Dissection of the Drosophila Pupal Retina for Immunohistochemistry, Western Analysis, and RNA Isolation
Summary
This paper presents a surgical method for dissecting Drosophila pupal retinas along with protocols for the processing of tissue for immunohistochemistry, western analysis, and RNA-extraction.
Abstract
The Drosophila pupal retina provides an excellent model system for the study of morphogenetic processes during development. In this paper, we present a reliable protocol for the dissection of the delicate Drosophila pupal retina. Our surgical approach utilizes readily-available microdissection tools to open pupae and precisely extract eye-brain complexes. These can be fixed, subjected to immunohistochemistry, and retinas then mounted onto microscope slides and imaged if the goal is to detect cellular or subcellular structures. Alternatively, unfixed retinas can be isolated from brain tissue, lysed in appropriate buffers and utilized for protein gel electrophoresis or mRNA extraction (to assess protein or gene expression, respectively). Significant practice and patience may be required to master the microdissection protocol described, but once mastered, the protocol enables relatively quick isolation of mainly undamaged retinas.
Introduction
The Drosophila retina is composed of approximately 750 ommatidia surrounded by pigment cells arranged in a honeycomb lattice1,2,3,4. Each ommatidium contains eight photoreceptor neurons, four lens-secreting cone cells, and two primary pigment cells. Surrounding each ommatidium are pigment-producing lattice cells and sensory bristle groups. Due to its post-mitotic nature and stereotypical hexagonal arrangement, the Drosophila pupal retina provides an excellent model system for the study of morphogenetic processes including cell adhesion5,6,7,8,9,10 and apoptosis11,12,13,14,15.
Several published protocols utilize air pressure to extract eye-brain complexes from Drosophila pupae16,17,18. The protocol described here instead utilizes microdissection tools to carefully and precisely isolate eye-brain complexes with the goal of obtaining undamaged retinal tissue. This is crucial if retinas are to be utilized for morphological, protein, or gene expression analyses since damage to retinas can result in cellular stress or death, which could modify the cellular phenotype or gene expression. Additionally, after practice, 6 to 10 eye-brain complexes can be isolated in 10 to 15 min, facilitating the goal of minimizing variability in the age and developmental stage of dissected eye tissue.
The fixation, immunostaining, and whole-mount protocol described below is suitable for the preparation of Drosophila eyes for fluorescent microscopy. Retinas can be incubated with antibodies targeting proteins of interest. For example, antibodies to adherens junction components can be utilized to visualize the apical circumferences of cells so that characteristics including cell type, shape and arrangement can be assessed19. Prior to fixation, eyes can instead be cleaved from the brain for the purpose of extracting protein for Western analysis, or RNA for use in qRT-PCR or RNA-sequencing.
Protocol
1. Tissue Preparation
- Set up Drosophila crosses (as described previously20) or culture specific Drosophila strains to obtain pupae of the desired genotype. To ensure that a large number of pupae emerge coincidently, establish these fly cultures in duplicate on nutrient-rich food media or standard food media generously supplemented with yeast-paste.
- Maintain Drosophila cultures at 25 °C. For crosses utilizing the UAS-GAL4 system, GMR-GAL4 is an ideal driver expressed in larval eye disc cells posterior to the morphogenetic furrow and throughout pupal development21,22,23,24. The cross UAS-lacZ X GMR-GAL4 serves as an ideal control cross as it drives expression of the non-endogenous and inert β-galactosidase protein.
- Use the tip of a 6” bamboo splint that has been wet with distilled water to gently lift white pre-pupae (Figure 1B) from the side of healthy culture vials (Figure 1A) and place into a 1.5 mL microcentrifuge tube (Figure 1C).
- Place the tubes into a plastic pipette tip box along with a small piece of tissue soaked in distilled water to maintain the chamber at sufficient humidity to protect the pupae from desiccation (Figure 1D). Incubate the pupae at 25 °C until dissection.
- Use a dry 6” bamboo splint to gently push the pupae out from the microcentrifuge tube and onto a black dissecting dish. Lay a fresh piece of double-sided tape onto the dissecting dish away from the pupae.
- Using a pair of forceps, carefully place the pupae dorsal side up (i.e., operculums facing up, Figure 1B) onto the tape (Figure 2A). Ensure that the pupae adhere well to the tape.
NOTE: Moisture on the pupal case will inhibit secure adhesion to the tape, in which case, allow the pupae to air-dry before placing onto the double-sided tape.
2. Dissection of Eye-brain Complexes
- Use forceps to remove the operculum of each pupa (Figure 2C).
- Use microdissection scissors to slice or cut open the pupal case of each pupa. Flap open the pupal case to reveal the head, thorax, and anterior abdominal segment, securing the edges of the pupal case to the double-sided tape (Figure 2C).
NOTE: It is not necessary to entirely reveal the pupae. - Pierce the abdomen of each pupa with sharp forceps, to grasp the animal, and remove it from its pupal case (Figure 2C).
- Place the pupae onto the black dissection dish, away from the tape, and cover with about 400 µL of ice-cold phosphate-buffered-solution (PBS, pH 7.4).
- Grasp each pupa by the abdomen with forceps, and with microdissection scissors, make a clean cross-sectional cut through the entire thorax, cutting the pupa in half (Figure 2D).
- Remove the posterior remnant of each carcass from the PBS and put to one side on the dissecting dish (do not discard, see step 3.2.1).
- Using two pairs of fine forceps, open the thorax and head of each pupa to reveal the eye-brain complex (Figure 2E). To do this, grasp the cut-edges of the thorax epithelium and gradually tear to open the thorax and then head capsule, exposing the eye-brain complex and surrounding fat tissue.
- Without grasping the tissue, use forceps to guide the eye-brain complex away from the remnants of the head capsule or scoop the eye-brain complex from the torn-open head capsule.
