Human Colonoid Monolayers to Study Interactions Between Pathogens, Commensals, and Host Intestinal Epithelium
Summary
Here, we present a protocol to culture human enteroid or colonoid monolayers that have intact barrier function to study host epithelial-microbiota interactions at the cellular and biochemical level.
Abstract
Human 3-dimensional (3D) enteroid or colonoid cultures derived from crypt base stem cells are currently the most advanced ex vivo model of the intestinal epithelium. Due to their closed structures and significant supporting extracellular matrix, 3D cultures are not ideal for host-pathogen studies. Enteroids or colonoids can be grown as epithelial monolayers on permeable tissue culture membranes to allow manipulation of both luminal and basolateral cell surfaces and accompanying fluids. This enhanced luminal surface accessibility facilitates modeling bacterial-host epithelial interactions such as the mucus-degrading ability of enterohemorrhagic E. coli (EHEC) on colonic epithelium. A method for 3D culture fragmentation, monolayer seeding, and transepithelial electrical resistance (TER) measurements to monitor the progress towards confluency and differentiation are described. Colonoid monolayer differentiation yields secreted mucus that can be studied by the immunofluorescence or immunoblotting techniques. More generally, enteroid or colonoid monolayers enable a physiologically-relevant platform to evaluate specific cell populations that may be targeted by pathogenic or commensal microbiota.
Introduction
Intestinal organoids, enteroids, and colonoids have led to many advances in understanding stem cell behavior, intestinal development, barrier/transport function, and cellular differentiation.1 However, 3D culture limits the study of host epithelial-pathogen interaction because the lumen is not directly accessible to bacteria or virulence factors unless the closed structures are subjected to microinjection.2,3 Additionally, secreted materials such as small molecules, proteins, or mucus cannot be easily sampled from 3D culture for the downstream analysis. The impact of pathogenic agents on epithelial barrier function4 and ion transport5 has been evaluated in 3D culture using fluorescent dyes and time-lapse microscopy, but monolayers grown on permeable tissue culture supports are amenable to additional techniques such as TER measurement and Ussing chamber/voltage clamp recording.6,7
Numerous publications have described protocols for 2D or monolayer culture of enteroids/colonoids. Materials found to promote epithelial cell attachment include collagen hydrogels,8,9 0.1% gelatin,10 thin-layer murine sarcoma-derived basement membrane matrix (BMM),11,12,13 and human collagen IV.6,7,14,15,16,17 Seeding approaches include mechanical fragmentation by pipetting6,14,15 and/or dissociation of cell adhesion factors using trypsin,10,11 dispase,13 or EDTA.9 A few protocols use non-porous tissue culture plasticware for seeding, but this restricts basolateral access, so most applications depend on permeable tissue culture inserts. Documentation of stable confluent monolayer formation and maintenance varies widely among the publications. Additionally, growth and differentiation media compositions for human cultures differ among various groups and continue to evolve as more researchers adopt and adjust the methodology to suit their application and available resources.
To address the limitations of 3D intestinal epithelial culture in host-pathogen interaction studies, we present a modified protocol for converting 3D human enteroids or colonoids to a monolayer. After achieving confluency in a crypt-like immature state, withdrawal of the growth factors WNT3A, RSPO1 and the inhibitors A-83-01 and SB 202190 leads to differentiation representative of small intestinal villus or surface colonic epithelium. We describe the ideal matrix, human collagen type IV, to coat the inserts and obtain uniform enteroid or colonoid fragments for plating. We demonstrate that this protocol produces a confluent monolayer with a high TER. Colonoid monolayers secrete a thick apical mucus layer, allowing for ex vivo studies of pathogen-mucus interaction via immunoblotting or immunostaining. A modified fixation procedure to preserve the colonic mucus layer for immunostaining is also described. This method aims to provide a tractable model to study the earliest intestinal host-pathogen interactions upon infection.
Protocol
This protocol is based on studies previously published by the authors.6,7,14,15,16 The following steps should be carried out in a sterile biosafety cabinet using proper aseptic techniques. All methods involving human specimens have been approved by the Institutional Review Board of Johns Hopkins University School of Medicine (IRB NA_00038329).
1. Coat cell culture inserts with extracellular matrix
- Prepare 5 mL of stock collagen IV solution (1 mg/mL) in 100 mM acetic acid. Let stand at 4 °C for approximately 4 h to fully hydrate/dissolve.
- Aliquot the collagen IV stock solution and store at 4 °C (≤ 1 month) or -20 °C (> 1 month).
- Immediately prior to coating, dilute the collagen IV stock solution in sterile tissue culture grade water to a final concentration of 34 μg/mL. For 24-well plate cell culture inserts, coat each insert with 100 μL of solution, which corresponds to 10 μg/cm2.
- Incubate the plate in a standard CO2 tissue culture incubator at 37 °C for ≥ 2 h, or for convenience, seal the plate edges with paraffin film and incubate at 4 °C overnight or up to 1 week.
2. Isolate enteroids/colonoids from 3D culture
NOTE: Enteroids or colonoids are established from donor biopsies and maintained in 3D culture according to standard protocols previously described.14,18 Briefly, crypts are harvested from intestinal biopsies or resections via chelation and mechanical agitation. The crypts are washed, harvested, and plated in BMM. Expansion media (described in step 4.2) is added to the culture and replaced every 2 days. 3D culture formation is visible within hours after plating.
- Aspirate the culture medium from the 24-well plate and replace with 1 mL ice-cold harvesting solution (see Table of Materials) per well.
