Microfluidic Assay for the Assessment of Leukocyte Adhesion to Human Induced Pluripotent Stem Cell-derived Endothelial Cells (hiPSC-ECs)
Summary
This step-by-step protocol provides a detailed description of the experimental setup and data analysis for the assessment of inflammatory responses in hiPSC-ECs and the analysis of leukocyte adhesion under physiological flow.
Abstract
Endothelial cells (ECs) are essential for the regulation of inflammatory responses by either limiting or facilitating leukocyte recruitment into affected tissues via a well-characterized cascade of pro-adhesive receptors which are upregulated on the leukocyte cell surface upon the inflammatory trigger. Inflammatory responses differ between individuals in the population and the genetic background can contribute to these differences. Human induced pluripotent stem cells (hiPSCs) have been shown to be a reliable source of ECs (hiPSC-ECs), thus representing an unlimited source of cells that capture the genetic identity and any genetic variants or mutations of the donor. hiPSC-ECs can therefore be used for modeling inflammatory responses in donor-specific cells. Inflammatory responses can be modeled by determining leukocyte adhesion to the hiPSC-ECs under physiological flow. This step-by-step protocol provides a detailed description of the experimental setup and data analysis for the assessment of inflammatory responses in hiPSC-ECs and the analysis of leukocyte adhesion under physiological flow.
Introduction
Inflammation plays a pivotal role in many pathological conditions, including cardiovascular and neurodegenerative disorders, sepsis and adverse drug responses (ADRs). Endothelial cells (ECs) play an essential role in regulating inflammatory responses via the induction of pro-adhesive receptors, such as E-selectin, intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) on their surface1,2. Microvascular ECs in different tissues are known to exhibit heterogeneity in inflammatory responses3,4. Furthermore, genetic background or certain genetic conditions might result in the differences in inflammatory responses between individuals; it is therefore important to have access to ECs from different individuals. More recently, human induced pluripotent stem cells (hiPSCs)5, that can be derived from virtually any individual, were shown to serve as a reliable and renewable source of ECs6,7,8,9. Therefore, the assessment of inflammatory responses and leukocyte recruitment in hiPSC-ECs is valuable, not only for modeling of certain genetic disorders, but also to provide indications of inter-individual variability and to use as a tool for personalized medicine in the future.
Flow assays provide a useful tool for studying endothelial-leukocyte interaction. Advances in microfluidic devices enable the reproduction of physiological fluid flow conditions with precise control of vascular bed-specific shear stress levels. Live imaging allows monitoring of the cascade of events of leukocyte capture, rolling, crawling, adhesion and transmigration. Several flow assays to study endothelial-leukocyte interaction have been developed, however, they all utilize primary ECs10,11,12,13. Here, we will describe in detail the assay for the assessment of human leukocyte adhesion to hiPSC-ECs under physiological flow. In this procedure, we describe optimized conditions of hiPSC-EC stimulation with pro-inflammatory stimuli, such as tumor necrosis factor alpha (TNFα), dissociation, seeding into a microfluidic chip with eight parallel channels. We describe a step-by-step protocol for the perfusion of hiPSC-ECs with the suspension of fluorescently labeled leukocytes in microfluidic chips, live cell imaging and automated counting of adherent leukocytes.
This protocol is useful for the assessment of inflammatory responses in hiPSC-ECs in drug screening, disease modeling and personalized regenerative medicine.
Protocol
1. Preparation of Solutions and Reagents
- Prepare complete Human Endothelial-SFM medium (EC-SFM FULL) by adding platelet-poor plasma-derived serum (1%), basic fibroblast growth factor (bFGF, 20 ng/mL) and vascular endothelial growth factor (VEGF, 50 ng/mL) to Human Endothelial-SFM (see Table of Materials). Filter the medium through a 0.22 µm pore filter and store it at 4 °C for up to 2 weeks.
- Prepare complete RPMI medium by adding fetal bovine serum (10%), 2-mercaptoethanol (0.05 nM), L-glutamine (1%), Penicillin-Streptomycin (25 U/mL) to RPMI medium (see Table of Materials). Store at 4 °C for up to 3 weeks.
2. Coating of Microfluidic Chips
- Place a sterile biochip into a 10 cm sterile Petri dish.
- Prepare fibronectin working solution by reconstituting the stock solution in phosphate-buffered saline (PBS) without Ca2+/Mg2+ to a final concentration of 50 µg/mL. Estimate 80 µL of working solution per biochip.
- Inject 10 µL of fibronectin working solution into each channel of the biochip. Use a pipette with small 10 µL tips and make sure to form a tight contact between the channel inlet and the pipette tip (Figure 1C, left).
