Isolation and Characterization of Human Umbilical Cord-derived Mesenchymal Stem Cells from Preterm and Term Infants
Summary
Human umbilical cord (UC) can be obtained during the perinatal period as a result of preterm, term, and postterm delivery. In this protocol, we describe the isolation and characterization of UC-derived mesenchymal stem cells (UC-MSCs) from fetuses/infants at 19-40 weeks of gestation.
Abstract
Mesenchymal stem cells (MSCs) have considerable therapeutic potential and attract increasing interest in the biomedical field. MSCs are originally isolated and characterized from bone marrow (BM), then acquired from tissues including adipose tissue, synovium, skin, dental pulp, and fetal appendages such as placenta, umbilical cord blood (UCB), and umbilical cord (UC). MSCs are a heterogeneous cell population with the capacity for (1) adherence to plastic in standard culture conditions, (2) surface marker expression of CD73+/CD90+/CD105+/CD45–/CD34–/CD14–/CD19–/HLA-DR– phenotypes, and (3) trilineage differentiation into adipocytes, osteocytes, and chondrocytes, as currently defined by the International Society for Cellular Therapy (ISCT). Although BM is the most widely used source of MSCs, the invasive nature of BM aspiration ethically limits its accessibility. Proliferation and differentiation capacity of MSCs obtained from BM generally decline with the age of the donor. In contrast, fetal MSCs obtained from UC have advantages such as vigorous proliferation and differentiation capacity. There is no ethical concern for UC sampling, as it is typically regarded as medical waste. Human UC starts to develop with continuing growth of the amniotic cavity at 4-8 weeks of gestation and keeps growing until reaching 50-60 cm in length, and it can be isolated during the whole newborn delivery period. To gain insight into the pathophysiology of intractable diseases, we have used UC-derived MSCs (UC-MSCs) from infants delivered at various gestational ages. In this protocol, we describe the isolation and characterization of UC-MSCs from fetuses/infants at 19-40 weeks of gestation.
Introduction
Mesenchymal stem cells (MSCs) are originally isolated and characterized from bone marrow (BM)1,2 but can also be obtained from a wide variety of tissues including adipose tissue, synovium, skin, dental pulp, and fetal appendages3. MSCs are recognized as a heterogeneous cell population that can proliferate and differentiate into adipocytes, osteocytes, and chondrocytes. In addition, MSCs possess the ability to migrate to sites of injury, suppress and modulate immune responses, and remodel and repair injury. Currently, MSCs from different sources have attracted growing interest as a source for cell therapy against a number of intractable diseases, including graft-versus-host disease, myocardial infarction, and cerebral infarction4,5.
Although BM is the most well-characterized source of MSCs, the invasiveness of BM aspiration ethically limits its accessibility. Proliferation and differentiation capacity of MSCs obtained from BM generally decline with the age of the donor. In contrast, fetal MSCs obtained from fetal appendages such as placenta, umbilical cord blood (UCB), and umbilical cord (UC) have advantages including less ethical concerns regarding sampling and robust proliferation and differentiation capacity6,7. Among fetal appendages that are usually discarded as medical waste, UCB and UC are considered a fetal organ, while placenta is considered fetomaternal. In addition, placenta and UCB need to be sampled and collected at the exact moment of newborn delivery, whereas placenta and UC can be collected and processed after newborn delivery. Accordingly, UC is a promising MSC source for cell therapy8,9.
Human UC starts to develop with progressive expansion of the amniotic cavity at 4-8 weeks of gestation, continues to grow until 50-60 cm in length, and can be isolated during the whole period of newborn delivery10. To gain insight into the pathophysiology of intractable diseases, we use UC-derived MSCs (UC-MSCs) from infants delivered at various gestational ages11,12. In this protocol, we describe how to isolate and characterize UC-MSCs from fetuses/infants at 19-40 weeks of gestation.
Protocol
The use of human samples for this study was approved by the ethical committee of Kobe University Graduate School of Medicine (approval no. 1370 and 1694) and conducted in accordance with the approved guidelines.
1. Isolation and Culture of UC-MSCs
NOTE: UC-MSCs have been successfully isolated, cultured, and expanded (more than passage number 4) from more than 200 UCs subjected to this protocol. Among more than 200 UCs, 100% have shown successful UC-MSC isolation, less than 5% have shown accidental contamination, less than 15% have shown growth arrest, and more than 80% have shown successful UC-MSC expansion.
- Collect UC.
- Prepare a 50 mL plastic tube, an aseptic scissor, and a 500 mL bottle of alpha modified Eagle's minimum essential medium (alpha MEM) at 4 °C.