- Use a mini cell scraper to dislodge and break-up the basement membrane matrix pellet, paying particular attention to any material near the well edges.
- Agitate the plate on an orbital shaker at approximately 200 rpm, 4 °C for 30-45 min.
3. Dissociate 3D enteroids/colonoids
- Use a P200 single channel pipette or a multi-channel pipette fitted with sterile filter tips to triturate the cell suspension.
- Pool the cell suspension(s) in a 15 mL conical vial. Use multiple vials if total suspension volume is > 6 mL.
- Add an equal volume of Advanced DMEM/F12 medium containing 10 mM HEPES, L-alanyl-L-glutamine dipeptide (1x), and penicillin-streptomycin (1x) (wash medium). Invert the tube 3-4 times to mix. Centrifuge at 300 x g, 10 min, 4 °C.
- Optionally, aspirate the wash medium and replace with 0.5 mL/well trypsin (see Table of Materials). Resuspend colonoids with a P200 pipette, cap the tube and place in a 37 °C water bath for approximately 2 min. Immediately remove the tube, add wash medium to a final volume of 10 mL, and repeat the centrifugation as in Step 3.3.
NOTE: This step may be preferable if trituration is not resulting in uniformly sized fragments.
- Optionally, aspirate the wash medium and replace with 0.5 mL/well trypsin (see Table of Materials). Resuspend colonoids with a P200 pipette, cap the tube and place in a 37 °C water bath for approximately 2 min. Immediately remove the tube, add wash medium to a final volume of 10 mL, and repeat the centrifugation as in Step 3.3.
4. Plating the enteroid/colonoid suspension
- If coated inserts were stored at 4 °C, equilibrate in a 37 °C/5% CO2 tissue culture incubator for at least 30 min prior to cell plating.
- For each insert to be plated, prepare 1 mL warm (between 25-37 °C) expansion medium (EM) and add 10 μM Y-27632 and 10 μM CHIR 99021.
NOTE: EM is composed of Advanced DMEM/F12 containing B27 (1x), 50% WNT3A conditioned medium, 15% RSPO1 conditioned medium, 10% Noggin conditioned medium, 50 ng/mL human EGF, 500 nM A-83-01, 10 μM SB 202190, antibiotic/antimycotic cocktail (1x), 10 mM HEPES, L-alanyl-L-glutamine dipeptide (1x), and penicillin-streptomycin (1x) (see Table of Materials). - Aspirate the wash medium from the tube containing the cell pellet and resuspend in a volume of EM sufficient to yield at least 100 μL/insert.
- Aspirate the collagen IV solution from each insert and wash twice with 150 μL of the wash medium per insert.
- Pipet 600 μL of EM into the space beneath each insert.
- Pipet 100 μL of cell suspension into each insert.
- Return the plate to the tissue culture incubator and leave undisturbed for at least 12 h.
NOTE: Avoid shaking or sharply tilting the plate to prevent uneven distribution of colonoid fragments on the insert membrane. - Monitor the cell attachment and spreading to form a confluent monolayer by placing the plate on a phase-contrast light microscope under a 2.5x-10x objective lens.
- After 1-2 days, most fragments should adhere to the collagen matrix. Refresh the culture medium and discontinue treatment with Y-27632 and CHIR 99021. Continue to refresh the medium every 2-3 days until confluent. Confluency is generally reached in 7-10 days.
5. Measure transepithelial electrical resistance
- Attach the electrode leads and power on the epithelial voltohmmeter (EVOM). Verify that the measurement function is set to ohms.
- Dip the electrode tips briefly in 70% ethanol and wipe dry with a laboratory tissue. Equilibrate the electrode in 5 mL of wash medium for approximately 5 min.
- Immerse the shorter electrode tip in the insert medium and orient the longer electrode tip into the lower well plate. Maintain the electrode in a full vertical orientation until the EVOM reading is relatively stable, or for no longer than 60 s.
NOTE: If the electrode assembly is tilted far from vertical, or the tip height is improperly adjusted, the shorter tip may come into contact with and disrupt the cell monolayer. - Compare EVOM measurements to the value obtained from a cell-free insert submerged in culture medium to evaluate progress toward confluency or differentiation.
6. Differentiation of confluent enteroid/colonoid monolayers
- Exchange EM for differentiation medium (DM), and continue to refresh media every two days until day 5. DM is EM that lacks WNT3A, RSPO1, A-83-01, and SB 202190.
- Continue to monitor TER to verify that differentiation is proceeding as expected. TER will continue to increase during days 1-5 of differentiation. Monolayers may continue to increase TER and retain viability beyond day 5-6 but exhibit maximal and most reproducible results when used at day 5-6.
7. Bacterial infection with commensal or pathogenic E. coli
- One day prior to carrying out the infection, wash and feed monolayers with antibiotic-free EM or DM.
- Use a sterile microbiology loop to gently scrape the surface of a frozen bacterial glycerol stock and inoculate 2 mL of LB broth. Transfer the tube to a standard shaking incubator at 37 °C for 12-16 h.
- Dilute 50 μL of the starter culture into 5 mL fresh LB broth (1:100) and continue incubation with shaking for 90 min. This will yield a log phase culture with a density of 105-106 colony-forming units (cfu)/mL.
- Optionally, confirm the bacterial concentration by optical density measurement at 600 nm (OD600) in a spectrophotometer.
- Spin down the bacterial culture at 12,000 x g for 10 min, remove the supernatant, and resuspend bacteria in antibiotic-free EM or DM to a final concentration of 107 cfu/mL.