NOTE: The eye-brain complex is dumbbell-shaped, off-white, and more translucent than the surrounding cream-colored fat (Figure 2B, E). - With forceps, carefully remove most fat associated with the eye-brain complexes without touching the eye tissue.
3. Processing of Tissue for Immunofluorescence
- Dissect at least 6-10 pupae as described (steps 2.1-2.9).
NOTE: Three to four independent replicates of each dissection tend to yield sufficient data to test a hypothesis. - Washing and Fixation
- Cut the tip of a P20 or P100 pipette tip with a clean razor blade to increase the tip-opening to ~1 mm in radius and lubricate by pipetting a mix of PBS and fat up and down. This fat can be obtained from the carcass remnants removed in step 2.6.
- Transfer the eye-brain complexes into ~400 µL of PBS in a 9-well glass dish, on ice. Use the lubricated tip and a P20 or P100 air displacement micropipette to transfer.
NOTE: Eye-brain complexes will adhere to unlubricated tips during pipetting and be damaged or lost. - Remove the remaining fat associated with the eye tissue by pipetting the PBS up and down to gently swirl the eye-brain complexes. In this and all subsequent washing steps, do not directly pipette eye-brain complexes up and down as this will damage the fragile eye tissue.
- Transfer the eye-brain complexes in a minimal volume (<20 µL) of PBS into at least 250 µL of fixative. Mix by pipetting the solution up and down. Incubate for 35 min, on ice.
NOTE: The same lubricated tip prepared in step 3.1.1 can be used for this transfer. - With the same pipette tip, transfer the eye-brain complexes into a well containing ~400 µL of PBS. Mix by pipetting the solution up and down and incubate for 5-10 min on ice, to wash. Repeat the washing step at least twice.
NOTE: Eye-brain complexes can be maintained for several hours in the final PBS wash, on ice, or at 4 °C, before proceeding to Step 3.3.1.
- Antibody Staining
- To block the tissue prior to exposure to antibody solutions, transfer the eye-brain complexes to ~400 µL of PBT. Use the lubricated tip and a P20 or P100 air displacement micropipette for the transfer. Incubate for 10 to 60 min, on ice.
- Prepare the primary antibody solution by diluting appropriate antibodies in PBT. Since eye-brain complexes are incubated in 10 µL aliquots of antibody solution, the volume prepared will be a replicate of 10.
- Aliquot 10 µL of antibody solution into a clean well of a 72-well microwell tray (see discussion).
- Cut the tip of a P10 pipette tip with a clean razor blade to increase the tip-opening to ~0.5 mm in radius and lubricate by pipetting PBT up and down.
- Transfer no more than 5 eye-brain complexes, in a volume of <3 µL of PBS, into each 10 µL well of antibody solution using a P10 air displacement micropipette and the lubricated tip.
- Homogenize the antibody solution by pipetting the solution up and down. Do not pipette the eye-brain complexes up and down as this will damage the tissue.
- To minimize evaporation of the antibody solution, place a small piece of tissue soaked in distilled water into the microwell tray and seal the tray (e.g., with the lid provided). Incubate overnight at 4 °C.
- Transfer the eye-brain complexes from the microwell tray into ~400 µL of PBT in a 9-welled glass dish, to wash. Use a P10 air displacement micropipette and PBT-lubricated tip (prepare as described in step 3.3.4). Mix by pipetting the solution up and down. Incubate for 5-10 min on ice.
- Repeat the washing step at least twice. Use a P20 or P100 air displacement micropipette and PBT-lubricated tip to transfer the eye-brain complexes between wash solutions.
- Prepare the secondary antibody solution by diluting appropriate fluorophore-tagged secondary antibodies in PBT as required. 100 µL of secondary antibody solution is sufficient per batch of 6-10 pupae. Aliquot the secondary antibody solution into a 9-welled glass dissection dish.
- Transfer the eye-brain complexes into secondary antibody solution. Use a P20 or P100 air displacement micropipette and PBT-lubricated tip for the transfer.
- Incubate the eye-brain complexes in secondary antibody solution for 1-2 h at room temperature, or 3-5 h at 4 °C. To prevent bleaching of fluorophores by light, cover the preparation with foil.
NOTE: Optimal duration of incubation in secondary antibody solution may differ according to the primary and secondary antibodies used. - Transfer the eye-brain complexes into ~400 µL of PBT in a 9-well glass dish, to wash. Mix by pipetting the solution up and down. Incubate for 5-10 min on ice. This washing step should be repeated at least twice.
- Secondary Fixation
- For a secondary fixation, transfer the eye-brain complexes to at least 200 µL of fixative and incubate for 20 min at room temperature or 35 min at 4 °C.
NOTE: Secondary fixation stiffens the eye tissue, which aids mounting (step 3.5). - Use a P20 or P100 air displacement micropipette and PBT-lubricated tip to transfer eye-brain complexes into ~400 µL of PBT in a 9-well glass dish, on ice, to wash for 5 min.
- Repeat the washing step at least once.
- Use a P20 or P100 air displacement micropipette and PBT-lubricated tip to transfer the eye-brain complexes into ~400 µL of PBS in a 9-well glass dish, on ice, to rinse for 1-2 min. Keep the tissue in motion by pipetting PBS, but not the tissue, up and down to prevent eye-brain complexes from settling and adhering to the glass dish during PBS-rinse.
- For a secondary fixation, transfer the eye-brain complexes to at least 200 µL of fixative and incubate for 20 min at room temperature or 35 min at 4 °C.
- Mounting
- Transfer the eye-brain complexes into ~200 µL of mounting media. Use a P20 or P100 air displacement micropipette and PBT-lubricated tip to transfer the eye-brain complexes. Allow the tissue to equilibrate in mounting media for 1-2 h.
- Place a 5-8 µL drop of fresh mounting media onto a clean microscope slide.
- Cut a P10 tip with a clean razor blade to increase the tip-opening to ~0.5 mm in radius and use this and a P10 air displacement micropipette to transfer the eye-brain complexes in 5-8 µL of mounting media into the drop of mounting media on the slide.
