Excitation-Scanning Hyperspectral Imaging Microscopy to Efficiently Discriminate Fluorescence Signals
Summary
Spectral imaging has become a reliable solution for identification and separation of multiple fluorescence signals in a single sample and can readily distinguish signals of interest from background or autofluorescence. Excitation-scanning hyperspectral imaging improves on this technique by decreasing the necessary image acquisition time while simultaneously increasing the signal-to-noise ratio.
Abstract
Several techniques rely on detection of fluorescence signals to identify or study phenomena or to elucidate functions. Separation of these fluorescence signals were proven cumbersome until the advent of hyperspectral imaging, in which fluorescence sources can be separated from each other as well as from background signals and autofluorescence (given knowledge of their spectral signatures). However, traditional, emission-scanning hyperspectral imaging suffers from slow acquisition times and low signal-to-noise ratios due to the necessary filtering of both excitation and emission light. It has been previously shown that excitation-scanning hyperspectral imaging reduces the necessary acquisition time while simultaneously increasing the signal-to-noise ratio of acquired data. Using commercially available equipment, this protocol describes how to assemble, calibrate, and use an excitation-scanning hyperspectral imaging microscopy system for separation of signals from several fluorescence sources in a single sample. While highly applicable to microscopic imaging of cells and tissues, this technique may also be useful for any type of experiment utilizing fluorescence in which it is possible to vary excitation wavelengths, including but not limited to: chemical imaging, environmental applications, eye care, food science, forensic science, medical science, and mineralogy.
Introduction
Spectral imaging may be performed in a variety of ways and is referred to by several terms1,2,3,4. In general, spectral imaging refers to data acquired in at least two spatial dimensions and one spectral dimension. Multispectral and hyperspectral imaging are most often distinguished by the number of wavelength bands or whether the spectral bands are contiguous1. For this application, hyperspectral data is defined as spectral data acquired with contiguous wavelength bands achieved by spacing of center wavelengths no less than half the full width at half maximum (FWHM) of each bandpass filter used for excitation (i.e., 5 nm center wavelength spacing for bandpass filters with 14-20 nm bandwidths). The contiguous nature of the data bands allows for an oversampling of the dataset, ensuring that Nyquist criteria are satisfied when sampling the spectral domain.
Hyperspectral imaging was developed by NASA in the 1970s and 1980s in conjunction with the first Landsat satellite5,6. Collecting data from several contiguous spectral bands allowed the generation of a radiance spectrum of each pixel. Identifying and defining the radiance spectrum of individual components made it possible to not only detect surface materials by their characteristic spectra, but it also allowed for the removal of intervening signals, such as variations in the signal due to atmospheric conditions. The concept of detecting materials using their characteristic spectra was applied to biological systems in 1996 when Schröck et al. used combinations of five different fluorophores and their known spectra to distinguish labeled chromosomes in a process termed spectral karyotyping7. This technique was elaborated upon in 2000 by Tsurui et al. for fluorescence imaging of tissue samples, using seven fluorescent dyes and singular value decomposition to achieve spectral separation of each pixel into linear combinations of spectra in the reference library8. Similar to their remote sensing counterparts, the contribution of each known fluorophore can be calculated from the hyperspectral image, given a priori information of the spectrum of each fluorophore.
Hyperspectral imaging has also been used in the areas of agriculture9, astronomy10, biomedicine11, chemical imaging12, environmental applications13, eye care14, food science15, forensic science16,17, medical science18, mineralogy19, and surveillance20. A key limitation of current fluorescence microscope hyperspectral imaging systems is that the standard hyperspectral imaging technology isolates fluorescence signals in narrow bands by 1) first filtering the excitation light to control sample excitation, then 2) further filtering emitted light to separate the fluorescence emission into narrow bands that can later be separated mathematically21. Filtering both the excitation illumination and emitted fluorescence reduces the amount of available signal, which lowers the signal-to-noise ratio and necessitates lengthy acquisition times. The low signal and lengthy acquisition times limit the applicability of hyperspectral imaging as a diagnostic tool.
