell Biol. 214 (3), 347-358 (2016).
- Greiss, F., Deligiannaki, M., Jung, C., Gaul, U., Braun, D. Single-Molecule Imaging in Living Drosophila Embryos with Reflected Light-Sheet Microscopy. Biophys J. 110 (4), 939-946 (2016).
- Robin, F. B., McFadden, W. M., Yao, B., Munro, E. M. Single-molecule analysis of cell surface dynamics in Caenorhabditis elegans embryos. Nat Methods. 11 (6), 677-682 (2014).
- Singh, A. P., et al. 3D Protein Dynamics in the Cell Nucleus. Biophys J. 112 (1), 133-142 (2017).
- Perillo, E. P., et al. Deep and high-resolution three-dimensional tracking of single particles using nonlinear and multiplexed illumination. Nat Commun. 6, 7874 (2015).
- Knodel, M. M., et al. Synaptic bouton properties are tuned to best fit the prevailing firing pattern. Front Comput Neurosci. 8, 101 (2014).
- Yang, Q., Karpikov, A., Toomre, D., Duncan, J. Estimation of 3D geometry of microtubules using multi-angle total internal reflection fluorescence microscopy. Med Image Comput Comput Assist Interv. 13 (Pt 2), 538-545 (2010).
- Boulanger, J., et al. Fast high-resolution 3D total internal reflection fluorescence microscopy by incidence angle scanning and azimuthal averaging. Proc Natl Acad Sci U S A. 111 (48), 17164-17169 (2014).