- Add 10 μL of bacterial suspension to the insert (final concentration 106 cfu/mL). Pipet slowly to mix and avoid disturbing the extracellular mucus layer.
- Return the plate to a tissue culture incubator for the desired infection period.
8. Fixation to preserve and immunostain mucus
- Lift the cell culture inserts from the 24-well plate, carefully invert on a laboratory tissue paper to remove the apical medium, and wipe away external liquid clinging to the insert.
- In a new 24-well plate, immerse the insert in a 1:3 solution of glacial acetic acid in absolute ethanol (Clarke’s solution) for 10 min at room temperature.
- Invert the insert to remove the fixative and rehydrate the cells in 1x PBS for 10 min. Proceed with standard immunostaining protocols.
- Use a razor blade to cut around the perimeter of the insert membrane. Transfer the membrane to a glass microscope slide using forceps and affix a coverglass with mounting medium.
9. Preparation of cell lysates for immunoblotting
NOTE: Perform the following steps on ice or in a cold room.
- Aspirate medium and wash insert once with PBS.
- Add 150 μL lysis buffer containing protease inhibitors (see Table of Materials) to the insert and let stand 5-10 min. Lysis buffer contains 50 mM Tris pH 8, 150 mM NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 1:100 protease inhibitor cocktail.
- Use a mini cell scraper to remove the cells from the insert, then use a P200 pipette to briefly triturate the suspension and transfer to a microcentrifuge tube.
- Sonicate the suspension with 5 x 1 s pulses at 20% amplitude using a microtip probe (see Table of Materials).
- Add SDS-PAGE loading buffer if immediately performing electrophoresis, or aliquot and store lysates at -20 °C.
Representative Results
Human enteroid and colonoid cultures are grown as 3D structures, then dissociated and fragmented for plating on human collagen IV-coated cell culture inserts. The progress of monolayer formation is easily monitored on a daily basis via bright field microscopy, immunofluorescence staining (Figure 1), and by a steady increase in transepithelial electrical resistance (TER) (Figure 2), which reflects the permeability of tight junctions to ions and correlates with monolayer confluency. The TER of an empty 24-well insert is approximately 50-100 Ω·cm2, and upon reaching confluency increases to approximately 400-500 Ω·cm2. All epithelial cells in confluent monolayers are connected by junctional complexes detected by the F-actin ring at the cell perimeter (Figure 1). Monolayers in full growth factor media represent the proliferative crypt-like epithelium composed primarily of actively dividing cells that incorporate the nucleoside analog EdU (Figure 2). Withdrawal of the growth factors WNT3A and RSPO1 promotes differentiation, leading to villus-like cultures that lack proliferating cells. Differentiated monolayers contain enterocytes (enteroids) or colonocytes (colonoids) and develop specialized intestinal epithelial cells as has been shown in 3D cultures,18 including goblet and enteroendocrine cells.15 Differentiated monolayers also demonstrate a significant increase in TER (> 1000 Ω·cm2) (Figure 2), indicating mature tight junctions.
In normal human physiology, the intestinal epithelial layer separates nutrients and the microbe-enriched luminal space from the sterile serosal environment. The epithelium tightly regulates the cross-talk between these two compartments. Enteroid and colonoid monolayers preserve this important compartmentalization property as shown by the proteomic analysis in Table 1. There are substantial differences between the protein composition in apical and basolateral conditioned media collected from differentiated enteroid monolayers. Access to both the apical and basolateral sides allows for collection of both supernatants in a time dependent manner for measurement of differential secretion of other molecules such as cytokines and chemokines.15,17
Monolayers are convenient and highly reproducible models to detect the changes in protein expression using both immunoblotting and immunostaining. Thus, changes in mucin 2 (MUC2) expression by shRNA knockdown (KD) can be detected using both techniques (Figure 3). The shRNA transduction was performed on 3D colonoids and maintained in antibiotic selection media. After verifying the KD, colonoids can be continuously maintained as 3D cultures and/or plated as monolayers for experimental purposes. Moreover, MUC2 KD does not affect the efficiency of monolayer formation compared to the wild type parental colonoid line. Collection of monolayers for immunoblotting is simple and similar to the protocols described for human epithelial adenocarcinoma-derived cell lines. Typically, approximately 50 µg or more of total protein can be extracted from a single 0.33 cm2 insert-grown monolayer. As shown in Figure 3, MUC2 is barely detectable in undifferentiated (UD) WT colonoid monolayers but is highly expressed in differentiated (DF) WT monolayers. MUC2 is below the level of detection in both UD and DF colonoid monolayers transduced with MUC2 shRNA.
Importantly, monolayers are a suitable model to study host-microbial interactions at the apical surface of the epithelia. Colonoid monolayers form a thick attached MUC2-positive mucus layer upon differentiation which is not easily permeated by commensal E. coli HS bacteria (Figure 4), similar to what has been suggested in the normal human colon.19 However, enterohemorrhagic E. coli (EHEC), a human colonic pathogen, has been shown to have the ability to destroy the MUC2-enriched attached mucus layer to reach the apical surface of the epithelium (Figure 4).14,20 Remaining MUC2 is only present inside the goblet cells.