An imaging modality has been developed that makes use of hyperspectral imaging but boosts the available signal, thereby reducing the necessary acquisition time21,22. This new modality, called excitation-scanning hyperspectral imaging, acquires spectral image data by varying the excitation wavelength and collecting a broad range of emitted light. It has been previously shown that this technique yields orders of magnitude increases in signal-to-noise ratio compared to emission scanning techniques21,22. The increase in signal-to-noise ratio is largely due to the wide bandpass (~600 nm) of emission light detected, while specificity is provided by filtering only the excitation light instead of the fluorescence emission. This allows all emitted light (for every excitation wavelength) to reach the detector21. Additionally, this technique can be used to discriminate autofluorescence from exogenous labels. Furthermore, the ability to reduce acquisition time due to increased detectable signal reduces the danger of photobleaching as well as allows spectral scans at an acquisition rate that is acceptable for spectral video imaging.
The goal of this protocol is to serve as a data acquisition guide for excitation-scanning hyperspectral imaging microscopy. In addition, descriptions are included that help to understand the light path and hardware. Also described is the implementation of open-source software for an excitation-scanning hyperspectral imaging microscope. Finally, descriptions are provided for how to calibrate the system to a NIST-traceable standard, adjust software and hardware settings for accurate results, and unmix the detected signal into contributions from individual components.
Protocol
1. Device set-up
- Light source: select a broad-band spectral light source with high power output and high collimation (a 300 W Xe arc lamp was used for these studies).
- Shutter (optional): add a shutter to the optical path to reduce photobleaching for time-lapse imaging.
- Tunable filter system: incorporate a mechanical tuning assembly and thin-film tunable filter (TFTF) set to enable the desired wavelength-adjustable excitation range (e.g., 360-485 nm).
- Microscope: use an inverted fluorescence microscope including a motorized dichroic filter turret and controller.
- Fluorescence filter cube: assemble a long-pass fluorescence filter cube. For best results, choose a dichroic mirror and long-pass emission filter at the same wavelength to achieve optimal separation of excitation from emission light (e.g., 495 nm).
- Objective: use an appropriate apochromatic objective to ensure uniform focus over the wavelength range used for the experiment (a 60x water objective was used for this study).
- Automated stage (optional): use an automated stage for rapid sampling of multiple fields of view and/or very large fields through image stitching. Calibrate the stage with the objective and the camera.
- Camera: select an appropriate camera to achieve the spatial resolution, sensitivity, and noise requirements of the experiment (this system utilized a high-sensitivity sCMOS camera).
2. Acquisition software
- Choose a software package that allows independent control of each hardware component, such as Micro-Manager.
- Creating the configuration file: the configuration file is a saved set of instruments and instructions that Micro-Manager will load to operate the system's hardware. To create a configuration file, click Tools > Hardware Configuration Wizard.
- In step 1, click Create new configuration > Next to continue. Note that the user may choose to modify or explore existing configuration if one is available already.
- In step 2, browse the Available Devices list to find the devices to control through Micro-Manager, such as the microscope, lamp, stage, shutter, and camera. When highlighted, click the Add… button to add the device to the installed devices list. This will open a new window.
- Designate the device's label in the Label box. (e.g., sCMOS camera)
- Under Value, select the appropriate COM port for each device. This will cause a list of device properties to appear in the bottom of the window.
- Leave the device properties at their default settings. Note that custom values for each device property may be entered as desired.
- When each device is added, click the Next button to continue to the next step. Note that if any devices are left out or need to be changed, the configuration file can be modified later as described in step 2.1.1.1.
- In step 3, use the drop-down menus to select the default camera, shutter, and focus stage. If auto-shutter is desired, click the Check box below the focus stage drop-down list. If the focus direction of the Z-stage is known, select it from the drop-down list under Stage focus directions (advanced). Otherwise, leave the default as Unknown.