- Close the lid of the Petri dish containing the biochip and place it in a humidified chamber (Figure 2B). Store the chamber at 4 °C overnight. To humidify the chamber, place a clean tissue paper at the bottom of the humidified chamber and add 5 mL of sterile H2O.
- Next day, before the assay, pre-warm the biochip at 37 °C in the incubator.
3. Preparation of hiPSC-ECs
NOTE: In the protocol described here, hiPSC-ECs were differentiated, purified, cryopreserved and thawed as previously described8.
- Thaw and plate hiPSC-ECs in complete EC-SFM FULL into one 0.1 % gelatin-coated T75 flask, three to four days prior the assay.
- The night prior to the assay, stimulate hiPSC-ECs with TNFα by adding EC-SFM FULL supplemented with 10 ng/mL TNFα and incubate overnight (~12 h) at 37 °C, as previously described 7.
4. hiPSC-ECs Dissociation and Seeding into Microfluidic Chip
- On the day of the assay, take the flask of hiPSC-ECs from the incubator and examine the cells under the microscope. Ensure that hiPSC-ECs are 80–100% confluent and have a typical EC-like morphology.
- Transfer the flask into the cell culture hood.
- Aspirate the medium from the flask of hiPSC-ECs.
- Briefly wash the cell culture by adding 10 mL of PBS without Ca2+/Mg2+. Aspirate the PBS.
- Add 4 mL per flask of 1x dissociation enzyme. Incubate for 5 min at room temperature (RT) (Figure 1B).
- Stop the dissociation reaction by adding 8 mL of Human Endothelial-SFM medium per flask. Gently detach and collect the hiPSC-ECs suspension in a 15 mL conical tube using a 5 mL pipette.
- Count the total number of the cells in the suspension.
- Centrifuge the cells at 300 x g for 3 min at RT.
- Aspirate the supernatant and resuspend the cell pellet in EC-SFM FULL to a final concentration of 1.5 x 107 cells/mL.
- Take out the Petri dish with the biochip from the incubator and transfer it to the cell culture hood.
- Aspirate the fibronectin solution from all the channels of the microfluidic chip.
- Inject 6 µL of the cell suspension into each channel using a pipette with a small 10 µL tip (Figure 1C, middle). Make sure that the cell suspension is mixed well before the injection and avoid cell precipitation.
- Examine the biochip under the microscope and ensure that the cells are uniformly distributed, and the cell density is high (Figure 2C).
- Incubate the biochip for 15 min at 37 °C.
- Add 40 µL of EC-SFM FULL into the medium reservoirs from both sides of each channel.
- Incubate the biochip for 1 h at 37 °C (Figure 1C, right).
- Add another 50 µL of EC-SFM FULL into the reservoirs from both sides of each channel.
5. Preparation of Human Leukocytes
NOTE: In the protocol described here, a commercially available monocytic cell line (THP-1) was used. Alternatively, peripheral blood mononuclear cells or neutrophils can be used. Isolation of peripheral blood monocytes and neutrophils can be performed using standard procedure11. Perform all steps described in this paragraph in a cell culture hood.
- Collect the leukocytes in a 50 mL conical tube.
- Wash the leukocytes with 10 mL of PBS without Ca2+/Mg2+. Centrifuge the cells at 300 x g for 3 min at RT (Figure 1D).
- Aspirate the supernatant and resuspend the cell pellet in 1 mL of PBS with DiOC6 dye (λex = 485 nm, λem = 501 nm) (1:5,000). Incubate the cells in the dark for 10 min at RT.
- Wash the leukocytes with 10 mL of PBS without Ca2+/Mg2+. Centrifuge the cells at 300 x g for 3 min at RT. Repeat the washing step one more time.
- Count the total number of the cells in the suspension.
- Aspirate the supernatant and resuspend the cell pellet in complete RPMI medium to a final concentration of 2.5 x 106 cells/mL.
6. Preparation of the Microfluidic Pump
- Assemble the microfluidic pump with the 8-channel manifold.
- Connect the pump to a PC with the installed operating software.
- Start the microfluidic software and load the protocol by clicking Open Protocol and navigate on the PC to select the microfluidic protocol.
- Connect the software and the pump by selecting the Setup folder and clicking Run Assay Step.
- Define the geometry of the channels for the correct flow rate control: select Update Geometry in the Geometry Setup folder and click Run Assay Step.