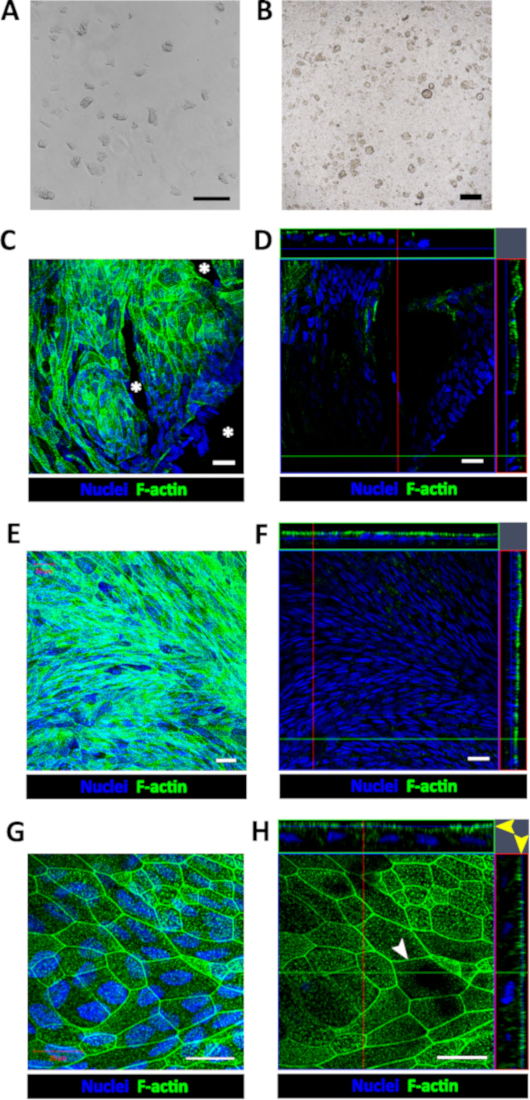
Figure 1: Establishment of human enteroid/colonoid monolayers. (A) Example of colonoid fragments after the dissolution of BMM and trituration. (B) Example of insert immediately after plating colonoid fragments. Scale bar (A-B) = 200 μm. Representative (C) maximum intensity projection and (D) confocal optical Z-section with the corresponding orthogonal projections show that colonoid fragments seeded onto human collagen IV-coated filters form multiple monolayer islands 2-4 days post-seeding. Cell-free areas (asterisk) are identifiable by the absence of both nuclear (Hoechst 33342, blue) and apical F-actin (phalloidin, green) staining in both C and D. Representative (E) maximum intensity projection and (F) confocal optical section with the corresponding orthogonal projections show a confluent colonoid monolayer with continuous apical surface detected by F-actin immunostaining approximately 1-week post-seeding. (G) High magnification of a representative maximum intensity projection and (H) confocal optical section with the corresponding orthogonal projections show that cells in confluent colonoid monolayers form the F-actin perijunctional rings (white arrowhead) and an immature apical brush border (yellow arrow heads). Scale bar (C-H) = 20 μm. Please click here to view a larger version of this figure.
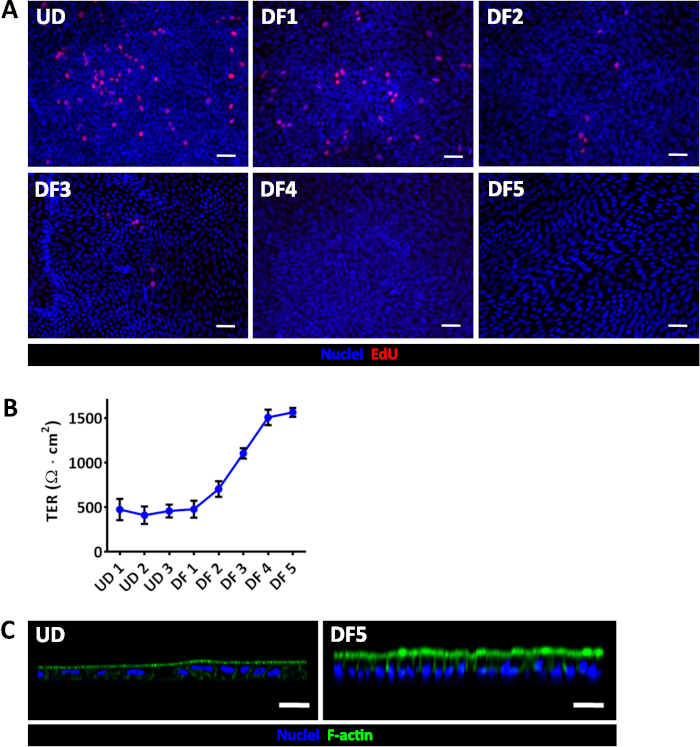
Figure 2: Evaluation of enteroid/colonoid monolayer differentiation. (A) EdU (red) incorporation demonstrates a progressive loss of proliferation during jejunal monolayer differentiation. (B) Average TER measurements in confluent jejunal monolayers in expansion or differentiation medium. Error bars represent SEM. UD, undifferentiated; DF, differentiated. Numbers correspond to days under the specified condition; UD1 was the first day of confluency, approximately 1 week after seeding. (C) UD jejunal monolayers have broad, shorter cells and a less-mature apical actin-based brush border than DF day 5 jejunal monolayers. Scale bar (A, C) = 50 μm. All monolayers were depicted at least 1-week post-seeding and were confluent prior to beginning differentiation. Please click here to view a larger version of this figure.