- Use two tungsten needles to separate the eyes from optic lobes. Pin an eye-brain complex to the slide with a sturdy tungsten needle and slice each eye away from its associated optic lobe with a fine tungsten needle (Figure 2F).
- Use the fine tungsten needle to maneuver each eye to the surface of the microscope slide with the basal surface of each eye adjacent to the slide. Gently lower a clean cover-slip over the tissue and secure with nail polish.
NOTE: Arranging the eyes on the slide so that they are close to each other or in a line will facilitate subsequent microscopy. - Image the retinas using fluorescent or confocal microscopy.
NOTE: Slides should be stored at 4 °C if not imaged immediately.
4. Preparation of Eye Tissue for Western Blotting:
- Dissect 10-16 pupae (steps 2.1-2.9) with the following modifications: a) gather and dissect the pupae in two batches, 10-15 min apart and b) dissect in PBS supplemented with protease and phosphatase inhibitors (PBS+pi).
- Transfer the eye-brain complexes using a P20 or P100 air displacement micropipette and lubricated tip (prepared as in step 3.2.1) into ~400 µL of PBS+pi in a 9-well glass dish, on ice, to rinse briefly.
NOTE: This lubricated tip can be used for subsequent steps 4.3 and 4.4. - Repeat the rinse step twice.
- Transfer all eye-brain complexes into ~400 µL of ice cold PBS+pi decanted onto a clean black dissecting dish.
- Remove each eye from the optic lobes using a sturdy tungsten needle (to steady the eye- brain complex) and a fine razor blade or microdissection scissors to cleanly cut each eye from the optic lobe.
NOTE: Eyes are ‘sticky’ at this stage and may adhere lightly to the dissecting dish. This is advantageous as it can help facilitate the removal of the eyes from the optic lobes (see discussion). - ‘Sweep’ all eyes into a ‘pile’ in the PBS+pi on the dissecting dish, using one blade of a fine pair of forceps.
- Cut a P10 tip with a clean razor blade to widen the tip-opening to ~0.5 mm in radius and lubricate the tip-interior by pipetting Western Tissue Lysis Buffer (WTLB) up and down.
- Transfer the eye-brain complexes in 5 µL of PBS+pi into a 500 µL microcentrifuge tube. Use this lubricated tip and a P10 air displacement micropipette to complete the transfer.
- Place the microcentrifuge tube on ice. Add 1 volume (5 µL) of WTLB to the sample, and 10 µL of 2x concentrated protein sample buffer (or 2.5 µL of 5x concentrated protein sample buffer). Mix by tapping.
- Vortex the microcentrifuge tube briefly and spin down sample using a desktop mini-centrifuge.
- Store the sample at -20 °C until analysis. Analyze using standard protocols.
5. Preparation of Eye Tissue for RNA Extraction:
- Wearing gloves, clean benchtop, microscope, dissection tools, and black dissecting dish first with 70% ethanol and then RNase decontamination reagent. Allow to dry.
- Dissect 30-40 pupae as described (steps 2.1-2.9) with the following modifications: a) gather and dissect 6 to 8 pupae in batches, 15-20 min apart; b) dissect in nuclease-free PBS and c) isolate each batch of eyes separately but accumulate all retinal tissue into the same microcentrifuge tube (step 5.5).
NOTE: Having two people dissect concurrently will accelerate efficient isolation of good eye tissue. - For each batch, isolate the eye-brain complexes (steps 2.1-2.9) and transfer into ~400 µL of nuclease-free PBS in a 9-well glass dish to rinse briefly, on ice, using a P20 or P100 air displacement micropipette and lubricated tip (prepared as described in step 3.1.2).
NOTE: This lubricated tip can be used for subsequent steps 5.4, 5.5, and 5.8. - Repeat the rinse step twice.
- Transfer the eye-brain complexes into a fresh 400 µL drop of ice-cold nuclease-free PBS on a clean black dissecting dish.
- Remove each eye from the optic lobes using a sturdy tungsten needle (to steady the eye brain complex) and a fine razor blade or microdissection scissors to cleanly cut each eye from the optic lobe.
NOTE: Eyes are ‘sticky’ at this stage and may adhere lightly to the dissecting dish. This is advantageous as it can help facilitate the removal of the eyes from the optic lobes (see Discussion). - ‘Sweep’ all eyes into a ‘pile’ in the nuclease-free PBS on the dissecting dish, using one blade of a fine pair of forceps.
- Transfer the eyes to a sterile RNase-free microcentrifuge tube and flash-freeze in liquid nitrogen.
- Repeat dissection (steps 5.3-5.8) of each batch of pupae. Transfer each batch of eyes into the same microcentrifuge tube that has been maintained in liquid nitrogen (step 5.5) to accumulate all eyes in one tube.
- Store the samples at -80 °C until RNA extraction.
Representative Results
The pupal eye is an easily-accessible tissue that serves as an excellent model to investigate developmental processes that drive morphogenesis. Here, we have dissected retinas and used immunofluorescence to detect the apical adherens junctions (Figure 3A, C) or the Dcp-1 caspase (Figure 3D) that is activated during apoptosis (Figure 3)25. These approaches allow one to clearly observe cells during key morphogenetic processes including the recruitment and morphogenesis of primary cells (from 18 h APF), the intercalation of lattice cells around each ommatidium (18-24 h APF), the establishment of the tertiary niche (21-24 h APF), changes in cell size and shape (from 18 h to 40 h APF), and apoptosis (from 18 to ~33 h APF). The timing of these morphogenetic events is temperature-dependent and will therefore vary modestly in response to minor differences in incubator temperatures in different laboratory settings. However, by around 40 h APF, the final arrangement of cells is usually achieved (Figure 3B, C) and this is an ideal age at which to assess the consequences of genetic mutation or modified gene expression. For example, following ectopic expression of Diap1, a core inhibitor of apoptosis caspase activation (Figure 3C)26, our approach allows one to quantify the consequent increase in the number of lattice cells when compared to a control GMR>lacZ retina. One can also easily assess apoptosis more directly by utilizing the anti-Dcp-1 antibody or other approaches to detect dying cells (Figure 3D).
