Induction of Endothelial Differentiation in Cardiac Progenitor Cells Under Low Serum Conditions
Summary
This protocol describes an endothelial differentiation technique for cardiac progenitor cells. It particularly focuses on how serum concentration and cell-seeding density affect the endothelial differentiation potential.
Abstract
Cardiac progenitor cells (CPCs) may have therapeutic potential for cardiac regeneration after injury. In the adult mammalian heart, intrinsic CPCs are extremely scarce, but expanded CPCs could be useful for cell therapy. A prerequisite for their use is their ability to differentiate in a controlled manner into the various cardiac lineages using defined and efficient protocols. In addition, upon in vitro expansion, CPCs isolated from patients or preclinical disease models may offer fruitful research tools for the investigation of disease mechanisms.
Current studies use different markers to identify CPCs. However, not all of them are expressed in humans, which limits the translational impact of some preclinical studies. Differentiation protocols that are applicable irrespective of the isolation technique and marker expression will allow for the standardized expansion and priming of CPCs for cell therapy purpose. Here we describe that the priming of CPCs under a low fetal bovine serum (FBS) concentration and low cell density conditions facilitates the endothelial differentiation of CPCs. Using two different subpopulations of mouse and rat CPCs, we show that laminin is a more suitable substrate than fibronectin for this purpose under the following protocol: after culturing for 2 – 3 days in medium including supplements that maintain multipotency and with 3.5% FBS, CPCs are seeded on laminin at <60% confluence and cultured in supplement-free medium with low concentrations of FBS (0.1%) for 20 – 24 hours before differentiation in endothelial differentiation medium. Because CPCs are a heterogeneous population, serum concentrations and incubation times may need to be adjusted depending on the properties of the respective CPC subpopulation. Considering this, the technique can be applied to other types of CPCs as well and provides a useful method to investigate the potential and mechanisms of differentiation and how they are affected by disease when using CPCs isolated from respective disease models.
Introduction
Recent studies support the existence of resident cardiac progenitor cells (CPCs) in the adult mammalian heart1,2,3, and CPCs could be a useful source for cell therapy after cardiac injury4,5. In addition, expanded CPCs may provide a fruitful model for drug screening and the investigation of disease mechanisms when isolated from patients with rare cardiomyopathies, or from respective disease models6,7.
CPCs isolated from the adult heart possess stem/progenitor cell characteristics1,2,3,8 as they are multipotent, clonogenic, and have the capacity for self-renewal. However, there are many different (sub)populations of CPCs exhibiting different surface marker profiles, including, for instance, c-kit, Sca-1, and others, or retrieved by different isolation techniques (Table 1). Several culture and differentiation protocols have been established1,2,8,9,10,11,12,13,14,15,16,17,18. These protocols vary mostly with respect to the growth factor and serum content, which are adjusted according to the purpose of the culturing and which can lead to differences in results and outcomes, including differentiation efficiency.
Marker-based Isolation Techniques:
CPCs can be isolated based on a specific surface marker expression1,2,8,9,10,11,12,13,14,15,16,17,18. Previous studies suggest that c-kit and Sca-1 may be the best markers to isolate resident CPCs1,11,14,19,20. Because none of these markers is truly specific for CPCs, combinations of different markers are usually applied. For example, whereas CPCs express low levels of c-kit21, c-kit is also expressed by other cell types, including mast cells22, endothelial cells23, and hematopoietic stem/progenitor cells24. An additional problem is the fact that not all markers are expressed across all species. This is the case for Sca-1, which expresses in mouse but not in human25. Therefore, using protocols that are independent of isolation markers may be advantageous in view of clinical trials and studies using human samples.
Marker-independent Isolation Techniques:
There are several major techniques of CPC isolation, which are primarily independent of surface marker expression, but which can be refined by the consecutive selection of specific marker-positive subfractions as needed (see also Table 1). (1) The side population (SP) technique has originally been characterized in a primitive population of hematopoietic stem cells based on the ability to efflux the DNA dye Hoechst 3334226 by ATP-binding cassette (ABC) transporters27. Cardiac SP cells have been isolated by different groups and reported to express a variety of markers with some minor differences between reports2,8,13,14. (2) Colony-forming unit fibroblast cells (CFU-Fs) have originally been defined based on a mesenchymal stromal cell (MSC)-like phenotype. Isolated MSCs are cultured on dishes to induce colony formation. Such colony-forming MSC-like CFU-Fs can be isolated from the adult heart and are capable to differentiate into cardiac lineages15. (3) Cardiosphere-derived cells (CDC) are single cells derived from clusters of cells grown from tissue biopsies or explants28,29,30,31. It was recently shown that mostly the CD105+/CD90–/c-kit– cell fraction exhibits cardiomyogenic and regenerative potential32.
Here, using SP-CPCs isolated from mice, we provide a protocol for the efficient induction of endothelial lineage based on a previous study in rat CPCs and mouse SP-CPCs33. The protocol contains specific adaptations to the culture and expansion technique with respect to the cell density, the serum content of the medium, and the substrate. It can be applied not only to mouse SP-CPCs but to different types of CPCs for the purpose to induce a fate switch from an amplifying to an endothelial-committed CPC, be it in view of transplantation of these cells or their use for mechanistic in vitro studies.
Protocol
The use of mice for cell isolation purpose was in accordance with the Guide for the Care and Use of Laboratory Animals and with the Swiss Animal Protection Law and was approved by the Swiss Cantonal Authorities.
NOTE: The isolation of Sca-1+/CD31– SP-CPCs from the mouse heart was essentially done as previously described34 with some modifications. For the materials and reagents used, see Table of Materials. For all experiments, cardiac SP-CPCs isolated from mice were amplified, passaged, and used in a cell line-like manner. Passages 7 – 20 were used for this study.
1. Tissue Preparation
NOTE: All experiments using mice must be carried out according to the guidelines and regulations. This protocol uses four mice. Culture plates that are 100 mm in diameter are described as P100 and culture plates that are 60 mm in diameter are described as P60 for the following parts of the protocol.
- Inject each mouse with 200 mg/kg of pentobarbital intraperitoneally (i.p.) and wait until it is fully anesthetized by checking its response to toe pinching.
- Wipe the chest with 70% ethanol. Cut the skin and thoracic wall with scissors to expose the thoracic cavity.
- Lift the heart with forceps and cut it at the base using scissors. Put the heart in a P100 with 25 mL (5 mL for P60) of cold phosphate-buffered saline (PBS) (three to five hearts per P100 or one to two hearts per P60).
- Pump the heart with forceps to eject the blood out of the cavities (Figure 1A).
- Put the heart in a P100 with 25 mL (5 mL for P60) of cold PBS for washing. Remove the atria using small scissors. Cut the heart into two longitudinal pieces and wash them again in ice-cold PBS (25 mL/P100, 5 mL/P60).
- Transfer the pieces to a new P100 with 25 mL (5 mL for P60) of cold PBS. Cut the pieces into smaller pieces using small scissors (Figure 1B). Add a drop of 1 mg/mL collagenase B diluted in Hank’s balanced salt solution (HBSS) and mince the small pieces thoroughly with a sterile razor blade (Figure 1C).
2. Digestion
- Add 10 mL (2.5 mL for P60) of the 1 mg/mL collagenase B solution to the dish from step 1.6.