NOTE: Make sure that the geometry details match the syringe and channels to be used (Geometry ID = 1, Number of channels = 8, Channel width = 800.0 µm, Channel depth = 120.0 µm, Syringe volume = 250 µL) and the viscosity of the sample (Liquid viscosity = 1.0). - Wash the pump by placing the inlet cable (orange) into the 50 mL conical tube containing 80% ethanol. Place the outlet cable (green) in the 50 mL conical empty tube (waste container).
- Select the Washout/Pump Washout folder and click Run Assay Step (Wash volume = 2000 µL, Wash volume step = 250 µL, Washout direction = Output). Repeat the washing step once more.
- Repeat the pump washing steps (Steps 6.6–6.7) with cell culture grade water, and Human Endothelial-SFM culture medium.
- Continue with the washing step of the manifold.
- Manually open the valve between the syringe and the manifold by placing the 3-way adapter into the correct position (indicator OFF is turned to the left).
- Select Set Current Channel/Open step in the Washout/Manifold Wash folder and click Run Assay Step to open all valves of the manifold.
- Wash the manifold manually by pushing 5 mL of 80% ethanol from the syringe, followed by 5 mL of cell culture grade water.
- Wash the manifold by pushing 5 mL of Human Endothelial-SFM culture medium from the syringe.
- Remove all large air bubbles that are trapped in the manifold. To do this, place the 8-well pin adapter into the 10 cm Petri dish with the medium and perform pushing/pulling with the syringe until all large air bubbles are gone (small bubbles are not a problem).
- Select Set Current Channel/Close step in the Washout/Manifold Wash folder and click Run Assay Step to close all valves of the manifold.
- Connect the green outlet cable from the pump to the manifold adapter.
- Switch the 3-way valve back to the OFF position to close the syringe.
- Select Washout/Cable Washout folder and click Run Assay Step to wash each port with RPMI medium individually, ensuring that all bubbles are removed from each cable (Dispense volume = 100 µL).
NOTE: Each valve of the manifold is opened one-by-one, 1 to 8, and 100 μL of the medium is dispensed through each individual channel while keeping the other channels closed. Make sure that the liquid runs from each pin. If there is no liquid coming out, it is an indication of an air bubble or a clog.
7. Preparation of the Microfluidic Chip for Live Imaging
- Ensure that the microfluidic setups are ready, and the live imaging chamber is pre-equilibrated to 37 °C and the level of CO2 reaches 5% (Figure 3A, B).
- Take the biochip from the incubator and position it in the microscope chip holder. Mount the chip holder on the microscope imaging stage (Figure 3C).
- In order to connect the biochip to the pump via the manifold, place the 8-pin adapter close to the outlet reservoirs of the chip and deepen all pins in the medium (but do not connect completely).
- Select the Washout | Connect to Chip folder and click Run Assay Step to dispense RPMI medium prior to connecting the chip (Dispense volume = 30 mL). Once the medium is dispensed, insert the 8-pin adapter into the outlet of the biochip.
NOTE: During this step, all channels are set to On (open) and 30 µL of the medium is dispensed through each channel and following this the channels are set to Off (closed). This ensures that no air bubbles will be introduced into the channels of the microfluidic chip. - Manually aspirate the medium from all inlet reservoirs of the biochip with a pipette.
- Select the Washout | Chip Washout folder and click Run Assay Step to perfuse 40 μL of EC-SFM FULL medium through each of the channels one-by-one to get rid of the dead cells/debris from the channel (Washout volume = 40 µL, Maximum washout shear = 40 dynes/cm2).
8. Flow Adhesion Assay and Image Acquisition
- Manually aspirate the medium from the inlet reservoir of the channel of interest with a pipette.
- Add 100 µL of leukocytes from Step 5.6 to the inlet reservoir of the channel of interest.
NOTE: Gently mix the cells to make a single-cell suspension. - In the Cell Assay | Step Description folder, select Set Current Channel/Step Description and, in the list of channels, select only the current channel of interest and set to On (open) and set all other seven channels to Off (closed).
- Select the Cell Assay | Step Description folder and click Run Assay Step.
NOTE: Make sure that Variable Flow Rate Dispensing applies constant negative pressure to pull the leukocyte suspension from the inlet reservoir through the channel for 5 min (Shear set = Constant, Shear stress = -0.5 dyne/cm2, Duration = 00:05:00, Scan delay = 00:00:00). The negative value of shear stress ensures the correct direction of the flow. - Aspirate the remaining leukocytes from the inlet reservoir.