- Aseptically cut out UC with a surgical scissor at approximately 5-10 cm in length and collect it soon after newborn birth by cesarean section.
- Immediately place the UC in a 50 mL plastic tube and add 20-30 mL of alpha MEM into the tube.
- Store the UC at room temperature (RT) until it is transported to the laboratory.
NOTE: Aseptic UC can be stored in serum-free medium at RT for up to 2 days. Therefore, UC obtained from neighboring hospitals can be collected if it is delivered to the lab within 2 days.
- Dissect UC.
- Prepare a plastic tray, sterilized scissors and forceps, 10 mL pipettes, 25 mL pipettes, two 60 mm tissue culture dishes, phosphate-buffered saline (PBS), purified enzyme blends (see Table of Materials, reconstituted with sterile PBS in a concentration of 13 Wünsch units/mL and stored at -80 °C), and a 500 mL bottle of reduced serum medium (see Table of Materials).
- Warm up the PBS, purified enzyme blends, reduced serum medium, and culture medium [alpha MEM containing 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic solution (AA) stored at 4 °C] to RT.
- Take out the UC from the tube and place it in a plastic tray.
- Weigh and dissect 5 g of UC with a sterilized scissor.
- Pour 10 mL of 70% ethanol over the UC for sterilization using a 10 mL pipette.
- Wash the UC with 10 mL of PBS twice using a 10 mL pipette.
- Place the UC in a sterilized 60 mm tissue culture dish (see Figure 1, step 1).
- Add 10 mL of reduced serum medium into the dish.
- Add 0.5 mL of purified enzyme blends to reach a final concentration of approximately 0.62 Wünsch units/mL.
NOTE: The use of purified enzyme blends instead of traditional collagenase has been shown to improve the yield and viability of UC-MSCs isolated from UC. - Cut the UC into 2-3 mm pieces with sterilized scissors and forceps (see Figure 1, step 2).
NOTE: This process takes about 30 min. - Incubate the UC pieces at 37 °C for 30 min in a 5% CO2 incubator.
- Cut the partially-digested UC pieces into smaller pieces that easily flow through a 25 mL pipette.
NOTE: This process takes about 30 min. - Incubate the smaller UC pieces at 37 °C for 15-45 min in a 5% CO2 incubator.
NOTE: Incubation needs to continue until the homogenates become viscous. The total digestion time is approximately 120 min. In the case of using UCs from preterm infants, however, the digestion time can be shortened because those are easily cut and digested.
- Isolate UC-MSCs.
- Prepare four 50 mL plastic tubes and a 500 mL bottle of culture medium.
- Divide the UC homogenate into two 50 mL plastic tubes using a 25 mL pipette, with approximately 7.5 mL in each tube.
- Add 20 mL of culture medium into each tube and mix well.
- Filter each UC homogenate through a 100 µm cell strainer placed on top of a new 50 mL collection tube, using a 25 mL pipette to collect UC-derived cells.
NOTE: This process takes about 15 min each, as the digested UC solution is sticky. - Centrifuge two tubes at 1,000 x g for 5 min.
- Carefully aspirate the supernatant and discard it.
- Resuspend the cell pellets in two tubes with 5 mL of culture medium.
- Transfer the cell suspension into a new 60 mm plate and culture at 37 °C in a 5% CO2 incubator (see Figure 1, step 3).
- Culture UC-MSCs.
- Warm up the PBS, trypsin-EDTA (0.25 w/v%), and culture medium (alpha MEM containing 10% FBS and 1% AA) to RT.
- When cells are attached to a 60 mm plate, remove the culture medium and wash with 5 mL of PBS twice to remove debris and red blood cells.
NOTE: Attached cells usually appear at 3-5 days after initial plating (see Figure 1, step 4). - Replace the culture medium twice per week and incubate at 37 °C in a 5% CO2 incubator until 90-100% confluency.
NOTE: This usually takes 7-14 days after initial plating. - Wash cells with 5 mL of PBS twice, add 0.5 mL of trypsin-EDTA, and incubate at 37 °C for 5-10 min.
- When cells become rounded and detached, add 9 mL of culture medium and mix well to inactivate trypsin.
- Transfer the cell suspension into a new 100 mm plate and culture at 37 °C in a 5% CO2 incubator (see Figure 1, step 5).
- Replace the culture medium twice per week and incubate at 37 °C in a 5% CO2 incubator until 90-100% confluency.
NOTE: This usually takes 4-8 days after initial plating. - Wash cells with 10 mL of PBS twice, add 1 mL of trypsin-EDTA, and incubate at 37 °C for 5-10 min.