- In step 4, double-click the boxes under the Delay [ms] heading to set delays associated with the automated devices such as delays of 100 ms for the lamp, stage, and shutter, and a 250 ms delay for commands to the mechanical tuning assembly to account for the mechanical switching time between filters.
- In step 5, assign labels for the state devices. The configuration for the excitation-scanning microscope system uses labels in the filter block to identify each fluorescence filter cube (e.g., state 0 corresponds to the label Bright, the empty filter cube used during bright-field imaging; while state 3 corresponds to the label 495 nm and custom 495 nm dichroic filter cube used to separate excitation and emission light at 495 nm). Labels are also used in the mechanical tuning assembly to store the switching commands for each excitation wavelength (e.g., state 0 corresponds to the 340 nm excitation wavelength tuning command for the mechanical tuning assembly).
- In step 6, click the Esplora button next to the Configuration file box to choose a save location and file name for the configuration file. Click finish to save the configuration file.
- Startup groups and presets: Micro-Manager can store different groups within each configuration file to activate or change a subset of the hardware. For example, a "system startup" group and preset combination will store default settings such as binning size and readout rates for the camera.
- Create a group (e.g., System) by clicking + in the main window in the Group subsection.
- Check any Use in Group? boxes inside the group that will require a default value to be set (e.g., binning in the associated default camera).
- Create a new preset (e.g., "Startup") by clicking + in the main window in the Preset subsection.
- Each box checked in step 2 will appear as a preset option here. Choose a default value to be loaded for each box checked.
- Group and presets for excitation wavelengths: Micro-Manager treats each excitation wavelength as its own channel with it its own wavelength switching command; therefore, each must be saved as its own preset.
- Create a new group to contain a set of desired excitation wavelengths (e.g., "495 nm dichroic")
- Check the Use in Group? box named Label corresponding to the VF-5 device.
- Create a new preset named for each excitation wavelength (e.g., "340 nm").
- Assign to each preset the appropriate property name and preset value (e.g., property name: "Label"; preset value: "340 nm").
- Multi-dimensional acquisition tool: click Multi-D Acq. near the top left of the Micro-Manager window to open a customizable tool to use for capturing an image of the sample at each excitation wavelength with the click of a single button. There are several customization options to construct the acquisition exactly as desired.
- Timepoints (not used in this example): for studies with no time-lapse component, leave the box unchecked. If time-lapse studies are desired, check this box. If checked, enter the number of times to perform the rest of the acquisition settings in the Number box. Set the time between successive acquisitions by entering the duration in the Interval box and choose the proper unit (milliseconds, seconds, or minutes; usually seconds).
- Multiple positions (XY; not used in this example): for studies of a single XY position, leave the box unchecked. If multiple XY positions are desired, check the box and click the Edit position list button to open a separate window. Move the stage to each desired location and click the Mark button to save that position in the software. Repeat until all desired XY positions are marked. Close this window to continue.
- Z-stacks (slides; not used in this example): for studies of a single Z position, leave the box unchecked. If multiple Z positions are desired, check the box. Move the stage to the desired beginning Z position and click the Set button next to the Z-start box. Move the stage to the desired ending Z position and click the Set button next to the Z-end box. Enter the desired step size (in microns) in the Z-step box.
- Channels: ensure that the Channels box is checked. Here, channels are the names given to the individual excitation wavelengths. Available "Channels" correspond to the "Groups" described in steps 2.1.2 and 2.1.3.
- Group: click the drop-down list to select a group from which to choose available excitation wavelengths (e.g., 495 nm dichroic).
- Click the Keep Shutter Open box to keep the shutter open between acquisitions for each channel. Note that the shutter will still close between successive spectral scans if this box is checked, assuming the autoshutter option is also selected in the main window.