- Add 100 µL of complete RPMI medium to the inlet reservoir. Select Cell Assay/Step description folder and click Run Assay Step to perfuse the channel for 5 min with RPMI medium in order to wash out all non-adherent leukocytes (Shear set = Constant, Shear stress = -0.5 dyne/cm2, Duration = 00:05:00, Scan delay = 00:00:00).
- Acquire images from at least three to four different positions of the channel to image the adhered leukocytes (to ensure that representative areas are captured).
- Repeat Steps 8.1–8.7 for functional assessment of all the rest of the channels.
NOTE: It is possible to run multiple channels at once. To do so, open all the channels of interest during the Step 8.3. and perform all aforementioned Steps 8.4 –8.7 for multiple channels of interest.
9. Completing the Assay and Cleaning the Microfluidic Pump
- After completing the experiment, export and save all images in a RAW format.
- While saving the results, run the microfluidic pump and the manifold cleaning protocol.
- For the microfluidic pump, run the Washout/Pump Washout folder first by perfusing 4 mL of cell culture grade water, followed by 4 mL of 80% ethanol as described in Steps 6.6–6.7 Dry the pump by running air for several minutes.
- In order to clean the manifold, run Washout/Manifold Wash step with the syringe. Follow the steps to open the syringe and the manifold valves as described in Steps 6.10–6.11 Wash first with 80% ethanol and next with cell culture grade water. Dry the manifold by pushing air via the syringe. Close the syringe valve and the manifold valves as described in Steps 6.10 and 6.15.
10. Counting of Adherent Leukocytes
- Download and install the image processing software14 from http://cellprofiler.org.
NOTE: Image processing in this study was performed using software version 2.1.1. If using a newer version, the pipeline might require minor adaptation. - Download the pipeline Count_leukocytes.cpipe. Click File | Import | Pipeline from the file and navigate on PC to choose the pipeline Count_leukocytes.cpipe.
- Drag-and-drop a folder with acquired RAW images of adherent leukocytes to the File list window in the Input modules | Images section for analysis.
- Specify the typical size of leukocytes via Analysis modules | IdentifyPrimaryObjects. Specify the minimal and maximal diameter of adherent leukocytes in pixels.
- Select the filtering criteria via Analysis modules | FilterObjects. Specify the minimally accepted area of objects in pixels.
- Start the analysis by pressing the Analyze images button.
NOTE: All the RAW images will be processed, and the results will be saved in a default output folder. The Image.txt output file contains the numbers of adherent leukocytes in each processed image (each number corresponds to one image, i.e., each acquired field of view).
Representative Results
First, the response of hiPSC-ECs to the stimulation with pro-inflammatory agent TNFα should be examined, as previously described7. TNFα treatment for 12 h triggers the upregulation of E-selectin with the peak at 6 h after starting treatment. Additionally, ICAM-1 is upregulated 6 – 12 h post-treatment. Although we did not observe the expected upregulation of VCAM-1, it is typically observed in primary human umbilical vein endothelial cells (HUVECs). We found overnight (12 h) TNFα treatment to be the most convenient for the leukocyte adhesion assay.
Optimal hiPSC-ECs dissociation should result in a single-cell suspension free of cell clumps. Figure 2C illustrates the optimal hiPSC-EC density right after injection. Typically, 15 min after injection, hiPSC-ECs attach to the bottom of the channel and start to spread (Figure 2D). 1 h after injection, hiPSC-ECs form a well-spread monolayer along the channel and will be ready to begin the flow assay (Figure 2E).
Labeling of leukocytes with fluorescent tracer is beneficial for phase contrast images in automated image quantification, as it makes the cell identification step easier, especially because the leukocytes adhere to the monolayer of ECs and it is often difficult for software to distinguish different cell types.
Free open-source image processing software is very useful for automated quantification of adherent leukocytes. The pipeline described in the protocol here is based on primary object identification using adaptive thresholding of fluorescent images of leukocytes (Figure 4A). Filtering of identified objects based on a minimal area additionally filters out falsely identified objects, such as cell debris, autofluorescence or any other non-specific fluorescent signal (Figure 4C, D).
One microfluidic chip with eight parallel channels allows the comparison of two independent groups, for instance, ECs differentiated from independent hiPSCs, such as healthy donors or patients with genetic disorders. Additional controls, such as untreated ECs, or pre-treatment with an ICAM-1 blocking antibody can be included as well10,11. Two to three microfluidic chips can be processed at a time. For the robustness of the conclusions, a minimum of three independent biological experiments performed on independent days is recommended.