- When cells become rounded and detached, add 9 mL of culture medium and mix well to inactivate trypsin.
- Transfer the cell suspension into two new 100 mm plates and culture at 37 °C in a 5% CO2 incubator (see Figure 1, step 5).
- Repeat subculture (1:2 splits) until tenth passage (see Figure 1, step 5).
NOTE: Because cells cultured under less confluent conditions tend to reach earlier proliferation arrest, it is important to passage cells with a split ratio of 1:2. - Use cells at fifth to eighth passage for cell surface marker analysis, cell differentiation assay, and other experiments.
NOTE: To keep the vigorous proliferation for as long as possible, cells are generally cultured under more than 70-80% confluent conditions.
2. Surface Marker Expression of UC-MSCs
- Prepare cells in a 100 mm plate and dissociate them with 1 mL of trypsin-EDTA at 37 °C for 5-10 min.
- Add 9 mL of culture medium and centrifuge at 1,000 x g for 5 min.
- Aspirate the supernatant and discard it.
- Resuspend the cell pellets with 10 mL of PBS and centrifuge at 1,000 x g for 5 min. Repeat this washing step twice.
- Resuspend cells with flow cytometry (FCM) buffer (PBS containing 2 mM EDTA and 10% blocking reagent) at ~1 × 106 cells/mL.
- Transfer 50-100 µL of cell suspension into a 1.5 mL tube; add phycoerythrin (PE)-conjugated antibodies against CD14, CD19, CD34, CD45, CD73, CD90, CD105, or HLA-DR; and incubate on ice for 45 min.
- Wash cells with FCM buffer twice and add 50-100 µL of 0.2% viability dye solution (see Table of Materials).
- Incubate at RT for 15 min, wash cells with FCM buffer twice, and filter cells through a 70 µm cell strainer.
- Run cells through the FCM and analyze the obtained results using FCM software according to the manufacturer's instructions.
3. Trilineage Differentiation of UC-MSCs
- Perform adipogenesis.
- Prepare a cell suspension in culture medium at a concentration of 5 × 103 cells/mL.
- Plate 1 mL of cell suspension in a 12-well plate and incubate at 37 °C in a 5% CO2 incubator for ~24 h.
- Replace the culture medium with adipogenic differentiation medium (see Table of Materials) and incubate at 37 °C in a 5% CO2 incubator for 2-3 weeks. Change adipogenic differentiation medium twice a week.
- Wash cells with PBS three times.
- Incubate cells in 4% formaldehyde solution for 30 min at RT.
- Wash cells with PBS three times and with 60% isopropanol three times.
- Prepare 0.5% oil red O solution by dissolving 84 mg of oil red O in 10 mL of 100% isopropanol and adding 6.7 mL of distilled water. Stain cells with 1 mL of 0.5% oil red O solution at RT for 20 min.
- Wash cells with 60% isopropanol three times and visualize under a microscope.
- Perform osteogenesis.
- Prepare a cell suspension in culture medium at a concentration of 1 × 104 cells/mL.
- Plate 1 mL of cell suspension in a 12-well plate and incubate at 37 °C in a 5% CO2 incubator for ~24 h.
- Replace the culture medium with osteogeneic differentiation medium (see Table of Materials) and incubate at 37 °C in a 5% CO2 incubator for 1-2 weeks. Change osteogenic differentiation medium twice a week.
- Wash cells with PBS three times.
- Incubate cells in 4% formaldehyde solution for 30 min at RT.
- Wash cells with PBS three times.
- Prepare 2% alizarin red S solution by dissolving 200 mg of alizarin red S with 10 mL of distilled water. Stain cells with 1 mL of 2% alizarin red S solution at RT for 3 min.
- Wash wells with PBS three times and visualize under a microscope.
- Perform chondrogenesis.
- Prepare a cell suspension in culture medium at a concentration of 1.6 × 107 cells/mL.
- Place a 5 µL droplet of cell suspension onto the center of well in a 12-well plate (micromass cultures) and incubate at 37 °C in a 5% CO2 incubator for 2-3 h.
- Gently add 1 mL of chondrogenic differentiation medium (see Table of Materials) and incubate at 37 °C in a 5% CO2 incubator for 4-7 days. Change chondrogenic differentiation medium twice a week.
- Wash cells with PBS three times.
- Incubate cells in 4% formaldehyde solution for 30 min at RT.
- Wash cells with PBS three times.
- Prepare 0.05% toluidine blue solution by dissolving 250 mg of toluidine blue with 500 mL of 0.1 M sodium acetate buffer with pH 4.1. Stain cells with 1 mL of 0.05% toluidine blue solution at RT for 30 min.