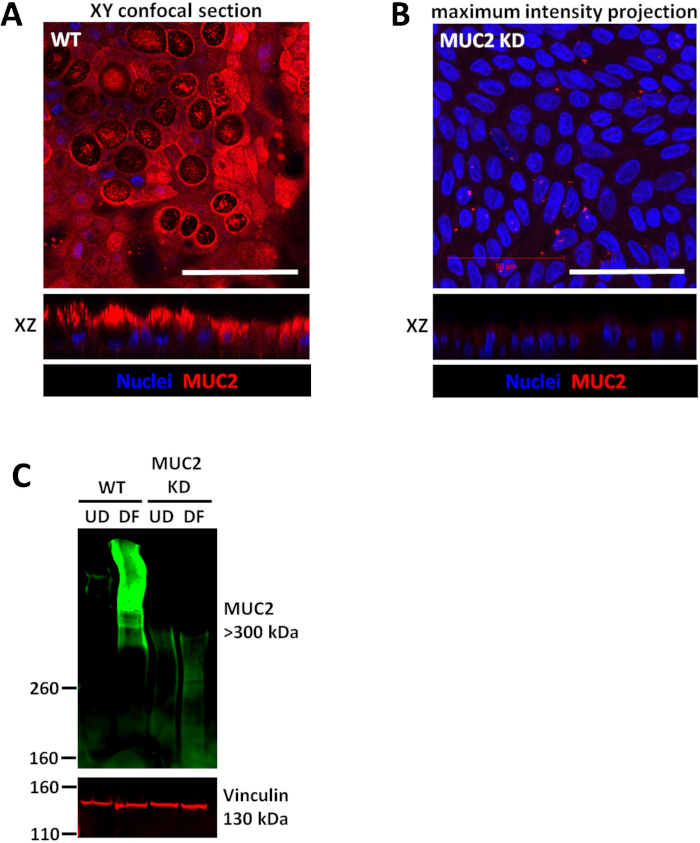
Figure 3: Wild type colonoids and shRNA transduced knockdown (KD) colonoids each form confluent monolayers. (A) Representative images of wild type (WT) human confluent colonoid monolayer differentiated for 5 days and (B) similarly grown monolayers derived from MUC2 KD colonoid cultures. Scale bar (A-B) = 50 μm (C) Representative immunoblot of colonoid cultures transduced with scrambled shRNA or MUC2 shRNA demonstrates that the changes in protein expression due to KD can be quantitated by immunoblot. Please click here to view a larger version of this figure.
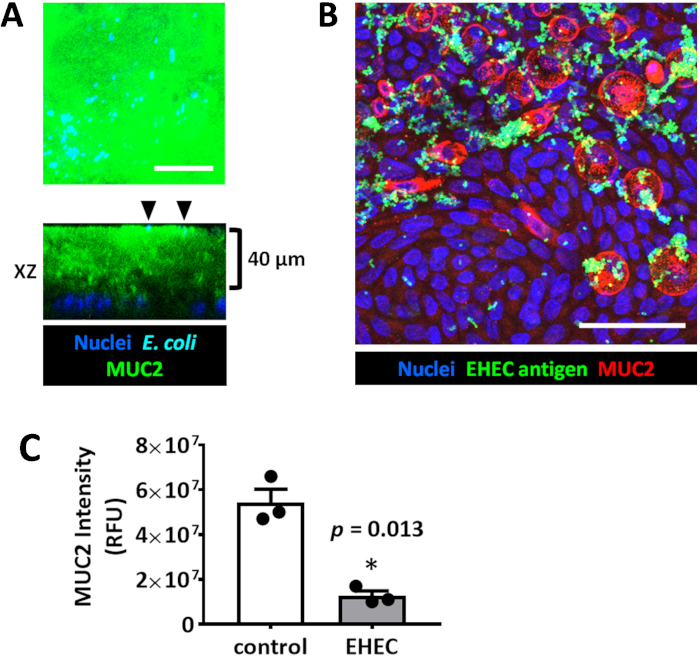
Figure 4: Enteroid/colonoid monolayers are suitable models to study luminal microbe-host interactions. (A) Representative maximum intensity projection and orthogonal optical section of a colonoid monolayer shows the thick apical MUC2-positive mucus layer which is not permeable to E. coli HS (black arrowheads) sitting at the apical mucus surface. The monolayer was infected for 6 hours with 106 cfu/mL HS. (B) Representative maximum intensity projection of human colonoid monolayer apically infected with EHEC (106 cfu/mL, 6 hours). Scale bar (A-B) = 50 μm. (C) MUC2 immunofluorescence intensity measurements show a significant decrease in MUC2 in EHEC-infected colonoid monolayers compared to uninfected controls. Error bars represent SEM. Please click here to view a larger version of this figure.
| Protein | Accession Number | Peptide Abundance |
| Basolateral Media | ||
| fatty acid-binding protein, liver [Homo sapiens] | NP_001434.1 | 1299 |
| profilin-1 [Homo sapiens] | NP_005013.1 | 797 |
| apolipoprotein A-IV precursor [Homo sapiens] | NP_000473.2 | 744 |
| ecto-ADP-ribosyltransferase 4 precursor [Homo sapiens] | NP_066549.2 | 633 |
| glyceraldehyde-3-phosphate dehydrogenase isoform 1 [Homo sapiens] | NP_002037.2 (+1) | 616 |
| serotransferrin precursor [Homo sapiens] | NP_001054.1 (+1) | 572 |
| vitamin D-binding protein isoform 3 precursor [Homo sapiens] | NP_001191236.1 (+2) | 567 |
| catalase [Homo sapiens] | NP_001743.1 | 427 |
| agrin isoform 1 precursor [Homo sapiens] | NP_940978.2 (+1) | 377 |
| lactotransferrin isoform 1 precursor [Homo sapiens] | NP_002334.2 (+1) | 262 |
| Apical Media | ||
| trefoil factor 3 precursor [Homo sapiens] | NP_003217.3 | 326 |
| trefoil factor 1 precursor [Homo sapiens] | NP_003216.1 | 304 |
| basement membrane-specific heparan sulfate proteoglycan core protein isoform a precursor [Homo sapiens] | NP_001278789.1 (+3) | 303 |
| filamin-B isoform 1 [Homo sapiens] | NP_001157789.1 (+2) | 240 |
| trefoil factor 2 precursor [Homo sapiens] | NP_005414.1 | 182 |
| deleted in malignant brain tumors 1 protein isoform b precursor [Homo sapiens] | NP_015568.2 (+3) | 169 |
| keratin, type II cytoskeletal 1 [Homo sapiens] | NP_006112.3 | 157 |
| myosin-9 [Homo sapiens] | NP_002464.1 | 150 |
| aminopeptidase N precursor [Homo sapiens] | NP_001141.2 (+1) | 145 |
| agrin precursor [Homo sapiens] | NP_940978.2 (+1) | 112 |
Table 1. A partial list of peptides identified by liquid chromatography/tandem mass spectrometry in apical and basolateral fluids sampled from differentiated jejunal monolayers.