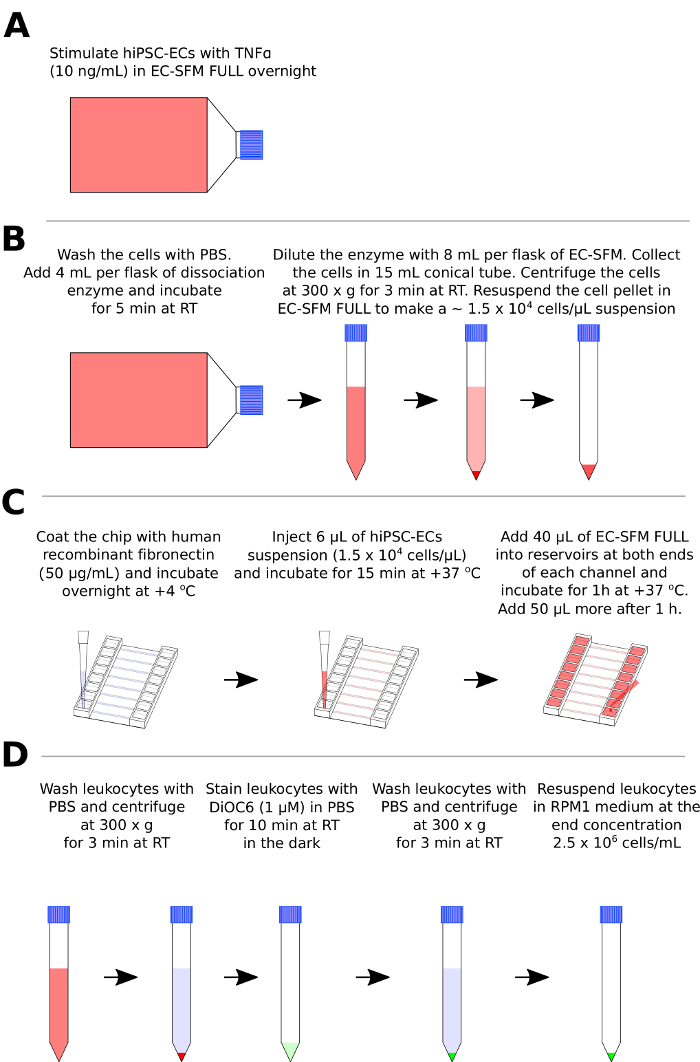
Figure 1: Preparation of hiPSC-ECs and leukocytes for the flow assay. (A) Pre-treatment of hiPSC-ECs with the pro-inflammatory stimulus (TNFα). (B) Schematic representation of hiPSC-ECs enzymatic dissociation, centrifugation, and resuspension. (C) Schematic representation of coating of the channels (C, left), injection of the hiPSC-EC suspension (C, middle) and cell culture medium addition after hiPSC-ECs attachment in the microfluidic chip. (D) Schematic representation of the leukocyte washing step, labeling with fluorescent dye DiOC6 and resuspension. Please click here to view a larger version of this figure.
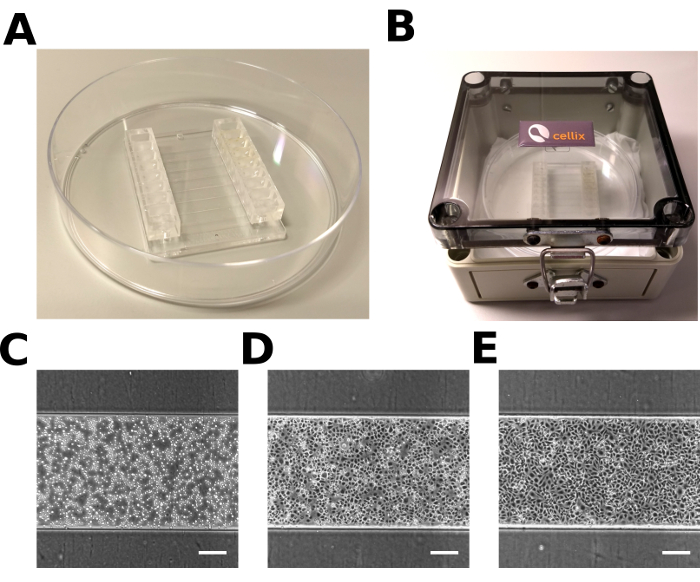
Figure 2: Microfluidic chip with hiPSC-ECs. (A) Microfluidic chip in a 10 cm Petri dish. (B) The humidified chamber used for incubation. (C, D, E) Representative images of hiPSC-ECs in the microfluidic channel 0 min (C), 15 min (D), 1 h (E) after injection. Scale bar = 200 µm. Please click here to view a larger version of this figure.