- Wash cells with PBS three times and visualize under a microscope.
Representative Results
The procedures from UC collection to MSC culture are summarized in Figure 1. UC of approximately 5-10 cm in length can be collected from all newborns delivered by cesarean section. UC starts to develop at 4-8 weeks of gestation and continues to grow until 50-60 cm in length, as shown in Figure 2. There are two arteries (A), one vein (V), cord lining (CL), and Wharton's Jelly (WJ) in UC, as depicted in Figure 3 and Figure 4. UC-MSCs can be isolated from all cord regions or whole cord13. Because UC from infants with early gestational ages is fragile and difficult for dissection into a single cord region, UC-MSCs are isolated from whole cord11,12. There are several methods to isolate MSCs from UC, which include the explant method14 and enzymatic digestion method15, plus their derivatives16. Due to the longer culture cycle, lower yield, and earlier proliferation arrest associated with the explant method17,18, UC-MSCs are cultured by enzymatic digestion method as illustrated in Figure 5 and Figure 6.
The minimal criteria for defining MSCs are established by the ISCT and include their surface marker characterization19. To determine surface marker expression, UC-MSCs from preterm and term infants are incubated with appropriate PE-conjugated antibodies and analyzed by flow cytometry. They are positive for MSC signature markers (CD73, CD90, CD105) but negative for monocyte/macrophage (CD14), endothelial (CD34), hematopoietic (CD19, CD45), and major histocompatibility complex (HLA-DR) markers, as shown in Figure 7.
According to the ISCT criteria19, MSC must possess mesodermal differentiation capacity assessed by a trilineage differentiation assay. To evaluate trilineage differentiation capacity, UC-MSCs from preterm and term infants are induced to differentiate into osteocytes, adipocytes, and chondrocytes under standard in vitro differentiation conditions. As shown in Figure 8, they are well-differentiated into all three types of mesodermal cells.
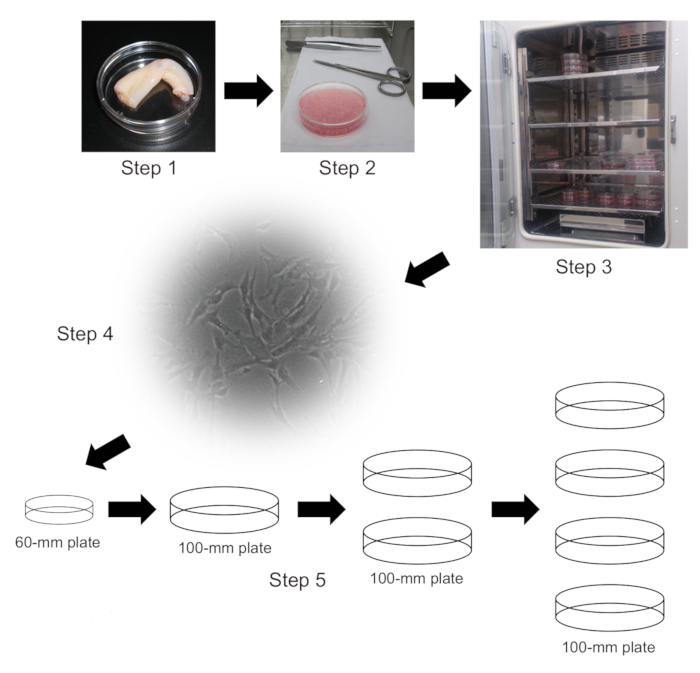
Figure 1: Schematic diagram of isolation and culture of UC-MSCs. Step 1. 5-10 cm of UC aseptically collected from placental tissue. Step 2. Purified enzyme blend-digested UC pieces. Step 3. Culture of UC-MSCs at 37 °C in a 5% CO2 incubator. Step 4. Attached UC-MSCs appeared at 3 days after initial plating. Step 5. Subculture of UC-MSCs. Please click here to view a larger version of this figure.
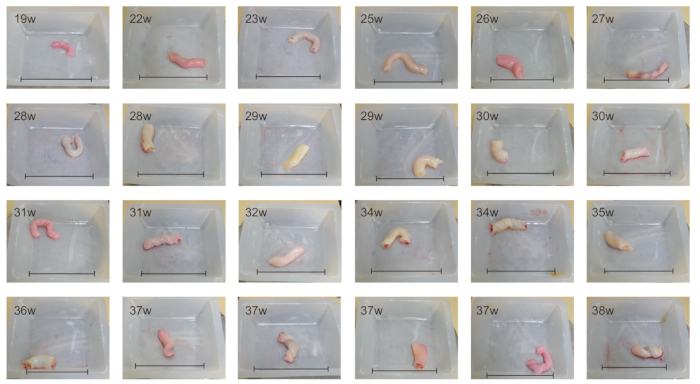
Figure 2: UC from fetuses/infants delivered at various gestational ages. UCs from fetuses/infants at 19-38 weeks of gestation are shown. The size of UC increases with gestational age. Scale bars = 8 cm. Please click here to view a larger version of this figure.