Eye lysate can be interrogated using Western analysis to determine the presence and/or relative expression of proteins of interest. Here, we show the detection of DE-cadherin, the core component of adherens junctions, in wild type Canton S retinas dissected at 21 and 40 h APF eyes (Figure 4A). Quantification of this sample Western blot (right, Figure 4A) revealed that the relative expression of DE-Cadherin, did not differ substantially at these two time-points, when band intensity is normalized according to expression of GAPDH (a core metabolic enzyme). Such analyses would usually be performed in triplicate rather than just once, as shown here.
It is essential to isolate high-purity mRNA from Drosophila pupal retinas if one’s aim is to analyze gene expression using advanced sequencing applications (e.g., Next-Gen, RNA-seq). We have found that dissecting more than 60 eyes per genotype or experimental condition is optimal if one’s goal is to isolate >1 mg of high-quality RNA (Figure 4B). Here, we show an example of an absorbance spectra of an RNA sample isolated from 57 wild type Canton S eyes, dissected at 40 h APF (Figure 4B, top panel). The peak absorbance at 260 nm corresponds to the absorbance wavelength of RNA. We also present a table reflecting the RNA yield and A260/A280 and A260/A230 purity ratios of six RNA samples, gathered at either 21 h or 40 h APF. These data reflect subtle differences in RNA yield when extracting RNA.
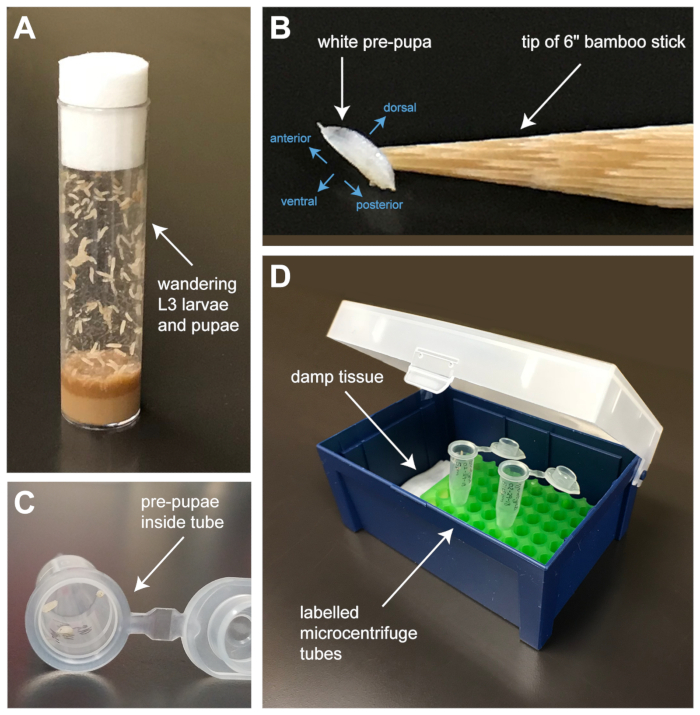
Figure 1: Selection and culture of pupae for dissection. (A) Wandering third larval instar (L3) larvae and pupae locate along the sides of healthy Drosophila cultures. (B) Pre-pupae can be identified by their translucent white color as pigment has yet to be generated in the protective pupal case. Anterior-posterior and dorsal-ventral axis of the pupa are shown in blue. A damp bamboo splint is used to dislodge and pick pre-pupae from the vial walls. (C) Pupae are placed inside 1.5 mL microcentrifuge tubes that are labelled appropriately (genotype, date of collection, and time of collection) and (D) cultured inside a humidified chamber assembled from an empty pipette-tip box. Humidity is maintained by placing a piece of damp tissue inside the box. Please click here to view a larger version of this figure.
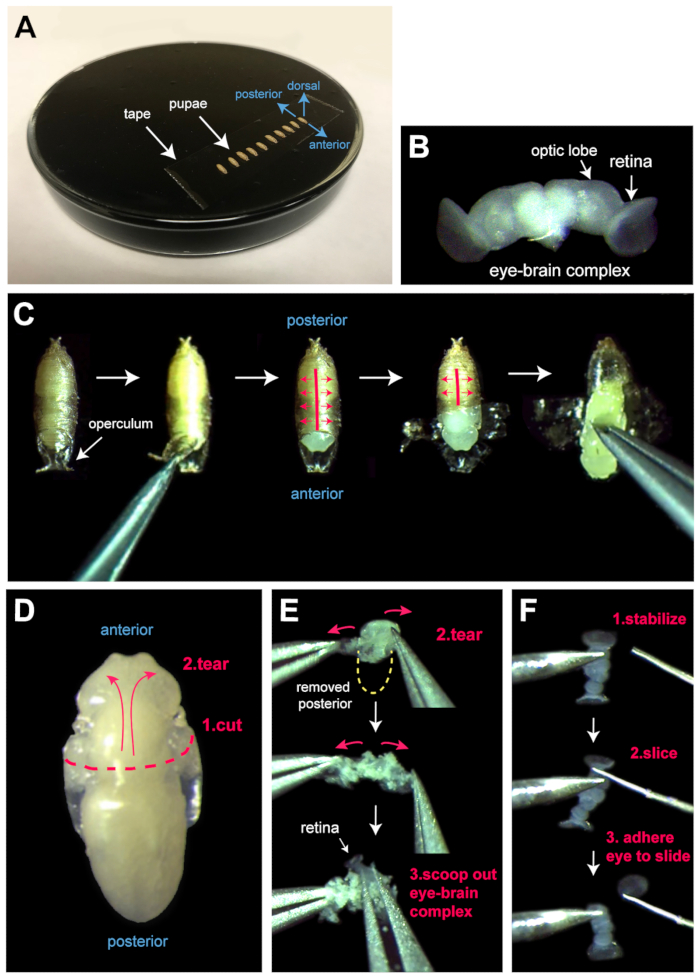
Figure 2: Dissecting the pupal eye. (A) Pupae are adhered to double-sided tape on a black dissecting dish. Anterior, posterior, and dorsal coordinates are indicated in blue. (B) To isolate eye-brain complexes (a single one is shown here), (C) the pupa is first removed from its pupal case. Important steps in this process are shown. Red lines indicate where to tear and open the pupal case after the operculum is removed. The pupae are then removed from the torn pupal case with forceps. (D) An exposed pupa is first cut along the thorax with microdissection scissors (position indicated with dashed red line) and the head epithelium then carefully torn open (red arrows), as shown in (E) to reveal the opaque eye-brain complex. (F) Following incubation with appropriate antibodies, retinas are sliced from eye-brain complexes. Important steps in this process are shown. The eye-brain is stabilized with a sturdy tungsten needle (left) and the retinas removed with a fine tungsten needle (right). For protein and RNA analyses, unfixed retinas can be cleanly cut from optic lobes using a fine razor blade or microdissection scissors, rather than a fine tungsten needle. Please click here to view a larger version of this figure.