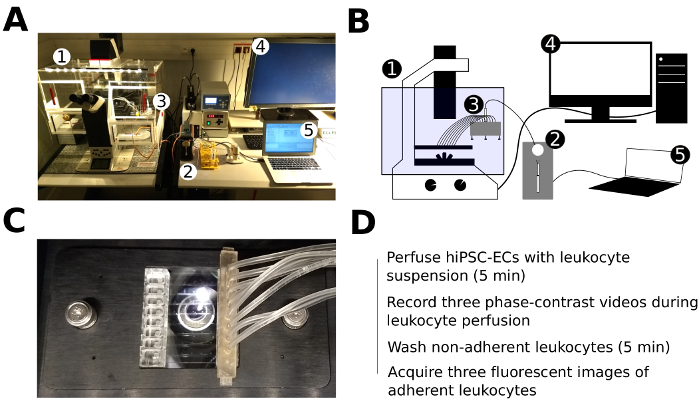
Figure 3: Experimental setup of the live cell imaging and leukocyte perfusion assay. (A, B) Photo (A) and schematic representation (B) of the experimental setup of the live cell imaging and flow assay: (1) – inverted fluorescent microscope with mounted live imaging chamber (5% CO2, 37 °C, humidified), (2) – microfluidic pump, (3) – 8-channel manifold, (4) – PC for microscope control and image acquisition, (5) – PC for microfluidic pump control. (C) Microfluidic chip with the inserted tubing of the 8-channel manifold fixed in a plate holder and mounted in the motorized stage of the microscope. (D) Schematic representation of the major steps of the flow assay. Please click here to view a larger version of this figure.
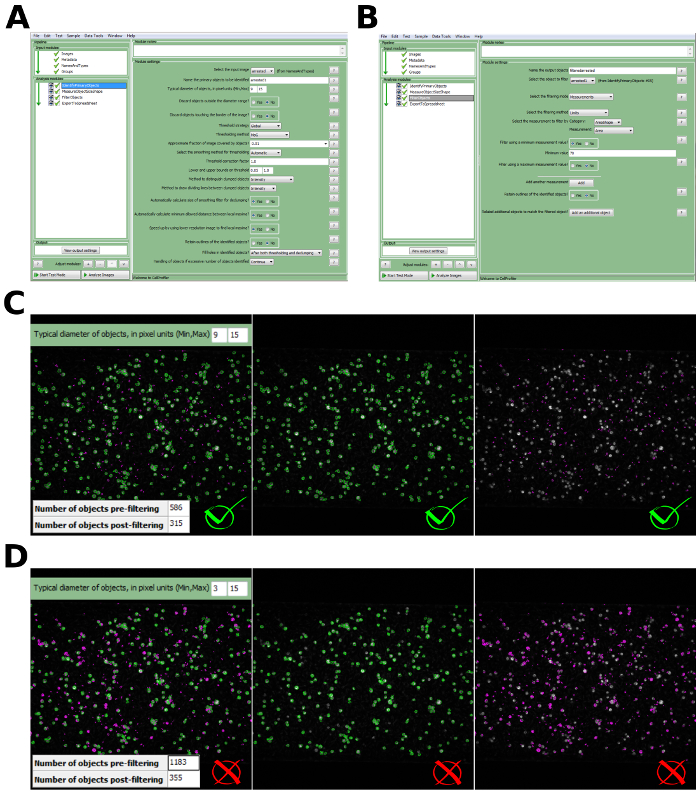
Figure 4: Image analysis and automated detection of adherent leukocytes. (A) The dialog window of the image processing software with specified settings for the identification of leukocytes. (B) The dialog window of the image processing software with specified filtering criteria of the identified objects based on minimally accepted surface area (SAmin = 70 px). (C) Example of correct identification of leukocytes and filtering of non-specific objects of the small surface area (SA) (typical diameter in pixel (Min, Max) = (9, 15); Filtering criteria SAmin = 70 px). Accepted objects are depicted in green and discarded objects are depicted in violet. (D) Example of incorrect identification of leukocytes due to the incorrect range of typical diameter (Typical diameter in pixel (Min, Max) = (3, 15); Filtering criteria SAmin = 70 px). Accepted objects are depicted in green and discarded objects are depicted in violet. Please click here to view a larger version of this figure.
Discussion
This protocol describes the characterization of hiPSC-EC treatment with pro-inflammatory stimuli, such as TNFα, functional assessment of leukocyte adhesion to hiPSC-ECs in the flow assay using live imaging and automated counting of adherent leukocytes.