Discussion
Critical parameters for successful formation of enteroid/colonoid monolayers include 1) healthy, proliferating 3D cultures as starting material; 2) coating the cell culture insert surface with human collagen IV prior to monolayer seeding; 3) fragmentation of 3D cultures either mechanically or enzymatically, but not to the single cell level.
During isolation of 3D enteroids/colonoids, the optimal shaking speed can vary depending on the rotational diameter of a given shaker model. The intent is to not only agitate the plate for an efficient mixing, but to also avoid splashing the cell suspension on the plate cover or into adjacent wells. Incubation periods of < 30 min may yield residual BMM clinging to cells, which can impede attachment and formation of uniform cell monolayers when plated on inserts. Extensive incubation (≥ 1 h) will yield significantly decreased cell viability.
The extent of trituration required to dissociate 3D enteroids/colonoids can vary with piston rigidity and user dexterity. Periodic examination of the well contents on a phase-contrast light microscope is recommended to determine fragment uniformity during trituration, with a goal of approximately 30 cells per fragment. In lieu of, or in addition to trituration, the suspension can be digested briefly with trypsin to obtain smaller uniform fragments. Trypsin digest may be desirable if monolayers contain a large proportion of 3D-like structures among an otherwise single epithelial layer. However, dissociation to single cells is not desirable as this significantly decreases cell viability. One well of enteroids/colonoids cultured in a 35 μL BMM droplet will generally be enough to populate 2-3 inserts, but this factor can vary depending on the number and average size of the enteroids/colonoids.
Colonoid monolayers may absorb a substantial volume of the apical medium as they differentiate. This can be circumvented by applying additional DM to the upper insert chamber (150-200 μL), although partial drying of the upper chamber does not appear to adversely affect monolayer viability or function in downstream assays. After approximately 4-5 days of differentiation, colonoid monolayers develop extracellular mucus that may be visible as thick/gelatinous material on the cell surface after careful aspiration of DM.
For EHEC interaction with the outer mucus layer, we routinely use the bacterial titer and incubation period specified in the protocol. However, unique bacterial strains should initially be assayed at multiple concentrations and incubation periods to determine the appropriate parameters for the desired effect.
During mucus fixation, aspirating the apical medium will likely remove most of the extracellular unattached mucus layer. Therefore, it is important to carefully pour out the medium. If the medium is retained in the insert by surface tension, use the corner of a folded laboratory tissue to break the surface tension and wick away most of the medium. After fixation and staining, the mucus layer can be unintentionally dislodged or flattened while mounting a coverglass over the insert filter. To preserve mucus height, place the filter on a drop of mounting media and carefully place the coverglass on the filter. Do not tap or press down on the coverglass as this may significantly flatten the mucus layer.
To form a polarized monolayer from cells in 3D culture, it is necessary to reproduce the interaction between basolateral membrane integrins of intestinal epithelial cells and extracellular matrix (ECM) proteins. Laminin, collagen IV, fibronectin, and an array of proteoglycans constitute the intestinal epithelial ECM.21,22 We compared human cell-derived laminin, fibronectin, collagen IV, and murine-derived BMM as insert coatings. Only collagen IV supported the formation of stable, long-term (up to 4 weeks) confluent enteroid/colonoid monolayers (Figures 1-2). Small patches of monolayer-like growth were obtained on each of the other tested matrices, but these regions failed to progress to confluency. Use of human collagen IV as the ECM surrogate has a number of advantages. BMM derived from Engelbreth-Holm-Swarm (EHS) murine sarcoma cells are not of human origin, whereas human collagen IV is commercially available and generally more cost-effective. Additionally, the BMM is a complex mixture of proteins with inherent variability, and may contain growth factors secreted by the sarcoma that affect gene expression.23
Interactions between intestinal epithelial cells and the underlying ECM in intestinal epithelial restitution is still incompletely understood. The significance of collagen IV, but not laminin, as an adhesion ligand for human colonocytes24 and enhancer of intestinal crypt epithelial cell restitution25 has been suggested using antibodies against collagen IV that prevented attachment. Epithelial-expressed β1-integrin appears to be important for ECM interaction, as an antibody specific for β1-integrin significantly blocked adhesion of colonocytes to type IV collagen.24 While collagen IV provides a reliable ECM for enteroid/colonoid monolayer formation on inserts, epithelial remodeling of the ECM over time has not yet been evaluated in this model, nor has the influence of more complex and defined ECM mixtures of collagen IV, laminin, and fibronectin.