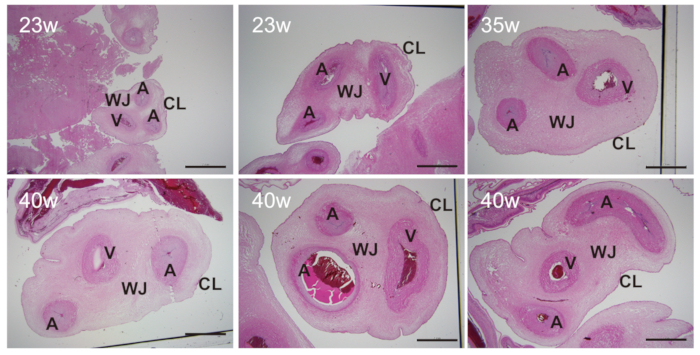
Figure 3: Sectional view of UC from preterm and term infants. There are two arteries (A), one vein (V), cord lining (CL), and Wharton's Jelly (WJ) in UC from preterm and term infants. The sizes of these tissues vary with gestational ages. Scale bars = 2 mm. Please click here to view a larger version of this figure.
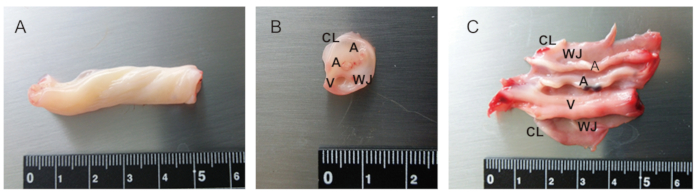
Figure 4: Anatomy of UC. (A) 5 cm of UC from term infant. (B) Cross section of UC. (C) Partially dissected UC. There are two arteries (A), one vein (V), cord lining (CL), and Whalton's Jelly (WJ). UC from infants of earlier gestational ages is particularly fragile and difficult to be dissected. Please click here to view a larger version of this figure.
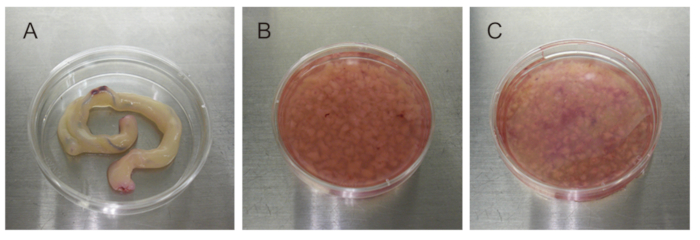
Figure 5: UC dissection using purified enzyme blends. (A) 5-10 cm of UC. (B) 2-3 mm pieces of UC. (C) Purified enzyme blend-digested UC pieces. Please click here to view a larger version of this figure.
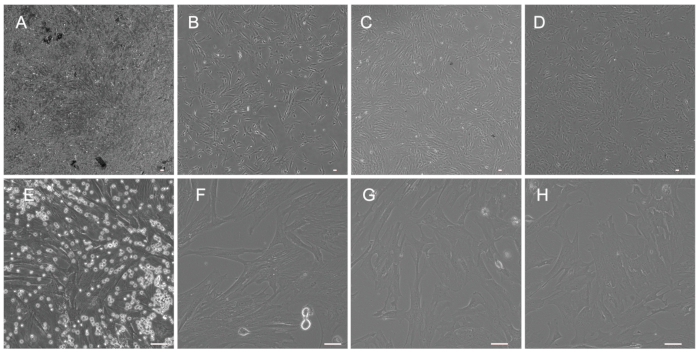
Figure 6: Morphology of UC-MSCs from preterm and term infants. (A and E) UC-MSCs at passage number 1 (P1) before the replacement of culture medium (A: X40; E: X200). (B and F) UC-MSCs at P1 after the replacement of culture medium (B: X40; F: X200). (C and G) UC-MSCs at P3 (C: X40; G: X200). (D and H) UC-MSCs at P5 (D: X40; H: X200). Scale bars = 50 µm. Please click here to view a larger version of this figure.