- Put the collagenase B solution containing the minced heart pieces in a tilted (about 30°) P100 (or P60) in a 37 °C incubator (Figure 1D).
- Incubate the minced heart pieces for a maximum of 30 min; homogenize by repeatedly passing them through a Pasteur pipette every 10 min during the incubation (Figure 1E).
NOTE: It is important to not exceed 30 min of incubation in total for step 1.3.
3. Filtration
- Add 10 mL (5 mL for P60) of cold HBSS supplemented with 2% FBS to the minced and homogenized heart pieces.
NOTE: HBSS supplemented with 2% FBS quenches collagenase B activity. - Filter the heart pieces through 100 µm filter to remove undigested tissue and centrifuge at 470 x g for 5 min at room temperature (RT) (Figure 1F, yellow filters).
- Discard the supernatant and resuspend the pellet in 5 mL (3 mL for P60) of red blood cell lysis buffer, and incubate the pellet for 5 min with occasional shaking on ice.
- Add 10 mL (5 mL for P60) of PBS (to stop the lysis reaction) and filter the sample through a 40 µm filter to exclude larger cells, including residual cardiomyocytes (Figure 1F, blue filters).
- Centrifuge the tube at 470 x g for 5 min at RT (without brake). Discard the supernatant and resuspend the pellet in 1 mL of DMEM including 10% FBS.
- Count the cells in an aliquot with a hemocytometer. Resuspend the cells with DMEM including 10% FBS, aiming at a final cell concentration of 1 x 106 cells/mL.
NOTE: Roughly 5 x 106 cardiomyocyte- and erythrocyte-depleted cells can be estimated per mouse. - Distribute the cells into two tubes (Figure 2): in tube A, add 1.5 mL for Hoechst 33342 staining with verapamil; in tube B, add 18.5 mL for Hoechst 33342 staining and proceed to stain and sort (steps 4.1 and 4.2) cardiac SP cells by flow cytometry.
4. Sorting of Cardiac SP Cells by Flow Cytometry
NOTE: Verapamil inhibits Hoechst efflux by blocking multidrug resistance (MDR) ABC transporter activity. Hoechst 33342 is a DNA-binding dye that can be used in living cells to detect the cell cycle as it correlates with the DNA content. Hoechst-33342-extruding cells appear in the Hoechst low part of both emission channels (450 nm, Hoechst blue; 650 nm, Hoechst red), that is, aside of the Hoechst-retaining “main population”, giving them their name “side population”. SP cells are enriched in cells with progenitor properties and show a high expression of multidrug-resistant ABC transporters (such as MDR1 and ABCG2). Hoechst 33342 is written as Hoechst for the following parts of the protocol. It is important to protect light-sensitive materials for ideal results.
- Staining with Hoechst in the presence and absence of verapamil
- Add verapamil (with a final concentration of 83.3 µM) and Hoechst (with a final concentration of 5 µg/106 cells) to the cell solution. For tube A, use Verapamil and Hoechst; for tube B, use Hoechst only. Incubate in a water bath (at 37 °C) for 90 min and revert the tubes every 20 min (Figure 1G).
- Centrifuge the tubes at 470 x g for 5 min at RT. Discard the supernatant and resuspend each pellet in HBSS (1 x 106 cells/mL).
- Take a 1.5 mL aliquot for single stainings and negative control from tube B (Figure 2): (i) fluorescein isothiocyanate (FITC)-conjugated anti-Sca-1; (ii) allophycocyanin (APC)-conjugated anti-CD31; (iii) nonstained cells (negative control).
NOTE: Isotype controls are used here for setting up the isolation protocol. - Centrifuge tubes A and B and the single-staining and negative control aliquots at 470 x g for 5 min at RT for washing out Hoechst and verapamil.
- Discard the supernatant and resuspend the pellets of tube A and (iii) the negative control in 250 µL of HBSS and keep them on ice in the dark until sorting.
- Resuspend the pellet of tube B in 200 µL of HBSS and the pellet of (i, ii) the single-staining aliquots in 100 µL of HBSS. Add FITC-conjugated anti-Sca-1 (0.6 µg/107 cells) and APC-conjugated anti-CD31 (0.25 µg/107 cells). Incubate the pellets for 30 min on ice and shake them from time to time in the dark.
- Add 2 mL of HBSS to the tubes. Centrifuge the tubes at 470 x g for 5 min at RT.
- Discard the supernatant and resuspend all pellets with 1 mL of HBSS and centrifuge as above. Discard the supernatants. Resuspend the pellet of the single-staining aliquots in 200 µL of HBSS. Resuspend the pellet of tube B in HBSS (20 x 106 cells/mL).
- Keep the samples on ice and protected from light until sorting.
- Sorting with flow cytometry
- Prepare sterile 1.5 mL sorting tubes with 500 µL of HBSS including 2% FBS.
- Stain the cells with 7-aminoactinomycin D (7-AAD) (0.15 µg/106 cells) on ice for 10 min to exclude dead cells.
- Sort the cardiac SP using the following settings: excite Hoechst using 350 nm (UV) excitation, collect fluorescence emission with a 450/50 nm band-pass filter (Hoechst Blue) and a 670/30 nm band-pass filter (Hoechst red), and use a nozzle size of 100 µm and pressure of 15 psi.
- For analysis, record 5 x 105 events for the sorting samples and 1 x 105 events for the negative control, verapamil control, and single-staining samples.
NOTE: The cardiac SP is around 0.5% – 2% (Figure 1H) but may vary between laboratories and isolates. The Sca-1+/CD31– fraction is around 1% – 11% of the total cardiac SP (Figure 1I) but may vary between laboratories and isolates. Sca-1+/CD31– SP-CPCs are written as SP-CPCs for the following parts of the protocol.
5. Primary Culture of Isolated SP-CPCs
NOTE: Three different types of media were used in this protocol. They are referred to as Medium 1 (according to Noseda et al.)8, Medium 2, and Medium 3 and are described in the Table of Materials regarding their composition.
- Warm up Medium 1 to 37 °C before use and cool down the centrifuge to 4 °C. Centrifuge the sorting tubes at 4 °C and 470 x g for 6 min and resuspend the cells in Medium 1.
- Put the cells on a gas-permeable P60 dish with 4 mL of Medium 1. Change the medium every 3 d until the cells have reached 70% – 80% confluence.
NOTE: Step 5.2 may take around 2 – 3 weeks.
6. Expansion and Differentiation of SP-CPCs
- Cell culture
- Culture 3 x 105 SP-CPCs in 8 mL of Medium 1 in a T75 flask.
- Incubate SP-CPCs at 37 °C with 5% CO2 until 70% – 80% confluence, changing the medium every 2 – 3 d.
NOTE: The cell number should be modified according to the type of CPCs used, the doubling time, and the cell size.
- SP-CPC growth and viability under different serum concentrations
- Culture SP-CPCs for 2 – 3 d in T75 flasks with 8 mL of Medium 1. Carefully aspirate the medium and rinse gently with 5 mL of warm (37 °C) HBSS.
- Treat the cells with 5 mL of Trypsin-EDTA for 5 min in the cell incubator, add 5 mL of Medium 1 to stop the Trypsin activity, and transfer the cell suspension to a 15 mL tube.