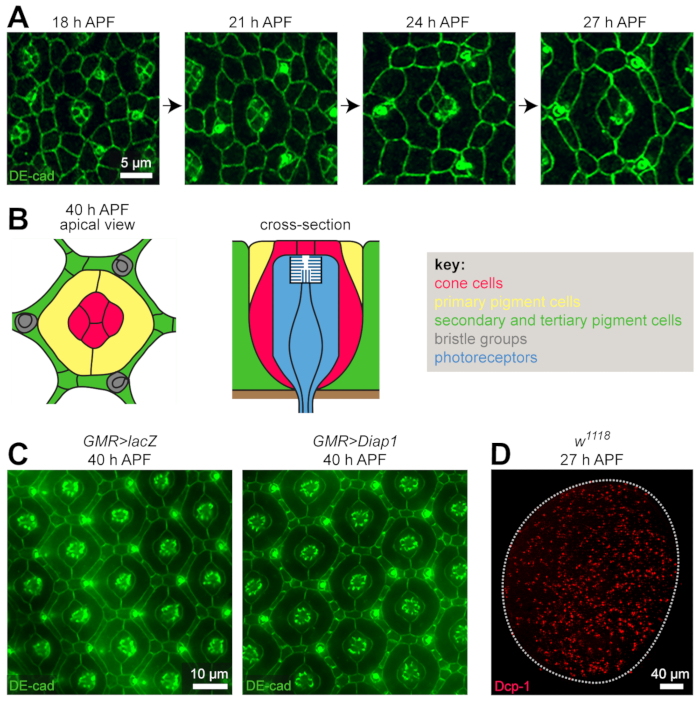
Figure 3: Immunofluorescence of the pupal eye. (A) Antibodies to DE-cadherin label apical adherens junctions of cells of the pupal eye. All images in panel A were gathered in the central region of a retina using confocal microscopy, minimally modified with an image processing software, and are presented at the same scale (scale bar = 5 µm). Note growth and rearrangement of cells from 18 h APF to 27 h APF. (B) Cartoon of the apical view of a single fully-patterned ommatidium at 40 h APF (left) and an ommatidium in cross-section. Cell types are color-coded as listed in the key. The photoreceptors are buried below the surface of the pseudostratified neuroepithlium and surrounded by cone and pigment cells. Three bristle groups surround each ommatidium. (C) Small regions of a control retina in which lacZ was expressed (left) or Diap1 (right) to inhibit apoptosis of secondary and tertiary pigment cells. Labeling of adherens junctions with anti-DE-cadherin enables the analysis of cell number, arrangement, and shape. Images were captured using standard fluorescent microscopy. (D) An entire retina incubated with antibodies to activated Dcp-1, a caspase activated during apoptosis, which prunes numerous cells from the eye. Image was captured using confocal microscopy. White dotted line outlines the eye. Please click here to view a larger version of this figure.
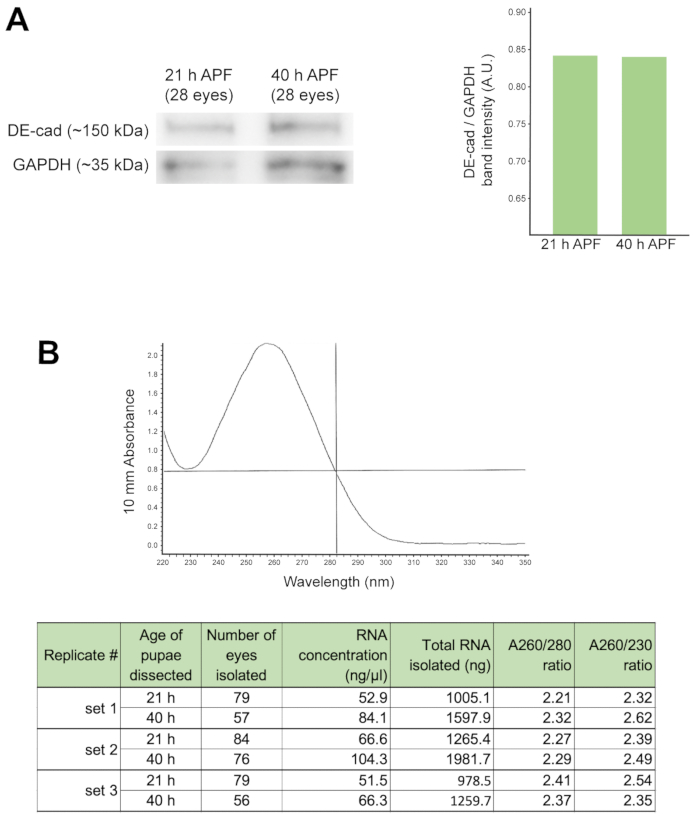
Figure 4: Protein and gene expression analyses of the pupal eye. (A) Western blot (left) of 28 eyes dissected at 21 h APF or 40 h APF, probed with antibodies to DE-cad and, as a loading control, GAPDH. Analyses of protein band intensities (right) reveals comparable concentrations of DE-cad present in these groups of retinas, relative to GAPDH. (B) Single absorbance spectrum of an RNA sample extracted from Canton S retinas dissected at 40 h APF (top). Note absorbance peak at 260 nm. Table documenting the amount and purity ratios of RNA extracted from Canton S pupal retinas isolated at 21 and 40 h APF (bottom). These data arise from three independent sample sets. Pupae for each sample set were raised and incubated in the same conditions and dissected on the same day. Please click here to view a larger version of this figure.