Human enteroids/colonoids in monolayer format (Figures 1-4) enable manipulations and sampling that would be cumbersome or impossible to achieve using 3D matrix-embedded cultures. 3D enteroids/colonoids vary in size, structural complexity, and luminal volume, and thus microinjection of microbes or small molecules are difficult to accurately quantitate. Additionally, in contrast to the monolayers (Table 1), 3D cultures prevent direct access to both the luminal and basolateral surfaces to measure ions, nutrients, cytokines or secreted factors associated with physiologic or pathophysiologic processes. Confluency (Figures 1-2) is one of the main properties of the enteroid/colonoid monolayer necessary for establishing not only the physiologic gradients of nutrients, ions, and other macromolecules,16 but also creating the proper barrier between luminal microbiota and sterile mesenchymal/immune cell-populated serosal environments. These facets are important to consider for future studies that incorporate enteroid/colonoid monolayers with mesenchymal, immune, or neuronal cell types to build more complex physiological models.
Noted limitations of the cell culture insert model described here include lack of physical forces such as fluid shear stress and mechanical stretch/compression (peristalsis), and absence of an anaerobic apical environment normally experienced by intestinal epithelia in vivo. These elements have the potential to be addressed with more sophisticated microphysiological platforms,26 but they also require additional expense, equipment, and expertise to implement. Both 3D and monolayer cultures are also without interactions with or contributions from the intestinal microbiome, stromal cell populations, and the immune system unless these components are purposefully added.
Future applications of the enteroid/colonoid monolayer on collagen IV-coated cell culture inserts may include other studies of pathogenic or commensal microbial interaction, drug or nutrient uptake, toxicity, metabolism, barrier function, and functional enhancement catalyzed by co-culture with additional intestinal cell types. Evaluations can be made not only within epithelia of healthy donors but also from individuals with genetic mutations or intestinal disorders, provided the in vivo phenotypes are established to be preserved in ex vivo enteroid/colonoid culture.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH grants P01 AI125181, K01 DK106323 (JGI), and K01 DK113043 (JFA). We thank James Kaper (University of Maryland, Baltimore, MD, USA) for providing E. coli strain HS and EHEC. We also acknowledge the Integrated Physiology and Imaging Cores of the Hopkins Conte Digestive Disease Basic and Translational Research Core Center (P30 DK089502) and the Johns Hopkins Mass Spectrometry and Proteomics Core.
Materials
| 2-Amino-2-(hydroxymethyl)-1,3-propanediol | Sigma | T4661 | Tris base, Component of lysis buffer |
| A-83-01 | Tocris | 2939 | ALK4/5/7 inhibitor |
| Acetic acid, glacial | Fisher Scientific | A38 | |
| Advanced DMEM/F12 | Life Technologies | 12634-010 | Component of growth medium |
| Alexa Fluor 488 phalloidin | Life Technologies | A12379 | Fluorescent probe for F-actin |
| Antibiotic/antimycotic cocktail | Invivogen | ant-pm-2 | Primocin (100x) |
| B27, 50x | Life Technologies | 17504-044 | Component of growth medium |
| Cell culture inserts | Corning | 3470 | Transwell, PET membrane, 0.4 μm pore, 24-well plate |
| CHIR 99021 | Tocris | 4423 | GSK3β inhibitor |
| Click-iT Plus EdU Alexa Fluor 594 Imaging Kit | Life Technologies | C10639 | |
| Collagen IV, from human placenta | Sigma | C5533 | |
| Cultrex Organoid Harvesting Solution | Trevigen | 3700-100-01 | Depolymerizes basement membrane matrix |
| Enterohemorrhagic Escherichia coli (EHEC) | Kaper lab, University of Maryland | ||
| Epithelial voltohmmeter | World Precision Instruments | EVOM2 | |
| Epithelial voltohmmeter electrode | World Precision Instruments | STX3 | |
| Escherichia coli strain HS | Kaper lab, University of Maryland | ||
| Ethanol, absolute | Pharmco | 111000200 | |
| FluorSave mounting medium | Millipore | 345789 | For mounting insert membrane on microscope slide |
| GlutaMAX | Life Technologies | 35050-061 | L-alanyl-L-glutamine dipeptide, 200 mM |
| HEK293T/Noggin-Fc cell line | van den Brink lab, Tytgat Institute for Liver and Intestinal Research | For production of Noggin conditioned medium | |
| HEK293T/RSPO1-Fc-HA cell line | Trevigen | 3710-001-K | For production of Rspondin-1 conditioned medium |
| HEPES, 1 M | Life Technologies | 15630-080 | Component of growth medium |
| Heraeus Multifuge X1R Centrifuge | Thermo Fisher Scientific | 75004250 | |
| Hoechst 33342 | Life Technologies | H3570 | Fluorescent nuclear dye |
| Human epidermal growth factor (EGF) | R&D Systems | 236-EG-01M | Component of growth medium |
| IGEPAL CA-630 | Sigma | I8896 | Component of lysis buffer |
| Inverted cell culture light microscope | Olympus | CKX51 | |
| LB broth | EMD Millipore | 1.10285.0500 | |
| L-WNT3A cell line | ATCC | CRL-2647 | For production of Wnt3a conditioned medium |
| Matrigel, growth factor reduced | Corning | 356231 | Basement membrane matrix for 3D culture |
| Mini cell scraper | United BioSystems | MCS-200 | |
| MUC2 antibody, mouse monoclonal | Abcam | ab11197 | Use at 1:100 for immunostaining, 1:500 for immunoblotting |
| MUC2 shRNA lentiviral particles | GE Dharmacon | RHS4531 | GIPZ lentiviral shRNA, ID: 4583 |
| Orbital shaker | Grant Instruments | PSU-10i | |
| Penicillin-streptomycin, 100x | Life Technologies | 15140-122 | Component of growth medium |
| Phosphate buffered saline | Corning | 21-031 | |
| Probe sonicator with microtip | Branson Ultrasonics | 450 | |
| Protease inhibitor cocktail for mammalian cells | Sigma | P8340 | Component of lysis buffer |
| SB 202190 | Tocris | 1264 | p38 MAPK inhibitor |
| Sodium chloride | Sigma | S3014 | Component of lysis buffer |
| Sodium dexoycholate | Sigma | D6750 | Component of lysis buffer |
| Sodium dodecyl sulfate | Sigma | L3771 | Component of lysis buffer |
| TrypLE Express, 1x | Life Technologies | 12605-010 | Trypsin, for digesting enteroid/colonoid fragments |
| Vinculin antibody, rabbit monoclonal | Abcam | ab129002 | Use at 1:1000 for immunoblotting |
| Water, tissue culture grade, sterile filtered | Corning | 25-055 | |
| Y-27632 | Tocris | 1254 | RhoA/ROCK inhibitor |
Riferimenti
- In, J. G., et al. Human mini-guts: new insights into intestinal physiology and host-pathogen interactions. Nature Reviews Gastroenterology & Hepatology. 13, 633-642 (2016).