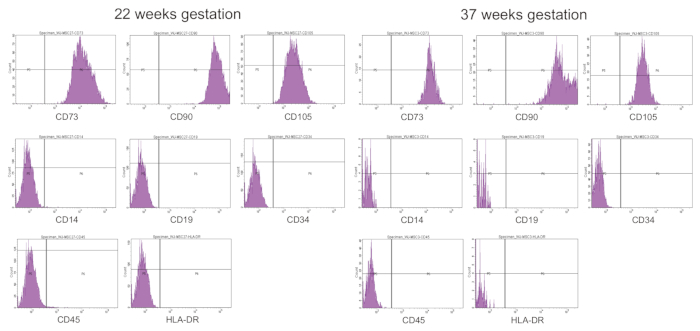
Figure 7: Surface marker expression of UC-MSCs from preterm and term infants. CD73/CD90/CD105-positive and CD14/CD19/CD34/CD45/HLA-DR-negative phenotypes are found in UC-MSCs from both preterm (22 weeks gestation) and term (37 weeks gestation) infants. Please click here to view a larger version of this figure.
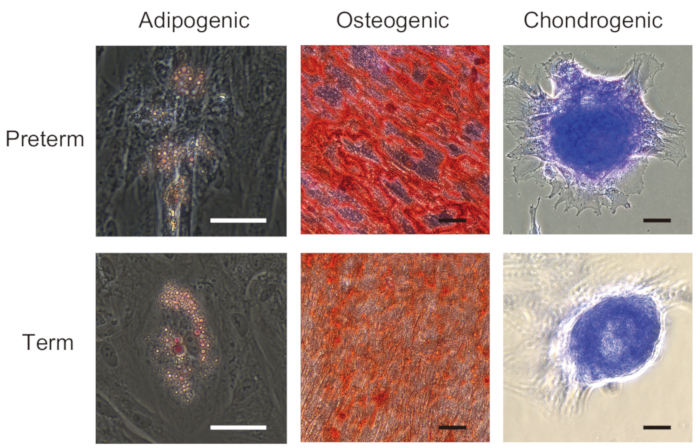
Figure 8: Trilineage mesenchymal differentiation of UC-MSCs from preterm and term infants. UC-MSCs from preterm (22 weeks gestation) and term (37-40 weeks gestation) infants are differentiated into adipocyte (visualized by oil red O), osteocyte (visualized by alizarin red S), and chondrocyte (visualized by toluidine blue). Images were taken at 200x. Scale bars = 50 µm. A part of this figure has been adapted from Iwatani et al.11. Please click here to view a larger version of this figure.
Discussion
MSCs can be isolated from a variety of tissues and are heterogeneous population of cells that do not all express the same phenotypic markers. Here, we outlined a protocol that guides the collection and dissection of UC from preterm and term infants and enables isolation and culture of UC-MSCs. Following this protocol, we have successfully isolated UC-MSCs that fulfill the ISCT criteria19 from fetuses/infants delivered at 19-40 weeks of gestation and demonstrated that they represent some aspects of intractable disease pathophysiology during prenatal development11,12.
In BM-derived MSCs (BM-MSCs), younger donor-derived BM-MSCs generally show greater proliferative and differentiative potential than older counterparts and hold more potential for cell therapy20,21. Similarly, preterm UC-MSCs isolated from fetuses aborted at 8-12 weeks of gestation were reported to exhibit more vigorous proliferation and differentiation compared to term UC-MSCs isolated from newborns delivered at 37-40 weeks of gestation22. Despite the differences in gestational age and cord region, UC-MSCs isolated with this protocol also revealed a more vigorous proliferation of preterm UC-MSCs than term UC-MSCs12.
A critical step within this protocol is the UC dissection with purified enzyme blends that are composed of collagenase I and II and dispase. As outlined in the representative results, the stiffness of UC dramatically varied with gestational age (Figure 2). To achieve optimal dissection of UC, the incubation time of purified enzyme blends needs to be adjusted to the stiffness of the individual UC. Generally, it usually takes longer for term UC than preterm UC. Among existing methods to isolate MSCs from UC, we choose the enzymatic digestion method15 over the explant method14 that may lead to a longer culture cycle, lower yield, and earlier proliferation arrest17,18. An important modification of the enzyme digestion method is the use of purified enzyme blends instead of traditional collagenase, which enables an efficient and consistent dissection of UC from fetuses/infants with variable gestational ages.
A limitation of this protocol is the use of whole cord to isolate MSCs from UC. Because UC-MSCs can be isolated from all cord regions or whole cord13, UC-MSCs are isolated from whole cord in this protocol. This is due to the feasibility of UC dissection from preterm infants. However, the potential variation in the proportion of each cord region may limit the strict comparison between UC-MSCs isolated by this protocol.