- Centrifuge the cells at 470 x g for 5 min at RT.
- Resuspend the cells in Medium 1 or Medium 2 (lineage induction medium) and plate 2.5 x 105 SP-CPCs on P60 dishes with 3 mL of medium containing different serum concentrations.
- Collect the medium from the dish of step 6.2.4 for the collection of dead cells into a 15 mL tube after 2 d of culturing in Medium 1 or Medium 2.
- Trypsinize adherent cells as in step 6.2.2 and collect the cell suspension into the 15 mL tube of step 6.2.5. Centrifuge the cells at 840 x g for 5 min at RT.
- Aspirate the supernatant and add 1 mL of Medium 1 or Medium 2 for cell counting. Stain SP-CPCs with trypan blue (0.4%) and count trypan blue-positive (dead) and trypan blue-negative (viable) cells.
NOTE: The cell viability (step 6.2.7) is given as the trypan-blue negative cell number in relation to the total cell number.
- Induction of endothelial differentiation
- Precoat a (6-well) culture plate with 10 µg/mL of laminin (LN) or fibronectin (FN).
- Make a substrate solution containing 10 µg/mL of LN or FN with F12 medium (or PBS). Add 2 mL of substrate solution to each well. Maintain the plate for 30 min at 37 °C.
- Aspirate the solution from the plate and add 2 mL of PBS to each well until using it.
- Aspirate the medium from the cells from step 6.1.2, rinse them gently with 5 mL of warm (37 °C) HBSS, treat them with 5 mL of Trypsin-EDTA for 5 min in the cell incubator, add 5 mL of Medium 1 to stop the Trypsin activity, and transfer the cell suspension to a 15 mL tube.
- Centrifuge the cells at 470 x g for 5 min at RT. Aspirate the supernatant and add Medium 2 for cell counting.
- Seed 8 x 104 cells per well on the coated plate with 3 mL of Medium 2 and keep it at 37 °C for 20 – 24 h. Change the medium to 3 mL of Medium 3.
NOTE: We recommend using medium containing a low serum concentration—and without supplements—for the first 20 – 24 h. We recommended using <60% cell confluence (i.e., the cell number of step 6.3.4 has to be adjusted depending on the cell size and growth rate). - Culture the cells for 21 d and change the medium every 3 d.
- Verify the endothelial nature of differentiated cells by staining them with an endothelial marker such as von Willebrand Factor (vWF) and performing fluorescence microscopy.
- Wash the cells with 1 mL of PBS and fix the cells in 3.7% formaldehyde for 2 min at RT.
- Permeabilize the cells with 0.1% Triton X in ddH2O (or PBS) for 30 min and block it with 10% goat serum for 1 h at RT.
- Incubate the cells with anti-von Willebrand factor antibody (1:100) for 48 h at 4 °C
- Wash the cells 3x with 1 mL of PBS, for 10 min each time. Incubate them with Alexa Fluor 546 goat anti-rabbit secondary antibody (1:500) for 1 h at RT in the dark.
- Wash again 3x, for 10 min each, with 1 mL of PBS. Stain the cell nuclei with 4’6-diamidino-2-phenylindole, dihydrochloride (DAPI; 1:500) for 5 min at RT in the dark.
- Wash the cells 3x, for 5 min each, with 1 mL of PBS. Mount the cells and store them at 4 °C
NOTE: The culturing conditions (substrate, medium, supplemental reagents, and FBS concentration) of steps 6.1 and 6.3 are described in Figure 3.
- Precoat a (6-well) culture plate with 10 µg/mL of laminin (LN) or fibronectin (FN).
- Tube formation assay
- Prepare a basement membrane matrix (e.g., Matrigel, henceforth referred to as matrix) plate.
- Thaw the matrix at 4 °C overnight.
- Coat a 96-well plate with 100 µL of the matrix on ice.
NOTE: It is important to avoid any bubbles in the matrix. The plates have to be coated on ice to avoid the jellification of the matrix. - Maintain the plate at 37 °C for 30 min.
- Aspirate the medium from the cells (after the completion of step 6.3.5), gently rinse the cells with 5 mL of warm (37 °C) HBSS, and treat them with Trypsin-EDTA for 5 min in the cell incubator. Add Medium 3 to stop the Trypsin activity and transfer the cell suspension to a 15 mL tube.
- Centrifuge the cells at 470 x g for 5 min at RT. Aspirate the supernatant, add 1 mL of Medium 3, and pipet gently.
- Filter the cells with a 35 µm cell strainer, if the cells are aggregated.
- Count the cells and seed 2 x 103 to 4 x 103 cells in 100 µL of Medium 3 in each matrix-coated well. Keep the plate at 37 °C in the cell incubator for 16 h.
- Take a picture with a bright-field microscope at 2X magnification.
NOTE: In the case of an incomplete trypsinization, prolong the exposure time to Trypsin-EDTA to a maximum of 7 – 8 min.
- Prepare a basement membrane matrix (e.g., Matrigel, henceforth referred to as matrix) plate.
Representative Results
Mouse SP-CPC Isolation:
In this study, we used mouse CPCs isolated according to the SP phenotype, whereas results from rat CPCs are modified and added from a previous report with permission (Figure 8)33.
Cell Proliferation Under High and Low Cell Densities and with Different Serum Concentrations:
Our previous study showed that the mRNA expression of cardiac lineage markers changed within the first 24 h culturing step. It is known that the extracellular matrix affects cell fate decisions, including endothelial differentiation35. To explore suitable conditions for the facilitation of endothelial lineage commitment of CPCs, we used FN and LN (both 10 µg/mL) on a 6-well plate (growth area: 9.6 cm2/well) in this study. Because cell cycle and cell fate decisions are closely linked, we are seeking the condition that exhibits a low cell proliferation rate in the absence of cell death, as such a condition may reflect the transition from cell proliferation to differentiation. We, therefore, applied the following conditions and compared cell proliferation rates: for cell density, high (80% – 90%) confluency and low (<60%) confluency, and for serum concentration, normal culture conditions (in this study 3.5% FBS) and low serum conditions (≤0.1% FBS). First, we tested the low cell density condition. There were no significant differences of cell viability and proliferation in the low cell density with 3.5% FBS between LN and FN, whereas a low cell density with 0.1% FBS showed a decreased cell proliferation on LN compared to FN but no increase in cell death (Figure 4). In contrast, under high cell density conditions, serum concentrations of both 3.5% and 0.1% showed no differences in cell proliferation and in cell death between the two substrates (Figure 5).
These results indicate that a low cell density on LN with 0.1% serum decreases proliferation without affecting the CPC viability.
Changes in Cell Shape in the Endothelial Differentiation Medium:
Although requiring further studies to understand the significance and underlying mechanisms, changes in the cell shape appear to be an indicator of suitable culture conditions as specific changes can be observed early on in cultures, in which endothelial differentiation is going to be successful (Figure 6). As shown in the white dashed circles in Figure 6, within 7 – 14 d in the endothelial differentiation medium, successful cultures contained cells that were larger and different in morphology to the other cells. Interestingly, these cells disappeared towards the end of the differentiation phase and also appeared in lower numbers and at later time points in high-density cultures on LN and FN. Whereas we did not further characterize these cells, this protocol suggests tracking the cell shape every 2 – 3 d until around day 14. If no such cells appear, the number of cells seeded should be decreased.