- Click the Nuovo button to add channels to the acquisition list. Note that the Remove button can be used to remove any channels no longer desired and that Up and Down can be used to reorder any selected channel.
- Ensure that the Use? checkbox is selected for each desired wavelength in the spectral scan.
- Configuration: click the drop-down list under Configuration and choose the first wavelength in the desired spectral range (e.g., 340 nm).
- Exposure: double-click the box under Exposure and input the desired exposure time for the selected wavelength (e.g., "100" for 100 ms). See section 5 ("Data Acquisition") for suggestions to select appropriate exposure times.
- Z-offset (not applicable in a spectral scan): leave this box blank (at 0) when performing a spectral scan.
- Z-stack: ensure that this box is checked for each wavelength in the spectral range if performing a Z-stack.
- Skip Fr. (not applicable in a spectral scan): leave this box blank (at 0) when performing a spectral scan. Note that it may be useful to selectively skip frames in the event that one or few excitation wavelengths are significantly more powerful than others or if individual excitation wavelengths prove to be especially phototoxic.
- Color: leave this box blank when performing a spectral scan. Note that colors can be chosen for individual wavelength bands for data visualization purposes, but color is inappropriate for subsequent spectral unmixing.
- Repeat these steps until each desired excitation wavelength within the given spectral scanning range is added.
- Acquisition order: click the drop-down list to choose in which order the above options (2.1.4.1-2.1.4.5) will be performed. For spectral scans such as the one shown here, the acquisition order is simply Channel. Note that additional options appear as additional boxes are checked within the Multi-Dimensional Acquisition tool [timepoints, Multiple Positions (XY), and Z-stacks (slices)] and allow the choice of either position or time first, followed by either slice or channel.
- Autofocus (not used in this example): If Multiple Positions (XY) is selected, several different styles of autofocusing are available. Click the Options button and select an autofocus method from the drop-down list beside the close button.
- Summary: review this window for a summary of number of timepoints, XY positions, Z slices, channels, total number of images, total memory, scan duration, and acquisition order to ensure the listed information matches expected acquisition settings.
- Save images: ensure that this box is checked to save the data collected through use of the Multi-Dimensional Acquisition tool.
- Click the … button next to Directory root to choose a directory root in which the files will be saved. Name the directory root in a fashion that describes relevant details of the experiment (e.g., GCaMP Airway Smooth Muscle Cells).
- Enter a name in the box next to Name prefix that describes the current image acquisition (e.g. FOV1_100ms_60X_495nmDichroicFilter). It is advisable to create a name prefix that identifies the field of view and exposure time, as well as other relevant information such as the object and dichroic filter used. Note that the first image stack taken using these settings will be saved with "_1" following the name prefix. Any subsequent stack taken with the same directory root and name prefix will be saved with "_n", in which "n" is the number of times a stack has been taken with this name in the directory.
- Click Separate image files to ensure the image generated at each excitation wavelength is saved.
- Save as: click the Save as… button near the top right of the Multi-Dimensional Acquisition tool to save these acquisition settings for easy future use. Note that the directory root and name prefixes will be saved, as well.
- Acquire: finally, click the Acquire! button to begin acquiring images according to the acquisition settings chosen above.
- Creating the configuration file: the configuration file is a saved set of instruments and instructions that Micro-Manager will load to operate the system's hardware. To create a configuration file, click Tools > Hardware Configuration Wizard.
3. Spectral response correction (optional):
- Spectral output correction can be performed to calibrate the spectral response of the system to a known standard, such as a NIST-traceable lamp or other NIST-traceable standard or instrument. This step is especially important if results are compared with other spectral imaging instruments, spectrometers, or between different laboratories. This process has been reported in detail previously21,23.