UC-MSCs are being increasingly used as a source of mesenchymal stromal cells for preclinical and clinical studies8,9. Despite this increased usage, a consensus on methods of UC-MSCs isolation is still missing, which may result in different cell populations to be deemed the same UC-MSCs. This protocol will ultimately assist researchers in better understanding the derivation and functional characteristics of UC-MSCs.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (C) (grant number: 25461644) and Young Scientists (B) (grant numbers: 15K19614, 26860845, 17K16298) of JSPS KAKENHI.
Materials
| 50mL plastic tube | AS One Coporation, Osaka, Japan | Violamo 1-3500-22 | |
| 12-well plate | AGC Techno Glass, Tokyo, Japan | Iwaki 3815-012 | |
| 60mm dish | AGC Techno Glass, Tokyo, Japan | Iwaki 3010-060 | |
| Cell strainer (100 μm) | Thermo Fisher Scientific, Waltham, MA | Falcon 35-2360 | |
| Cell strainer (70 μm) | Thermo Fisher Scientific, Waltham, MA | Falcon 35-2350 | |
| Alpha MEM | Wako Pure Chemical, Osaka, Japan | 135-15175 | |
| Fetal bovine serum | Sigma Aldrich, St. Louis, MO | 172012 | |
| Reduced serum medium | Thermo Fisher Scientific, waltham, MA | OPTI-MEM Gibco 31985-070 | |
| Antibiotic-antimycotic | Thermo Fisher Scientific, Waltham, MA | Gibco 15240-062 | |
| Trypsin-EDTA | Wako Pure Chemical, Osaka, Japan | 209-16941 | |
| PBS | Takara BIO, Shiga,Japan | T900 | |
| Purified enzyme blends | Roche, Mannheim, Germany | Liberase DH Research Grade 05401054001 | |
| PE-conjugated mouse primary antibody against CD14 | BD Bioscience, Franklin Lakes, NJ | 347497 | Lot: 3220644, RRID: AB_400312 |
| PE-conjugated mouse primary antibody against CD19 | BD Bioscience, Franklin Lakes, NJ | 340364 | Lot: 3198741, RRID: AB_400018 |
| PE-conjugated mouse primary antibody against CD34 | BD Bioscience, Franklin Lakes, NJ | 555822 | Lot: 3079912, RRID: AB_396151 |
| PE-conjugated mouse primary antibody against CD45 | BD Bioscience, Franklin Lakes, NJ | 555483 | Lot: 2300520, RRID: AB_395875 |
| PE-conjugated mouse primary antibody against CD73 | BD Bioscience, Franklin Lakes, NJ | 550257 | Lot: 3057778, RRID: AB_393561 |
| PE-conjugated mouse primary antibody against CD90 | BD Bioscience, Franklin Lakes, NJ | 555596 | Lot: 3128616, RRID: AB_395970 |
| PE-conjugated mouse primary antibody against CD105 | BD Bioscience, Franklin Lakes, NJ | 560839 | Lot: 4339624, RRID: AB_2033932 |
| PE-conjugated mouse primary antibody against HLA-DR | BD Bioscience, Franklin Lakes, NJ | 347367 | Lot: 3219843, RRID: AB_400293 |
| PE-conjugated mouse IgG1 k isotype | BD Bioscience, Franklin Lakes, NJ | 555749 | Lot: 3046675, RRID: AB_396091 |
| PE-conjugated mouse IgG2a k isotype | BD Bioscience, Franklin Lakes, NJ | 555574 | Lot: 3035934, RRID: AB_395953 |
| PE-conjugated mouse IgG2b k isotype | BD Bioscience, Franklin Lakes, NJ | 555743 | Lot: 3098896, RRID: AB_396086 |
| Viability dye | BD Bioscience, Franklin Lakes, NJ | Fixable Viability Stain 450 562247 | |
| Blocking reagent | Dainippon Pharmaceutical, Osaka, Japan | Block Ace UKB80 | |
| FCM | BD Bioscience, Franklin Lakes, NJ | BD FACSAria III Cell Sorter | |
| FCM software | BD Bioscience, Franklin Lakes, NJ | BD FACSDiva | |
| Adipogenic differentiation medium | Invitrogen, Carlsbad, CA | StemPro Adipogenesis Differentiation kit A10070-01 | |
| Osteogenic differentiation medium | Invitrogen, Carlsbad, CA | StemPro Osteogenesis Differentiation kit A10072-01 | |
| Chondrogenic differentiation medium | Invitrogen, Carlsbad, CA | StemPro Chondrogenesis Differentiation kit A10071-01 | |
| Formaldehyde | Polyscience, Warrigton, PA | 16% UltraPure Formaldehyde EM Grade #18814 | |
| Oil Red O | Sigma Aldrich, St. Louis, MO | O0625 | |
| Arizarin Red S | Sigma Aldrich, St. Louis, MO | A5533 | |
| Toluidine Blue | Sigma Aldrich, St. Louis, MO | 198161 | |
| Microscope | Keyence, Osaka, Japan | BZ-X700 |
Riferimenti
- Friedenstein, A. J., Petrakova, K. V., Kurolesova, A. I., Frolova, G. P. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 6 (2), 230-247 (1968).