Evaluation of the Endothelial Ability with a Tube Formation Assay:
The tube formation assay is a useful technique to evaluate the efficiency of endothelial differentiation of CPCs by measuring the tube formation capacity of differentiated cells. We, therefore, performed the tube formation assay with cells differentiated according to the described conditions. Interestingly, successful tube formation was consistently shown by cells plated at low density and differentiated on LN, whereas tube formation mostly failed in cells plated on LN at a high density (Figure 7A). Similarly, tube formation was mostly unsuccessful in cells cultured on FN irrespective of cell density, although cells differentiated on FN at a low density sometimes formed rudimentary tubes, depending on the cell condition (e.g., isolate and/or passage number, Figure 7B). To confirm the endothelial nature of the cells, cells plated on coverslips were differentiated according to the described protocol and stained for vWF. Again, the vWF expression was more homogeneous and more pronounced in CPCs differentiated on LN compared to FN (Figure 7C,D). Figure 8 shows the results from the tube formation assay as performed for previous studies using rat CPCs under the same protocol as here described33. These results show that tube formation is more efficient in cells differentiated on LN as compared to FN when plated under low cell density conditions and with low serum (0.1% FBS) for 20 – 24 h before differentiation in endothelial differentiation medium. These results suggest that this protocol could be useful for various cell types and independent of species.
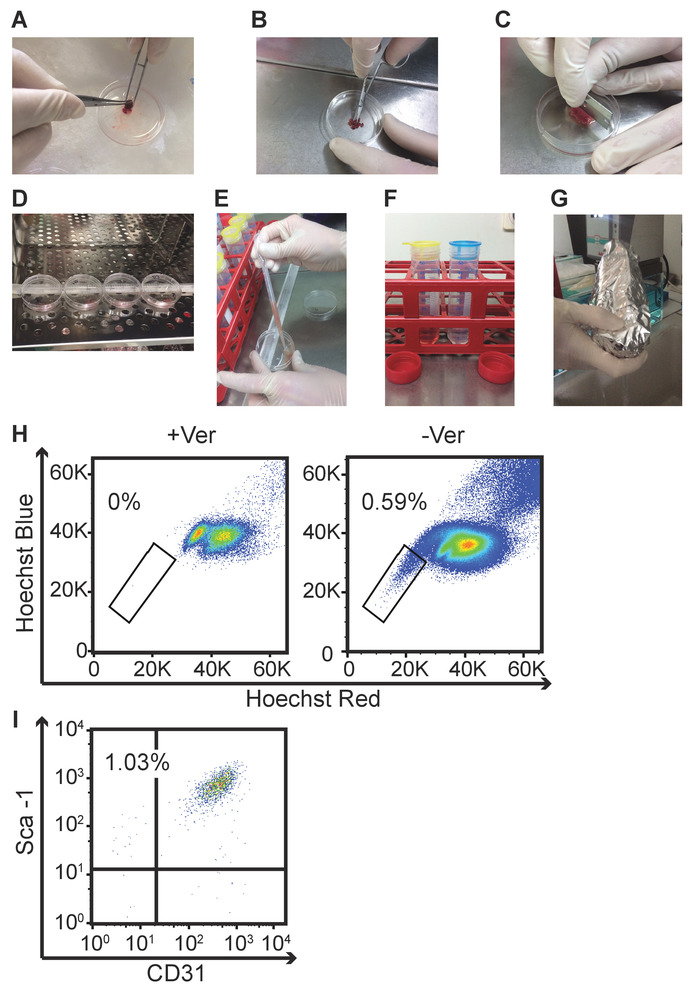
Figure 1: Illustration of specific isolation steps to obtain a cardiomyocyte-depleted cell suspension from isolated mouse hearts and representative flow cytometry readouts of the cardiac SP. (A) This panel shows the ejection of residual blood from the heart cavities through repeated slight pressure applied using small forceps. (B) This panel shows the cutting of the hearts into small pieces using small scissors. (C) This panel shows the mincing of the heart pieces using a razor blade. (D) This panel shows the incubation of the minced hearts with collagenase B in tilted plates at 37 °C. (E) This panel shows the homogenization of the minced hearts using a Pasteur pipette during the incubation step. (F) This panel shows the filtering of the digested tissue through a 100 µm filter (yellow) and a 40 µm filter (blue) for the removal of undigested tissue residues and cardiomyocytes. (G) This panel shows the gentle reversal of a 50 mL conical tube containing the cardiomyocyte-depleted cell suspension and wrapping in tin foil for light protection after the addition of Hoechst. (H) This panel shows representative flow cytometry readouts of Hoechst-stained cells in the presence and absence of the ABC transporter inhibitor verapamil for the identification of the SP. (I) This panel shows a representative dot plot of SP cells according to CD31 and Sca-1 positivity for the identification of the Sca-1+/CD31– subfraction. Please click here to view a larger version of this figure.
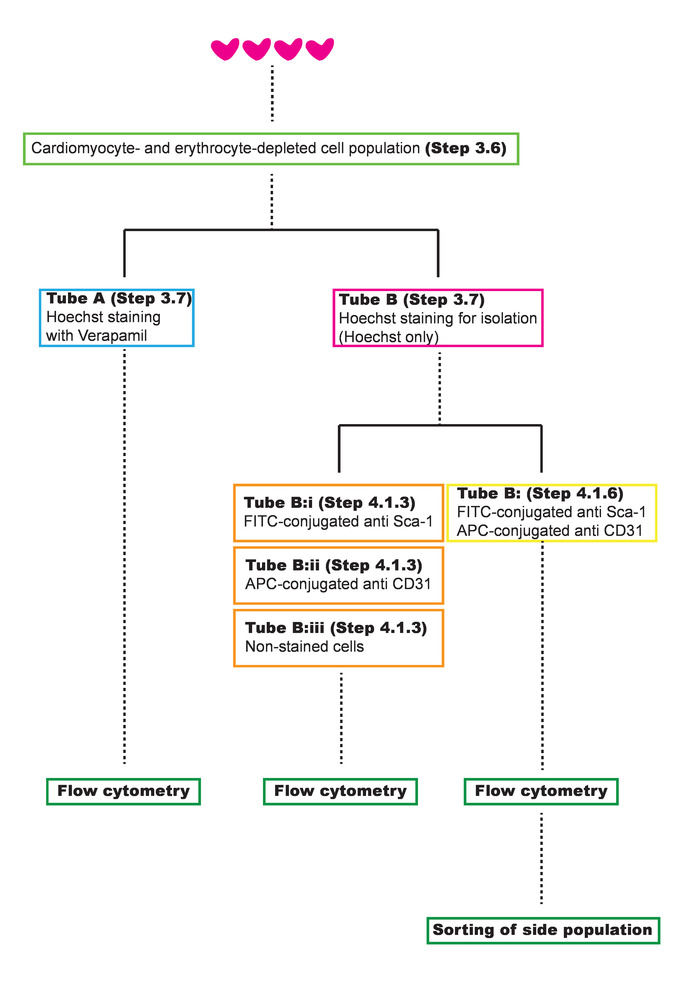
Figure 2: Schematic overview of the sample preparation leading up to the cardiac SP sorting. Please click here to view a larger version of this figure.