Overnight stimulation of hiPSC-ECs with TNFα results in the elongated appearance that typically indicates an activated pro-inflammatory phenotype in ECs. During the optimization stage, expression levels of E-selectin, ICAM-1, VCAM-1 should be verified by FACs7,8, and primary human umbilical vein ECs (HUVECs) are advisable to be included as positive controls.
Optimal hiPSC-ECs dissociation and seeding density should be reached to achieve a confluent monolayer without forming cell clumps and channel clogs. It is critical to re-suspend leukocytes right before perfusion in order to avoid cell precipitation and maintain a constant concentration when assaying each channel. Human peripheral blood leukocytes, such as monocytes and neutrophils can be used, or established monocytic cell lines, such as THP-1 or HL-60 could be used as an alternative.
During the assembly of the microfluidic setup and during the flow assay, it is critical to prevent air bubbles from being trapped in the microfluidic tubing and channels as they can result in the detachment of hiPSC-ECs and/or adherent leukocytes. Fluid-to-fluid interface or incorporation of the additional washing steps during the preparation of the microfluidic setup could help to prevent air bubbles.
It is advisable that the protocol is run by two operators where one prepares the cells and the second prepares the microfluidic system and flow assay. This ensures that ECs are plated into microfluidic chips within a constant time-window prior to processing and improves the accuracy and reproducibility of the results. In addition, this also allows setting up blinded experiments to avoid bias in the results.
For successful cell detection with the automated image processing pipeline, it is important to acquire images in focus with a high signal-to-noise ratio and to avoid autofluorescence of non-specific objects, such as cell clumps. Of crucial importance is the correct specification of minimal and maximal limits of the typical size of adherent leukocytes that need to be identified. One can estimate the typical size of leukocytes using a line measurement tool in ImageJ. For instance, in our analysis, we used the range of 9–15 pixels which corresponds to the physical size of 10.3–17.1 µm (Figure 4C). When using a wrong range, e.g., 3–15 pixels (3.4–17.1 µm), a single leukocyte is identified not as one cell, but rather its subparts are identified as separate objects (Figure 4D). This leads to the false number of objects to be identified.
Many objects of a low intensity and smaller surface area than leukocytes may be detected using the provided adaptive thresholding method. This might take place due to autofluorescence or any other non-specific fluorescent signals. Nevertheless, these non-specific objects, if their area is lower than the typical area of a single leukocyte, can be filtered by defining the minimally accepted surface area (Figure 4C).
The flow assay described here allows direct assessment of leukocyte adhesion to an EC monolayer, elucidating the interaction of these two cell types, but does not take into account other cell types, such as pericytes that are present in the vascular wall and might also participate in the regulation of leukocyte extravasation15. Integrating a 3D culture microenvironment with other cell types could improve the physiological relevance of the system. Despite these limitations, the flow assay for the assessment of inflammatory responses described here provides a valuable tool for basic characterization and disease modeling applications of hiPSC-ECs.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge the following grants: European Research Council (ERCAdG 323182 STEMCARDIOVASC); Netherlands Organ-on-Chip Initiative, an NWO Gravitation project funded by the Ministry of Education, Culture and Science of the government of the Netherlands (024.003.001).
Materials
| Vena8 Endothelial+ biochip | Cellix Ltd | V8EP-800-120-02P10 | |
| Mirus Evo Nanopump | Cellix Ltd | MIRUS-EVO-PUMP | 1 x syringe pump; 1 x VenaFluxAssay Software; 1 x tubing kit; power supply and cables. |
| 8-channel manifold MultiFlow8 | Cellix Ltd | MIRUS-MULTIFLOW8 | |
| Humidified box | Cellix Ltd | HUMID-BOX | |
| Fluorescence imaging system Leica AF6000 | Leica Microsystems | ||