Discussion
The method of Drosophila pupal eye dissection described here allows for the isolation of 6 to 10 eye-brain complexes within 10 to 15 min. However, patience and practice are essential in order to master the dissection technique and improve the quality and speed of dissections. This short dissection time ensures that each eye is approximately the same developmental stage, reducing variability in the phenotype or gene expression of retinas in a data set. Whilst alternative protocols may require less practice to master, our protocol is designed to methodically isolate delicate retinal tissue while minimizing the risk of tearing or shearing, because damage to the eye could induce stress pathway activation or even cell death. We advise beginners to first master the dissection of pupae cultured to 40 h APF before attempting to dissect younger eyes. We recommend setting up replicate crosses for all experiments (step 1.1) which will ensure that there are always sufficient pupae available for collection (step 1.3). Retinas can be dissected at a variety of timepoints during pupal development, depending on the distinct morphogenetic events of interest to the researcher. These may include the recruitment and morphogenesis of primary cells (from 18 h APF), intercalation of lattice cells (18-24 h APF), establishment of the tertiary niche (21-24 h APF), changes in cell size and shape (from 18 h to 40 h APF) and apoptosis (from 18 to ~33 h APF) (Figure 3A).
If during initial opening of the pupal case (step 2.2, Figure 2C), the thorax of a pupa is punctured, the researcher should still be able to proceed with the dissection. However, if the head is initially punctured, extracting undamaged eye-brain complexes may be difficult. When removing a pupa from its pupal case (step 2.3, Figure 2C, right panel), the abdominal pupal case can sometimes dislodge from the double-sided tape and remain wrapped about the pupa’s abdomen. This is unlikely a hindrance, although the pupa may float when surrounded by PBS until the abdomen and attached pupal case are severed from the pupa (step 2.6). To preserve the integrity of the retinal tissue, it is imperative that it is fixed or frozen as quickly as possible after the initial puncture of the pupae. In addition, using ice-cold solutions and maintaining the tissue on ice (or at 4 °C) wherever possible throughout the protocol will preserve the tissue and cellular integrity, leading to better immunofluorescence, protein-analysis or RNA-analysis. For the latter applications, it is important to remove all brain and fat tissue from eyes as these tissues will contaminate analyses (step 2.9 and 4.3 or 5.4).
The pupal eye-brain complex can adhere to unlubricated pipette tips, glass and dissection tools when unfixed or during PBS washes/rinses, leading to tissue damage. To prevent this, we recommend meticulous lubrication of pipette tips and not washing 9-well glass dishes with ethanol (dish soap should also be avoided). Instead, simply clean glass dishes with distilled H2O and lubricate, if needed, with a PBT rinse prior to use. Dissection forceps, scissors and needles should also be mainly cleaned with distilled H2O, except when preparing these for RNA-extraction dissections (step 5.1). However, light adhesion between retinas and glass microscope slides can assist in unfurling eyes and positioning them on slides when separating eyes from optic lobes during mounting (step 3.5.4 and 3.5.5). Similarly, light adhesion of eyes to black dissecting dishes can be used to flatten and very lightly stretch the tissue which facilitates clean severing of eye-optic lobe connections when isolating retinas for protein or RNA analyses (step 4.5 and 5.6). We have found that microdissection scissors or a fine razor blade are excellent tools to quickly and cleanly cleave unfixed eyes from optic lobes.
During primary antibody staining, rather than incubating eye-brain complexes in 72-well micro-well trays (step 3.3.5), a 9-well glass dish can instead be used. To prevent evaporation of the antibody solution overnight, be sure to cap the glass dish wells with a cover slip or slide. However, this approach will require 40-50 µL of primary antibody solution for a batch of 6-10 eye-brain complexes incubated in one well of 9-well glass dish, rather than 20 µL of primary antibody solution distributed between 2 wells of a 72-well micro-well tray.
Secondary fixation of eye-brain complexes (step 3.4) prior to mounting eyes on microscope slides (step 3.5) will marginally stiffen the tissue so that it is easier to cleave the eyes from optic lobes and the eyes are less likely to fold or adhere to dissection needles. After a 1-2 h incubation in mounting medium (step 3.5.1), the eye-brain complexes become slightly opaque and hence easier to see whilst mounting. In addition, after 1-2 h in mounting medium, most secondary antibodies will be suitably protected from photo-bleaching during fluorescent imaging. Incubating retinas overnight in mounting media before mounting is not recommended as this renders retinas soft, negating the stiffening effect of secondary fixation.