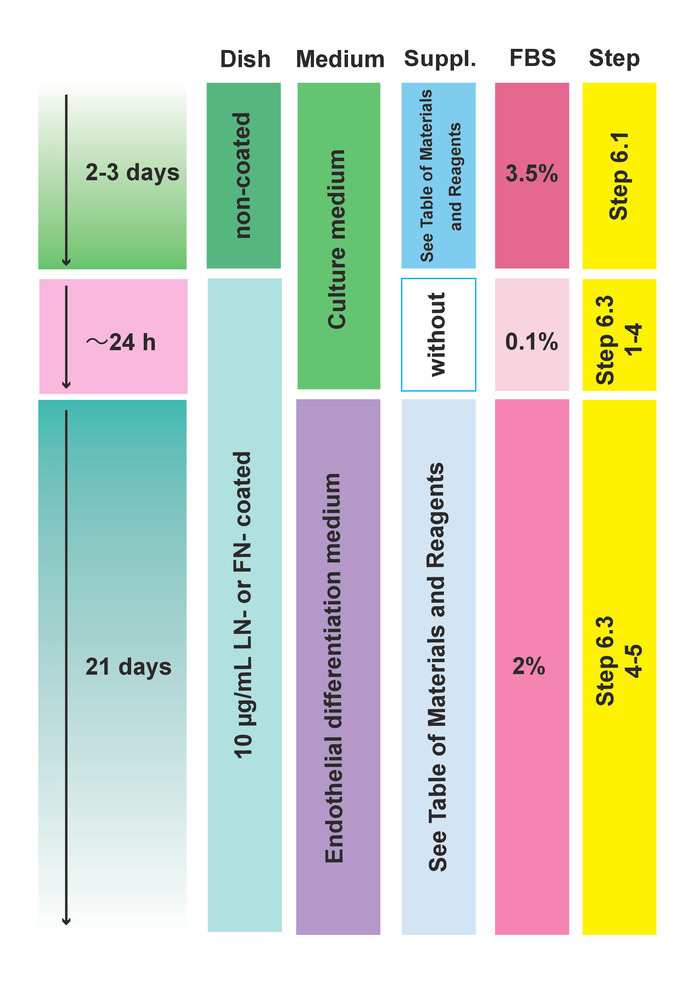
Figure 3: Protocol for endothelial lineage induction and differentiation. Please click here to view a larger version of this figure.

Figure 4: Mouse SP-CPC proliferation when plated at a low cell density under different serum concentrations on LN and FN. (A and B) These panels show the cell numbers at day 2. (C and D) These panels show the viability at day 2. The data are shown as the mean ± the standard error of the mean (SEM); N = 5; different passages; * p < 0.05 by Student's t-test. Please click here to view a larger version of this figure.
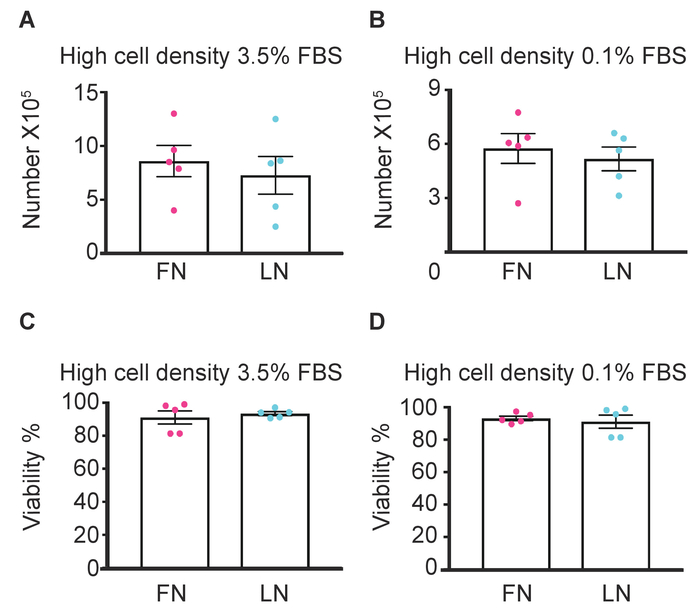
Figure 5: Mouse SP-CPC proliferation when plated at a high cell density under different serum concentrations on LN and FN. (A and B) These panels show the cell numbers at day 2. (C and D) These panels show the viability at day 2. The data are shown as the mean ± the SEM; N = 5; different passages. Please click here to view a larger version of this figure.
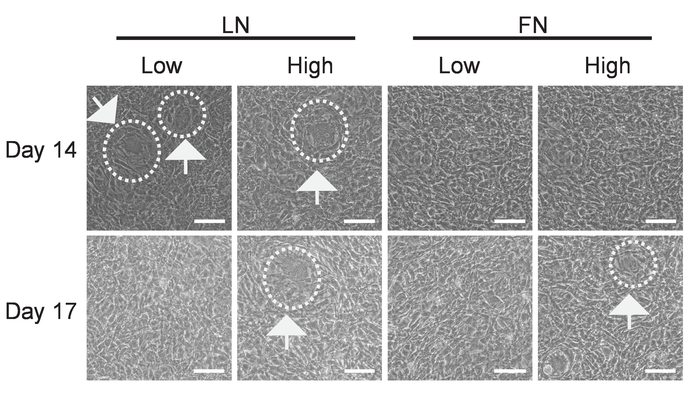
Figure 6: Cell morphology at 14 and 17 d during the differentiation process. This figure shows bright-field (BF) images of mouse SP-CPCs seeded on LN- or FN-coated dishes with a low (Low) or a high (High) cell density and with Medium 2 for 20 h, followed by Medium 3 for 14 or 17 d. White dashed circles mark the areas containing round-shaped cells with a larger cell size. The imaging was performed with bright-field microscopy. The magnification = 2X; the scale bar = 100 µm. Please click here to view a larger version of this figure.