- Hill, D. R., et al. Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. eLife. 6, e29132 (2017).
- Williamson, I. A., et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cellular and Molecular Gastroenterology and Hepatology. 6, 301-319 (2018).
- Leslie, J. L., et al. Persistence and Toxin Production by Clostridium difficile within Human Intestinal Organoids Result in Disruption of Epithelial Paracellular Barrier Function. Infection and Immunity. 83, 138 (2015).
- Foulke-Abel, J., et al. Human Enteroids as a Model of Upper Small Intestinal Ion Transport Physiology and Pathophysiology. Gastroenterology. 150, (2016).
- Yin, J., et al. Molecular Basis and Differentiation-Associated Alterations of Anion Secretion in Human Duodenal Enteroid Monolayers. Cellular and Molecular Gastroenterology and Hepatology. 5, 591-609 (2018).
- Tse, C., et al. Enterohemorrhagic E. coli (EHEC)—Secreted Serine Protease EspP Stimulates Electrogenic Ion Transport in Human Colonoid Monolayers. Toxins. 10, 351 (2018).
- Jabaji, Z., et al. Use of collagen gel as an alternative extracellular matrix for the in vitro and in vivo growth of murine small intestinal epithelium. Tissue engineering. Part C, Methods. 19, 961-969 (2013).
- Wang, Y., et al. Self-renewing Monolayer of Primary Colonic or Rectal Epithelial Cells. Cellular and Molecular Gastroenterology and Hepatology. 4, 165-182 (2017).
- Moon, C., VanDussen, K. L., Miyoshi, H., Stappenbeck, T. S. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunology. 7, 818-828 (2014).
- VanDussen, K. L., et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 64, 911 (2015).
- Ettayebi, K., et al. Replication of human noroviruses in stem cell–derived human enteroids. Science. 353, 1387 (2016).
- Kozuka, K., et al. Development and Characterization of a Human and Mouse Intestinal Epithelial Cell Monolayer Platform. Stem Cell Reports. 9, 1976-1990 (2017).
- In, J., et al. Enterohemorrhagic Escherichia coli Reduces Mucus and Intermicrovillar Bridges in Human Stem Cell-Derived Colonoids. Cellular and Molecular Gastroenterology and Hepatology. 2, 48-62 (2016).
- Noel, G., et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Scientific Reports. 7, 45270 (2017).
- Vernetti, L., et al. Functional Coupling of Human Microphysiology Systems: Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle. Scientific Reports. 7, 42296 (2017).
- Noel, G., et al. Enterotoxigenic Escherichia coli is phagocytosed by macrophages underlying villus-like intestinal epithelial cells: modeling ex vivo innate immune defenses of the human gut. Gut Microbes. 9, 382-389 (2018).
- Sato, T., et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology. 141, 1762-1772 (2011).
- Johansson, M. E. V., Sjövall, H., Hansson, G. C. The gastrointestinal mucus system in health and disease. Nature Reviews Gastroenterology & Hepatology. 10, 352-361 (2013).
- Hews, C. L., et al. The StcE metalloprotease of enterohaemorrhagic Escherichia coli reduces the inner mucus layer and promotes adherence to human colonic epithelium ex vivo. Cellular Microbiology. 19, e12717 (2017).
- Laurie, G. W., Leblond, C. P., Martin, G. R. Localization of type IV collagen, laminin, heparan sulfate proteoglycan, and fibronectin to the basal lamina of basement membranes. The Journal of Cell Biology. 95, 340 (1982).
- Timpl, R. Macromolecular organization of basement membranes. Current Opinion in Cell Biology. 8, 618-624 (1996).
- Hughes, C. S., Postovit, L. M., Lajoie, G. A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics. 10, 1886-1890 (2010).
- Ishii, S., et al. Normal colonic epithelium adheres to carcinoembryonic antigen and type IV collagen. Gastroenterology. 106, 1242-1250 (1994).
- Moore, R., Madri, J., Carlson, S., Madara, J. L. Collagens facilitate epithelial migration in restitution of native guinea pig intestinal epithelium. Gastroenterology. 102, 119-130 (1992).
- Bein, A., et al. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cellular and Molecular Gastroenterology and Hepatology. 5, 659-668 (2018).