For Western analyses, the lysate of at least 20 eyes is required to reliably detect proteins using antibodies that robustly recognize Drosophila proteins (e.g., Figure 4A). However, a larger number of eyes may be required if there is low expression of the target protein of interest. If lysis of the tissue for subsequent Western blotting is incomplete (step 4.7), the volume of WTLB added to the sample can be marginally increased and the volume of concentrated sample buffer adjusted accordingly (step 4.9). For RNA extraction, rapid dissection of a significant number of eyes may be required if the goal is to isolate a large amount of high-quality RNA (step 5.2, see Figure 4B). This potential drawback can be overcome if two scientists who have mastered Drosophila pupal-eye dissection collaborate to dissect these large numbers of fly retinas simultaneously. However, the total amount of RNA required will depend on the requirements of the institution or facility performing RNA analysis, the type of RNA analysis or sequencing, and the RNA extraction kit that is used.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank Zack Drum and our Reviewers for helpful comments on the manuscript. This work was supported by R15GM114729.
Materials
| Adobe Photoshop | Adobe | Image processing software | |
| Bamboo splints, 6" | Ted Pella Inc | 116 | |
| Beta mercaptoethanol | Sigma-Aldrich | M3148 | |
| Beta-glycerol phosphate | Sigma-Aldrich | 50020 | |
| Black dissecting dish | Glass petri dish filled to rim with SYLG170 or SYLG184 (colored black with finely ground charcoal powder). Leave at room temperature for 24-48 h to polymerize. | ||
| Blade holder | Fine Science Tools | 10053 | |
| Bovine serum albumin | Sigma-Aldrich | A7906 | |
| cOmplete, EDTA-free protease inhibitor cocktail tablets | Roche | 4693132001 | |
| Confocal microscope (Zeiss LSM 501) | Carl Zeiss | or similar microscope | |
| Diethyl Pyrocarbonate (DEPC) | Sigma-Aldrich | 40718 | |
| Double-sided tape | 3M | 665 | |
| Drosophila food media, nutrient-rich | 7.5% sucrose, 15% glucose, 2.5% agar, 20% brewers yeast, 5% peptone, 0.125% MgSO4.7H2O, 0.125% CaCl2.2H20 | ||
| Drosophila food media, standard | Bloomington Drosophila Stock center cornmeal recipe. (https://bdsc.indiana.edu/information/recipes/bloomfood.html) | ||
| Ethylenediaminetetraacetic acid | Sigma-Aldrich | E6758 | |
| Fixative solution | 4% formadehyde in PBS, pH 7.4. | ||
| Fluorescence microscope (TCS SP5 DM microscope) | Leica Microsystems | or similar microscope | |
| Forceps | Fine Science Tools | 91150-20 | Forceps should be sharpened frequently. |
| Formaldehyde | Thermo Scientific | 28908 | |
| Glass 9-well dishes | Corning | 7220-85 | Also known as 9-well dishes |
| Glass coverslips (22 x 22 mm) | Fisher Scientific | 12-542-B | |
| Glass microscope slides (25 x 75 x 1 mm) | Fisher Scientific | 12-550-413 | |
| Glass petri dish | Corning | 3160-100BO | |
| Glycerol | Sigma-Aldrich | G5516 | |
| Image Studio software version 5.2.5 | LI-COR Biosciences | Image processing software for quantitation of Western blots. | |
| Laemmli sample buffer | Bio-Rad | 161-0737 | 2X concentrated protein sample buffer, supplement with beta mercaptoethanol as per manufacturer's instructions. |
| Lane marker reducing sample buffer | ThermoFisher Scientific | 39000 | 5X concentrated protein sample buffer. |
| Microcentrigure tubes | Axygen | MCT-175-C | |
| Microdissection scissors | Fine Science Tools | 15000-03 | |
| Microwell trays (72 x 10 µL wells) | Nunc | 438733 | |
| Mounting media | 0.5% N-propylgallate and 80% glycerol in PBS | ||
| N-propylgallate | Sigma-Aldrich | P3130 | |
| Nuclease-free PBS (PBS in 0.1% DEPC, pH 7.4) | Add appropriate volume of DEPC to PBS, mix well and incubate overnight at room temperature with constant stirring. Autoclave for at least 20 minutes. Store at 4°C | ||
| PBS (phosphate buffered saline pH 7.4) | Sigma-Aldrich | P5368 | Prepare according to manufacturer's instructions |
| PBS+pi (PBS plus protease and phoshatase inhibitors) | 10mM NaF, 1mM beta-glycerol phosphate and 1mM Na3VO4 in PBS, pH 7.4. | ||
| PBT | 0.15% TritonX and 0.5% bovine serum albumin in PBS, pH 7.4 | ||
| Pin holder | Fine Science Tools | 26016-12 | |
| Primary antibody: goat anti-GAPDH | Imgenex | IMG-3073 | For Western blotting. Used at 1:3000 |
| Primary antibody: rabbit anti-cleaved Dcp-1 | Cell signaling | 9578S | For immunofluorescence. Used at 1:100 |
| Primary antibody: rat anti-DEcad | Developmental Studies Hybridoma Bank | DCAD2 | For immunofluorescence. Used at 1:20 |
| Primary antibody: rat anti-DEcad | DOI: 10.1006/dbio.1994.1287 | DCAD1 | Gift from Tadashi Uemura. Used at 1:100. |
| RNA extration kit: Relia Prep RNA tissue Miniprep kit | Promega | Z6110 | |
| Rnase decontamination reagent (RNase Away) | Molecular BioProducts | 7002 | |
| Scalpel blades | Fine Science Tools | 10050 | Break off small piece of scapel blade and secure in blade holder. |
| Secondary antibody: 488-conjugated donkey anti-rat IgG (H+L) | Jackson ImmunoResearch | 712-545-153 | For immunofluorescence. Used at 1:200 |
| Secondary antibody: cy3-conjugated goat anti-rabbit IgG (H+L) | Jackson ImmunoResearch | 111-165-144 | For immunofluorescence. Used at 1:100 |
| Secondary antibody: HRP-conjugated goat anti-rat IgG (H+L) | Cell Signaling Technology | 7077 | For Western blotting. Used at 1:3000 |
| Secondary antibody: HRP-conjugated rabbit anti-goat IgG (H+L) | Jackson ImmunoResearch | 305-035-003 | For Western blotting. Used at 1:3000 |
| Sodium Chloride | Sigma-Aldrich | S3014 | |
| Sodium Fluoride | Sigma-Aldrich | 215309 | |
| Sodium vanadate | Sigma-Aldrich | 50860 | |
| Spectrophotometer (NanoDrop) | ThermoFisher Scientific | 2000c | |
| Stereo dissecting microscope (M60 or M80) | Leica Microsystems | or similar microscope | |
| Sylgard (black) | Dow Corning | SYLG170 | |
| Sylgard (transparent) | Dow Corning | SYLG184 | Color black with finely ground charcol powder |
| Tissue: Kimwipes | KIMTECH | 34120 | |
| TritonX | Sigma-Aldrich | T8787 | |
| Trizma hydrochloride pH7.5 | Sigma-Aldrich | T5941 | |
| Tungsten needle, fine | Fine Science Tools | 10130-10 | Insert into pin holder |
| Tungsten needle, sturdy | Fine Science Tools | 10130-20 | Insert into pin holder |
| WTLB (western tissue lysis buffer) | 150mM NaCl, 1.5% Triton X-100, 1mM EDTA, 20% glycerol, 10mM NaF, 1mM beta-glycerol phosphate and 1mM Na3VO4 in 50mM Tris-HCl (pH 7.5). Supplement with one cOmplete protease cocktail table per 10 mL solution. | ||
| Yeast paste | (local supermarket) | Approximately 2 tablespoons Fleischmann's ActiveDry Yeast (or similar) dissolved in ~20 mL distilled H2O |
Riferimenti
- Cagan, R. L., Ready, D. F. The emergence of order in the Drosophila pupal retina. Developmental biology. 136 (2), 346-362 (1989).