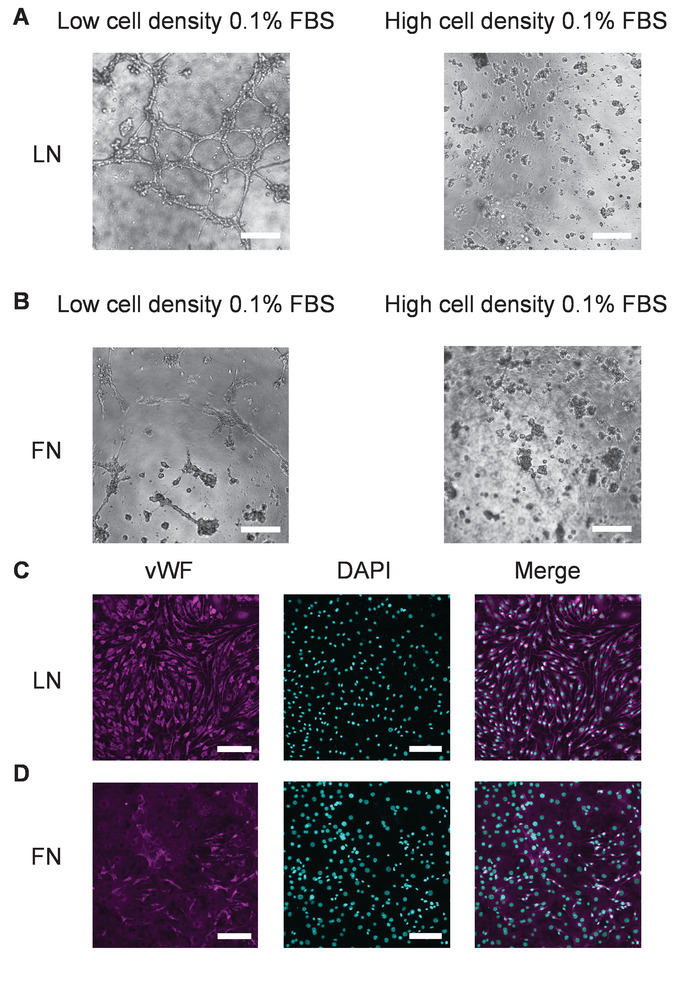
Figure 7: Tube formation and vWF staining after endothelial differentiation of SP-CPCs on LN and FN. (A and B) These panels show the tube formation. (C and D) These panels show the vWF staining. Cells were seeded on 10 µg/mL of LN- or FN-coated dishes with a low or high cell density and with Medium 2 for 20 h, followed by Medium 3 for 21 d, and then harvested with Trypsin and seeded on the basement membrane matrix. All pictures were taken after 16 h. The imaging was performed with bright-field microscopy and the magnification = 2X for panels A and B. The imaging is performed with fluorescent microscopy and the magnification = 10X for panels C and D. Panels A and C show LN-coated dishes. Panels B and D show FN-coated dishes. The scale bar = 100 µm. Please click here to view a larger version of this figure.
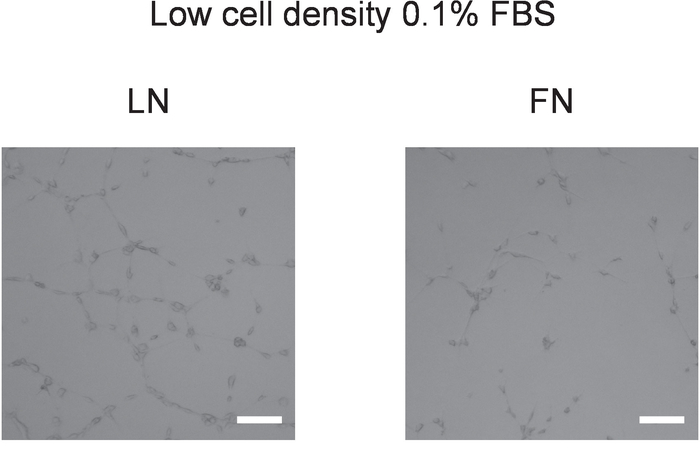
Figure 8: Tube formation after the endothelial differentiation of rat CPCs. Rat CPCs were seeded at a low cell density on 10 µg/mL of LN- or FN-coated dishes with 0.1% FBS-containing F12 medium for 20 h, followed by another 21 d in Medium 3, and then harvested with Trypsin. On the basement membrane matrix on a 96-well plate, 4 x 104 cells were seeded in 100 μL of Medium 3. The imaging was performed with bright-field microscopy. The magnification = 2X; the scale bar = 50 µm. This figure is modified based on a previous study33. Please click here to view a larger version of this figure.
| Species | Isolation, detection marker | Reference |
| Human, rat, mouse | c- kit+/Lin–, c-kit+/CD45–, c-kit+/CD45–/CD31– | Beltrami, Cell 2003; Bolli, Lancet 2011; Ellison, Cell 2013; Smith, Nat Protoc 2014; Vicinanza, Cell Death Differ 2017 |
| Mongrel dogs | Lin–, plus: c-kit+, MDR1+ or Sca-1+ | Linke, PNAS 2005 |
| Mouse | Sca-1+ | Oh, PNAS 2003 |
| Mouse | Side population, plus: Abcg2+, Sca-1+/CD31–, Sca-1+/PDGFRa+ | Hierlihy, FEBS Lett 2002; Martin, Dev Biol 2004; Pfister, Circ Res 2005; Noseda, Nat Commun 2015 |
| Mouse | Colony forming unit-fibroblast, plus: CD45–, CD31–, Sca-1+, PDGFRa+ | Pelekanos, Stem Cell Res 2012 |
| Mouse, rat, human | Isl-1+ *, plus: CD31–, c-kit–, Sca-1–, Gata4+, Nkx2.5+ | Laugwitz, Nature 2005; Bu, Nature 2009 |
| * Isl-1 is found in the embryonic or fetal myocardium, not in the adult heart. | ||
Table 1: Isolation techniques and markers of cardiac progenitor cells.
Discussion
Advantages of this Protocol:
This protocol provides an endothelial differentiation technique of CPCs. We found that a low serum concentration and low cell density could improve the efficiency of endothelial differentiation, whereby LN proved to be a more suitable substrate than FN under these conditions. We used two distinct types of CPCs: rat CPCs, which were used in a cell line-like manner, and mouse SP-CPCs, which were isolated and expanded. Notably, the protocol was applicable to both types of CPCs. Current techniques allow scientists to isolate and expand primary CPCs from genetically modified mice and from preclinical disease models, as well as from humans28,36. Using this protocol on isolated CPCs could be helpful to the investigation of not only disease mechanisms but also to the exploration of novel therapeutic targets.
CPCs are currently in clinical testing for cell therapy4,5, whereby cells are isolated from patients and transplanted back after in vitro amplification. In this regard, the protocol presented here could help identify strategies to enhance the differentiation potential of such CPCs before transplantation. However, cell therapy still suffers from limited efficacy due to low retention, survival, and engraftment of transplanted cells. Mechanistic in vitro studies based on this protocol or on adaptations thereof have the potential to contribute to a better understanding of the molecular mechanisms driving the differentiation process-in particular, mechanisms related to cell density, substrate, and growth factors. New knowledge retrieved from such studies may ultimately be used for cell reprogramming and applied to directly influence resident CPCs to differentiate after injury.
Limitations of this Protocol:
Isolated primary CPCs are heterogeneous populations that show different phenotypes with respect to cell size and cell growth rate depending on the isolation, even when using the same markers and identical isolation technique. Therefore, the following three points need to be defined: (1) the cell seeding numbers according to cell size, (2) the ideal serum concentration (<0.5%), and (3) the incubation time for the first step (induction of lineage) with a very low serum concentration. Here, we show differences in differentiation efficiency depending on cell density. However, although we compared low serum (0.1% FBS) versus culture serum concentrations (3.5% FBS), we did not examine other concentrations within the <0.5% FBS range. In addition, depending on the isolation markers used and the species, the efficiency of the induction of lineage commitment may vary. Therefore, the protocol should be optimized for each cell type.
Pharmacological and genetic inhibition/induction techniques could be used for examining disease mechanisms with this protocol, instead of genetically modified or disease animal models. In this case, the protocol should be redesigned: before the treatment with low-serum-containing medium, the cells will be treated with specific reagents or genetic tools. As a consequence, the cells may be more vulnerable than nontreated cells. Using low serum in the first step is the key point in this protocol. Therefore, a careful validation of the serum concentration and incubation time for the first step to allow for slowing the cell proliferation while maintaining viability under serum deprivation is critical to whether this protocol can be a success or not.