| Electron-multiplying charge-coupled device camera | Hamamatsu | C9100 | |
| Biological safety cabinet/laminar flow-hood | Cleanair | ||
| CO2 cell-culture incubator | Panasonic | MCO-170AICUV | |
| Centrifuge | Hitachi | himac-CT6EL | |
| Handheld pipetman (P-10 (10 mL), P-200 (200 µL), P-1000 (1,000 µL) | Gilson international | 4807 (10µl), 4810 (200µl), 4809 (1000µl) | |
| Sterile plastic pipette | Greiner Bio-One | 606180 (5ml), 607180 (10ml) | |
| Petri dish | VWR/ Duran Group | 391-0860 | |
| Culture flasks (75 cm2) | CELLSTAR | 658,170 | |
| Centrifuge tube (15 mL) | CELLSTAR | 188271 | |
| Name | Company | Catalog Number | Comments |
| Gelatin from porcine skin, type A | Sigma-Aldrich | G1890 | |
| Fibronectin (Bovine plasma) | Sigma-Aldrich | F1141 | |
| DPBS, no calcium, no magnesium | Life Technologies | 14190-169 | |
| Human Endothelial-SFM | Life Technologies | 11111-044 | |
| Human VEGF 165 IS, premium grade | Miltenyi Biotec | 130-109-386 | |
| Human FGF-2, premium grade | Miltenyi Biotec | 130-093-842 | |
| Platelet-poor Plasma Derived Serum, bovine | Biomedical Technologies | BT-214 | |
| Recombinant Human TNF-α | Tebu-bio | 300-01A-A | |
| TrypLE Select | Life Technologies | 12563029 | |
| DiOC6(3) | Sigma-Aldrich | 318426 | |
| RPMI 1640 Medium | Life Technologies | 21875-034 | |
| 2-Mercaptoethanol (50 mM) | Life Technologies | 31350010 | |
| FBS | Life Technologies | 10270-106 | |
| L-glutamine | Life Technologies | 25030-024 | |
| Penicillin-Streptomycin (5,000 U/mL) | Life Technologies | 15070-063 | |
| Distilled water | Life Technologies | 15230-089 | |
| Ethanol absolute | Merck | 1.00983.2500 | |
| VCAM-1 | R&D systems | FAB5649P | |
| E-Selectin | R&D systems | BBA21 | |
| ICAM-1 | R&D systems | BBA20 | |
| Name | Company | Software version | Comments |
| Cellrofiler | CellProfiler | 2.1.1 | |
| VenaFluxAssay Software | Cellix Ltd | 2.3.a |
Riferimenti
- Vestweber, D. How leukocytes cross the vascular endothelium. Nature Reviews Immunology. 15 (11), 692-704 (2015).
- Hajishengallis, G., Chavakis, T. Endogenous modulators of inflammatory cell recruitment. Trends in Immunology. 34 (1), 1-6 (2013).
- Molema, G. Heterogeneity in endothelial responsiveness to cytokines, molecular causes, and pharmacological consequences. Seminars in thrombosis and hemostasis. 36 (3), 246-264 (2010).
- Aird, W. C. Endothelial Cell Heterogeneity. Cold Spring Harbor Perspectives in Medicine. 2 (1), a006429 (2012).
- Shi, Y., Inoue, H., Wu, J. C., Yamanaka, S. Induced pluripotent stem cell technology: a decade of progress. Nature Reviews Drug Discovery. 16 (2), 1-16 (2016).
- Lin, Y., Gil, C. H., Yoder, M. C. Differentiation, Evaluation, and Application of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 37 (11), 2014-2025 (2017).
- Halaidych, O. V., Freund, C., et al. Inflammatory Responses and Barrier Function of Endothelial Cells Derived from Human Induced Pluripotent Stem Cells. Stem Cell Reports. , (2018).
- Orlova, V. V., vanden Hil, F. E., Petrus-Reurer, S., Drabsch, Y., ten Dijke, P., Mummery, C. L. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nature Protocols. 9 (6), 1514-1531 (2014).
- Orlova, V. V., Drabsch, Y., et al. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arteriosclerosis, Thrombosis, and Vascular Biology. 34 (1), 177-186 (2014).
- Shetty, S., Weston, C. J., Adams, D. H., Lalor, P. F. A Flow Adhesion Assay to Study Leucocyte Recruitment to Human Hepatic Sinusoidal Endothelium Under Conditions of Shear Stress. Journal of Visualized Experiments. (85), 1-6 (2014).
- Muller, W. A., Luscinskas, F. W. Assays of transendothelial migration in vitro. Methods in enzymology. 443, 155-176 (2008).
- Ganguly, A., Zhang, H., Sharma, R., Parsons, S., Patel, K. D. Isolation of Human Umbilical Vein Endothelial Cells and Their Use in the Study of Neutrophil Transmigration Under Flow Conditions. Journal of Visualized Experiments. (66), 1-5 (2012).
- Vajen, T., Heinzmann, A. C. A., et al. Laminar Flow-based Assays to Investigate Leukocyte Recruitment on Cultured Vascular Cells and Adherent Platelets. Journal of Visualized Experiments. (134), (2018).
- Carpenter, A. E., Jones, T. R., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 7 (10), R100 (2006).
- Nourshargh, S., Alon, R. Leukocyte Migration into Inflamed Tissues. Immunity. 41 (5), 694-707 (2014).