- Wolff, T., Ready, D. . The development of Drosophila melanogaster. , 1277-1325 (1993).
- Carthew, R. W. Pattern formation in the Drosophila eye. Current opinion in genetics & development. 17 (4), 309-313 (2007).
- Kumar, J. P. Building an ommatidium one cell at a time. Developmental Dynamics. 241 (1), 136-149 (2012).
- Hayashi, T., Carthew, R. W. Surface mechanics mediate pattern formation in the developing retina. Nature. 431, 647 (2004).
- Bao, S., Cagan, R. Preferential Adhesion Mediated by Hibris and Roughest Regulates Morphogenesis and Patterning in the Drosophila Eye. Developmental Cell. 8 (6), 925-935 (2016).
- Cordero, J. B., Larson, D. E., Craig, C. R., Hays, R., Cagan, R. Dynamic Decapentaplegic signaling regulates patterning and adhesion in the Drosophila pupal retina. Development (Cambridge, England). 134 (10), 1861-1871 (2007).
- Larson, D. E., Liberman, Z., Cagan, R. L. Cellular behavior in the developing Drosophila pupal retina. Mechanisms of development. 125 (3-4), 223-232 (2008).
- Martín-Bermudo, M. D., Bardet, P. L., Bellaïche, Y., Malartre, M. The vav oncogene antagonises EGFR signalling and regulates adherens junction dynamics during Drosophila eye development. Development. 142 (8), 1492-1501 (2015).
- Chan, E. H., Chavadimane Shivakumar, P., Clément, R., Laugier, E., Lenne, P. F. Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye. eLife. 6, e22796 (2017).
- Lin, H. V., Rogulja, A., Cadigan, K. M. Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development. 131 (10), 2409-2418 (2004).
- Cordero, J., Jassim, O., Bao, S., Cagan, R. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mechanisms of development. 121 (12), 1523-1530 (2004).
- Mendes, C. S., et al. Cytochrome c‐d regulates developmental apoptosis in the Drosophila retina. EMBO reports. 7 (9), 933-939 (2006).
- Monserrate, J., Brachmann, C. B. Identification of the death zone: a spatially restricted region for programmed cell death that sculpts the fly eye. Cell Death & Differentiation. 14 (2), 209-217 (2007).
- Bushnell, H. L., et al. JNK is antagonized to ensure the correct number of interommatidial cells pattern the Drosophila retina. Biologia dello sviluppo. 433 (1), 94-107 (2018).
- Wolff, T. Dissection techniques for pupal and larval Drosophila eyes. CSH Protoc. 2007, (2007).
- Hsiao, H. Y., et al. Dissection and Immunohistochemistry of Larval, Pupal and Adult Drosophila Retinas. Journal of visualized experiments : JoVE. (69), e4347 (2012).
- Tea, J. S., Cespedes, A., Dawson, D., Banerjee, U., Call, G. B. Dissection and Mounting of Drosophila Pupal Eye Discs. Journal of Visualized Experiments : JoVE. (93), e52315 (2014).
- Johnson, R. I., Cagan, R. L. A Quantitative Method to Analyze Drosophila Pupal Eye Patterning. PLoS ONE. 4 (9), e7008 (2009).
- Greenspan, R. J. . Fly pushing : the theory and practice of Drosophila genetics. , (2004).
- Brand, A. H., Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118 (2), 401-415 (1993).
- Ellis, M. C., O’Neill, E. M., Rubin, G. M. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 119 (3), 855-865 (1993).
- Duffy, B. J. GAL4 system in drosophila: A fly geneticist’s swiss army knife. genesis. 34 (1-2), 1-15 (2002).
- Li, W. Z., Li, S. L., Zheng, H. Y., Zhang, S. P., Xue, L. A broad expression profile of the GMR-GAL4 driver in Drosophila melanogaster. Genet Mol Res. 11 (3), 1997-2002 (2012).
- Song, Z., McCall, K., Steller, H. DCP-1, a Drosophila Cell Death Protease Essential for Development. Science. 275 (5299), 536-540 (1997).
- Hay, B. A., Wassarman, D. A., Rubin, G. M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 83 (7), 1253-1262 (1995).