Summary:
In summary, isolated CPCs from animal models provide a valuable tool for cardiac disease and regeneration studies. Upon expansion, human CPCs may also be directly used for cell therapy. We, here, provide a protocol for the efficient endothelial lineage induction and differentiation of CPCs based on careful adaptations of cell density and serum concentrations. The advantage of this protocol is its applicability to different types of CPCs and its potential to contribute a basis for the study of and/or the establishment of other novel cardiac regeneration tools.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors thank Vera Lorenz for her helpful support during the experiments and the staff from the Flow Cytometry Facility from the Department of Biomedicine (DBM), University and University Hospital Basel. This work was supported by the Stay-on track program from the University of Basel (to Michika Mochizuki). Gabriela M. Kuster is supported by a grant from the Swiss National Science Foundation (grant number 310030_156953).
Materials
| Culture medium | |||
| Iscove's Modified Dulbecco's Medium (IMDM) | ThermoFisher | #12440 | |
| Dulbecco's Modified Eagle's Medium (DMEM)/Nutrient Mixture F12 Ham | Merck | #D8437 | |
| Penicillin-Streptomycin (P/S) | ThermoFisher | #15140122 | |
| Fetal Bovine Serum (FBS) | Hyclone | #SH30071 | 3.5% (0.1% for lineage induction) |
| L-Glutamine | ThermoFisher | #25030 | Final concentration 2 mM |
| Glutathione | Merck | #G6013 | |
| Recombinant Human Epidermal Growth Factor (EGF) | Peprotech | #AF-100-15 | |
| Recombinant Basic Fibroblast Growth Factor (FGF) | Peprotech | #AF-100-18B | |
| B27 Supplement | ThermoFisher | #17504044 | |
| Cardiotrophin 1 | Peprotech | #250-25 | |
| Thrombin | Diagontech AG, Switzerland | #100-125 | |
| Hanks' Balanced Salt Solution (HBSS) CaCl2(-), MgCl2(-) | ThermoFisher | #14170 | |
| 0.05 % Trypsin-EDTA | ThermoFisher | #25300 | |
| T75 Flask | Sarstedt | #83.3911 | |
| Endothelial differentiation | |||
| Endothelial Cell Growth Medium (EGM)-2 BulletKit | Lonza | #CC-3162 | |
| Ham's F-12K (Kaighn's) Medium | ThermoFisher | #21127 | |
| Laminin | Merck | #L2020 | |
| Fibronectin | Merck | #F4759 | Dilute in ddH2O |
| 6 Well Plate | Falcon | #353046 | |
| Formaldehyde Solution | Merck | #F1635 | Diluite 1:10 in PBS (3.7% final concentration) |
| Triton X-100 | Merck | #93420 | 0.1% in ddH2O |
| Normal Goat Serum (10%) | ThermoFisher | #50062Z | |
| Anti-von Willebrand Factor antibody | Abcam | #ab6994 | 1:100 in 10% goat serum |
| Goat anti-Rabbit IgG, Alexa Fluor 546 | ThermoFisher | #A11010 | 1:500 in 10% goat serum |
| 4',6-diamidino-2-phenylindole, dihydrochloride (DAPI) | ThermoFisher | #62247 | 1:500 in ddH2O |
| SlowFade Antifade Kit | ThermoFisher | #S2828 | |
| BX63 widefield microscope | Olympus | ||
| Tube formation | |||
| 96 Well Plate | Falcon | #353072 | |
| 5 ml Round Bottom Tube with Strainer Cap | Falcon | #352235 | |
| Matrigel Growth Factor Reduced | Corning | #354230 | |
| IX50 widefield microscope | Olympus | ||
| Sca-1+/CD31- cardiac side population isolation34 | |||
| Reagents | |||
| Pentobarbital Natrium 50 mg/ml ad usum vet. | in house hospital pharmacy | #9077862 | Working solution: 200 mg/kg |
| Phosphate Buffered Saline (PBS) CaCl2(-), MgCl2(-) | ThermoFisher | #20012 | |
| Hanks' Balanced Salt Solution (HBSS) CaCl2(-), MgCl2(-), phenol red (-) | ThermoFisher | #14175 | Prepare HBSS 500 mL + 2% FBS for quenching Collagenase B activity |
| Dulbecco's Modified Eagle's Medium (DMEM) 1g/L of D-Glucose, L-Glutamine, Pyruvate | ThermoFisher | #331885 | Prepare DMEM + 10% FBS + 25 mM HEPES+ P/S for Hoechst stanining |
| Penicillin-Streptomycin (P/S) | ThermoFisher | #15140122 | |
| HEPES 1 M | ThermoFisher | #15630080 | Final concentration 25 mM |
| Fetal Bovine Serum (FBS) | Hyclone | #SH30071 | |
| RBC LysisBuffer (10X) | BioLegend | #420301/100mL | Dilute to 1X in ddH2O and filter through a 0.2 µm filter |
| Collagenase B | Merck | #11088807001 | Final concentration 1 mg/mL in HBSS, filtered through a 0.2 µm filter |
| bisBenzimide H33342 Trihydrochloride (Hoechst) | Merck | #B2261 | Prepare 1 mg/mL in ddH2O |
| Verapamil-hydrochloride | Merck | #V4629 | Final concentration 83.3 µM |
| APC Rat Anti-Mouse CD31 | BD Biosciences | #551262 | 0.25 µg/107cells |
| FITC Rat Anti-Mouse Ly-6A/E (Sca-1) | BD Biosciences | #557405 | 0.6 µg/107cells |
| 7-Aminoactinomycin D (7-ADD) | ThermoFisher | #A1310 | 0.15 µg/106cells |
| APC rat IgG2a k Isotype Control | BD Biosciences | #553932 | 0.25 µg/107cells |
| FITC Rat IgG2a k Isotype Control | BD Biosciences | #554688 | 0.6 µg/107cells |
| Material | |||
| Needles 27G | Terumo | #NN-2719R | |
| Needles 18G | Terumo | #NN-1838S | |
| Single Use Syringes 1 mL sterile | CODAN | #62.1640 | |
| Transferpipette 3.5 mL | Sarstedt | #86.1171.001 | |
| Cell Strainer 40 µm blue | BD Biosciences | #352340 | |
| Cell Strainer 100 µm yellow | BD Biosciences | #352360 | |
| Lumox Dish 50 | Sarstedt | #94.6077.305 | |
| Culture Dishes P100 | Corning | #353003 | |
| Culture Dishes P60 | Corning | #353004 | |
| Mouse | |||
| Line | Age | Breeding | |
| C57BL/6NRj / male | 12 weeks | in house | |
| Product Name | Company | Catalogue No. | |
| Reagents | |||
| Iscove's Modified Dulbecco's Medium (IMDM) | ThermoFisher | #12440 | |
| Dulbecco's Modified Eagle's Medium (DMEM)/Nutrient Mixture F12 Ham | Merck | #D8437 | |
| Penicillin-Streptomycin (P/S) | ThermoFisher | #15140122 | |
| Fetal Bovine Serum (FBS) | Hyclone | #SH30071 | |
| L-Glutamine | ThermoFisher | #25030 | |
| Glutathione | Merck | #G6013 | |
| B27 Supplement | ThermoFisher | #17504044 | |
| Recombinant Human Epidermal Growth Factor (EGF) | Peprotech | #AF-100-15 | |
| Recombinant Basic Fibroblast Growth Factor (FGF) | Peprotech | #AF-100-18B | |
| Cardiotrophin 1 | Peprotech | #250-25 | |
| Thrombin | Diagontech AG, Switzerland | #100-125 | |
| Endothelial Cell Growth Medium (EGM)-2 BulletKit | Lonza | #CC-3162 | |
| Overview of medium compositions. Some of this infomation is identical with the one provided above, but sorted according to the composition of Media 1-3. | |||
| Product Name | Medium 18 | Medium 2 | Medium 3 |
| Reagents | Culture | Lineage induction | Endothelial diff. |
| Iscove's Modified Dulbecco's Medium (IMDM) | 35% | 35% | |
| Dulbecco's Modified Eagle's Medium (DMEM)/Nutrient Mixture F12 Ham | 65% | 65% | |
| Penicillin-Streptomycin (P/S) | 1% | 1% | |
| Fetal Bovine Serum (FBS) | 3.5% | ≤0.1% | |
| L-Glutamine | 2 mM | 2 mM | |
| Glutathione | 0.2 nM | 0.2 nM | |
| B27 Supplement | 1.3% | ||
| Recombinant Human Epidermal Growth Factor (EGF) | 6.5 ng/mL | ||
| Recombinant Basic Fibroblast Growth Factor (FGF) | 13 ng/mL | ||
| Cardiotrophin 1 | 0.65 ng/mL | ||
Riferimenti
- Beltrami, A. P., et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 114 (6), 763-776 (2003).