- Caplan, A. I. Mesenchymal stem cells. Journal of Orthopaedic Research. 9 (5), 641-650 (1991).
- Crisan, M., et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 3 (3), 301-313 (2008).
- Bianco, P., Robey, P. G., Simmons, P. J. Mesenchymal Stem Cells: Revisiting History, Concepts, and Assays. Cell Stem Cell. 2 (4), 313-319 (2008).
- Bianco, P. 34;Mesenchymal" stem cells. Annual Review of Cell and Developmental Biology. 30 (1), 677-704 (2014).
- Baksh, D., Yao, R., Tuan, R. S. Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cells. 25 (6), 1384-1392 (2007).
- Manochantr, S., et al. Immunosuppressive properties of mesenchymal stromal cells derived from amnion, placenta, Wharton’s jelly and umbilical cord. Internal Medicine Journal. 43 (4), 430-439 (2013).
- Arutyunyan, I., et al. Umbilical Cord as Prospective Source for Mesenchymal Stem Cell-Based Therapy. Stem Cells International. 6901286, (2016).
- Davies, J. E., Walker, J. T., Keating, A. Concise Review: Wharton’s Jelly: The Rich, but Enigmatic, Source of Mesenchymal Stromal Cells. Stem Cells Translational Medicine. 6 (7), 1620-1630 (2017).
- Zhu, D., Wallace, E. M., Lim, R. Cell-based therapies for the preterm infant. Cytotherapy. 16 (12), 1614-1628 (2014).
- Iwatani, S., et al. Gestational Age-Dependent Increase of Survival Motor Neuron Protein in Umbilical Cord-Derived Mesenchymal Stem Cells. Frontiers in Pediatrics. 5, 194 (2017).
- Iwatani, S., et al. Involvement of WNT Signaling in the Regulation of Gestational Age-Dependent Umbilical Cord-Derived Mesenchymal Stem Cell Proliferation. Stem Cells International. , 8749751 (2017).
- Mennan, C., et al. Isolation and characterisation of mesenchymal stem cells from different regions of the human umbilical cord. BioMed Research International. 916136, (2013).
- Capelli, C., et al. Minimally manipulated whole human umbilical cord is a rich source of clinical-grade human mesenchymal stromal cells expanded in human platelet lysate. Cytotherapy. 13 (7), 786-801 (2011).
- Lu, L. L., et al. Isolation and characterization of human umbilical cord mesenchymal stem cells with hematopoiesis-supportive function and other potentials. Haematologica. 91 (8), 1017-1026 (2006).
- Tong, C. K., et al. Generation of mesenchymal stem cell from human umbilical cord tissue using a combination enzymatic and mechanical disassociation method. Cell Biology International. 35 (3), 221-226 (2011).
- Han, Y. F., et al. Optimization of human umbilical cord mesenchymal stem cell isolation and culture methods. Cytotechnology. 65 (5), 819-827 (2013).
- Paladino, F. V., Peixoto-Cruz, J. S., Santacruz-Perez, C., Goldberg, A. C. Comparison between isolation protocols highlights intrinsic variability of human umbilical cord mesenchymal cells. Cell Tissue Bank. 17 (1), 123-136 (2016).
- Dominici, M., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 8 (4), 315-317 (2006).
- Mareschi, K., et al. Expansion of mesenchymal stem cells isolated from pediatric and adult donor bone marrow. Journal of Cellular Biochemistry. 97 (4), 744-754 (2006).
- Choumerianou, D. M., et al. Comparative study of stemness characteristics of mesenchymal cells from bone marrow of children and adults. Cytotherapy. 12 (7), 881-887 (2010).
- Hong, S. H., et al. Ontogeny of human umbilical cord perivascular cells: molecular and fate potential changes during gestation. Stem Cells and Development. 22 (17), 2425-2439 (2013).