- Hierlihy, A. M., Seale, P., Lobe, C. G., Rudnicki, M. A., Megeney, L. A. The post-natal heart contains a myocardial stem cell population. FEBS Letters. 530 (1-3), 239-243 (2002).
- Sandstedt, J., et al. Left atrium of the human adult heart contains a population of side population cells. Basic Research in Cardiology. 107 (2), 255 (2012).
- Bolli, R., et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 378 (9806), 1847-1857 (2011).
- Makkar, R. R., et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 379 (9819), 895-904 (2012).
- Wu, S. M., Chien, K. R., Mummery, C. Origins and fates of cardiovascular progenitor cells. Cell. 132 (4), 537-543 (2008).
- Smits, A. M., et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nature Protocols. 4 (2), 232-243 (2009).
- Noseda, M., et al. PDGFRalpha demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nature Communications. 6, 6930 (2015).
- Linke, A., et al. Stem cells in the dog heart are self-renewing, clonogenic, and multipotent and regenerate infarcted myocardium, improving cardiac function. Proceeding of the National Acadademy of Sciences of the United States of America. 102 (25), 8966-8971 (2005).
- El-Mounayri, O., et al. Serum-free differentiation of functional human coronary-like vascular smooth muscle cells from embryonic stem cells. Cardiovascular Research. 98 (1), 125-135 (2013).
- Ellison, G. M., et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 154 (4), 827-842 (2013).
- Oh, H., et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proceeding of the National Acadademy of Sciences of the United States of America. 100 (21), 12313-12318 (2003).
- Martin, C. M., et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Biologia dello sviluppo. 265 (1), 262-275 (2004).
- Pfister, O., et al. CD31- but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circulation Research. 97 (1), 52-61 (2005).
- Pelekanos, R. A., et al. Comprehensive transcriptome and immunophenotype analysis of renal and cardiac MSC-like populations supports strong congruence with bone marrow MSC despite maintenance of distinct identities. Stem Cell Research. 8 (1), 58-73 (2012).
- Laugwitz, K. L., et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 433 (7026), 647-653 (2005).
- Bu, L., et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 460 (7251), 113-117 (2009).
- Smith, A. J., et al. Isolation and characterization of resident endogenous c-Kit+ cardiac stem cells from the adult mouse and rat heart. Nature Protocols. 9 (7), 1662-1681 (2014).
- Matsuura, K., et al. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. Journal of Biological Chemistry. 279 (12), 11384-11391 (2004).
- Liang, S. X., Tan, T. Y., Gaudry, L., Chong, B. Differentiation and migration of Sca1+/CD31- cardiac side population cells in a murine myocardial ischemic model. International Journal of Cardiology. 138 (1), 40-49 (2010).
- Vicinanza, C., et al. Adult cardiac stem cells are multipotent and robustly myogenic: c-kit expression is necessary but not sufficient for their identification. Cell Death and Differentiation. 24 (12), 2101-2116 (2017).
- Galli, S. J., Tsai, M., Wershil, B. K. The c-kit receptor, stem cell factor, and mast cells. What each is teaching us about the others. American Journal of Pathology. 142 (4), 965-974 (1993).
- Konig, A., Corbacioglu, S., Ballmaier, M., Welte, K. Downregulation of c-kit expression in human endothelial cells by inflammatory stimuli. Blood. 90 (1), 148-155 (1997).
- Ogawa, M., et al. Expression and function of c-kit in hemopoietic progenitor cells. Journal of Experimental Medicine. 174 (1), 63-71 (1991).
- Bradfute, S. B., Graubert, T. A., Goodell, M. A. Roles of Sca-1 in hematopoietic stem/progenitor cell function. Experimental Hematology. 33 (7), 836-843 (2005).
- Goodell, M. A., Brose, K., Paradis, G., Conner, A. S., Mulligan, R. C. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. Journal of Experimental Medicine. 183 (4), 1797-1806 (1996).
- Pfister, O., et al. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circulation Research. 103 (8), 825-835 (2008).
- Messina, E., et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circulation Research. 95 (9), 911-921 (2004).
- Davis, D. R., et al. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS One. 4 (9), e7195 (2009).
- Carr, C. A., et al. Cardiosphere-derived cells improve function in the infarcted rat heart for at least 16 weeks–an MRI study. PLoS One. 6 (10), e25669 (2011).
- Martens, A., et al. Rhesus monkey cardiosphere-derived cells for myocardial restoration. Cytotherapy. 13 (7), 864-872 (2011).
- Cheng, K., et al. Relative roles of CD90 and c-kit to the regenerative efficacy of cardiosphere-derived cells in humans and in a mouse model of myocardial infarction. Journal of the American Heart Association. 3 (5), e001260 (2014).
- Mochizuki, M., et al. Polo-Like Kinase 2 is Dynamically Regulated to Coordinate Proliferation and Early Lineage Specification Downstream of Yes-Associated Protein 1 in Cardiac Progenitor Cells. Journal of the American Heart Association. 6 (10), (2017).
- Pfister, O., Oikonomopoulos, A., Sereti, K. I., Liao, R. Isolation of resident cardiac progenitor cells by Hoechst 33342 staining. Methods in Molecular Biology. 660, 53-63 (2010).
- Discher, D. E., Mooney, D. J., Zandstra, P. W. Growth factors, matrices, and forces combine and control stem cells. Science. 324 (5935), 1673-1677 (2009).
- Plaisance, I., et al. Cardiomyocyte Lineage Specification in Adult Human Cardiac Precursor Cells Via Modulation of Enhancer-Associated Long Noncoding RNA Expression. JACC: Basic to Translational Science. 1 (6), 472-493 (2016).