- Use a spectrometer calibrated to a NIST-traceable light source (such as LS-1-CAL-INT, Ocean Optics) or other NIST-traceable standard to acquire the spectroradiometric power of the illumination as measured at the sample stage. Perform a separate correction for each combination of light source setting, dichroic mirror, and objective. Use an integrating sphere coupled to the spectrometer to accurately measure wide-angle illumination from the microscope objective.
- Use an integration method, such as the trapezoidal rule, to integrate the spectroradiometric data over wavelengths illuminated. A 40 nm bandwidth for integration centered around the center wavelength is sufficient for most filters that have a nominal bandwidth of between 14-20 nm full-width at half-maximum (FWHM). The integrated value represents the intensity of the spectral illumination at each excitation wavelength band.
- Plot the integrated intensity of each wavelength band as a function of excitation center wavelength to allow visualization of the spectral illumination intensity profile.
- Determine the wavelength band with the lowest integrated intensity.
- Normalize the spectral illumination intensity profile by dividing the lowest integrated intensity by the integrated intensity of each wavelength band to generate a wavelength-dependent correction factor.
4. Sample preparation
- Prepare a "blank" sample (e.g., place a glass coverslip in the cell chamber and add 1 mL of buffer). This buffer has been described previously24.
- Prepare an unlabeled sample to determine any sample autofluorescence (e.g., place a glass coverslip containing unlabeled airway smooth muscle cells25,26 in the cell chamber and add 1 mL of buffer).
- Prepare a separate, single-labeled sample for each fluorescent label used in the experiment as follows:
- Add diluted mitochondrial label to buffer to achieve a 100 nM concentration. Add this buffer to the coverslip containing airway smooth muscle cells. Incubate for 20 min at 20-25 ˚C. Move the coverslip to the cell chamber and add 1 mL of buffer. Note that the optimal concentration of mitochondrial label will vary by factors such as manufacturer, associated color, and desired specificity.
- Move a coverslip containing airway smooth muscle cells transfected with the GCaMP probe27 to the cell chamber and add 1 mL of buffer.
- Prepare one or more experimental samples containing a mixture of the desired fluorescent labels.
- Add 1 mL of buffer (100 nM concentration of mitochondrial label) to a coverslip containing airway smooth muscle cells transfected with the GCaMP probe and incubate for 20 min at 20-25 ˚C. Move the coverslip to the cell chamber and add 1 mL of buffer.
5. Data acquisition:
- Check that the proper dichroic beamsplitter (e.g., 495 nm dichroic filter cube), objective (e.g., 60x water objective), and camera (sCMOS camera) are selected.
- Load the sample on the stage.
- Double-click the box next to Exposure [ms] and type "100" to set the exposure time at 100 ms. Note that the exposure time may need to be increased or decreased depending on the fluorescence intensity of the sample.
- Select 475 nm from the drop-down menu of the Micro-Manager main window for initial sample viewing. Note that 475 nm may not be the optimal wavelength for sample viewing or determining whether oversaturation will occur throughout the image stack.
- Click Live to view the sample.
- Click the automatic intensity viewing range button Auto next to the histogram on the bottom of the window to bring the minimum and maximum values into meaningful visual ranges.
- Use the microscope's focus knobs to focus on the sample. It is often useful to find the edge of the sample to aid in focusing. The sample will be focused when the edge features in the image appear sharp. Note that it may be necessary to click the Auto button several times during focusing to aid in viewing the fluorescence image. Additionally, some users may find it easier to focus on the sample if the microscope is in transmission mode.
- If focusing in transmission mode, for safety, first ensure that the spectral light source is not transmitting light to the eyepieces before adjusting the light path for transmission imaging. Also note that there may be small deviations between the focus of the transmission and fluorescence images. When acceptable focus has been achieved, reconfigure the light path for spectral fluorescence imaging.
- Click the Multi-D Acq. button near the top left of the window to open the Multi-Dimensional Acquisition tool (described and configured in section 2).
- Choose an appropriate spectral range for spectral acquisition (e.g., 360-485 nm for the 495 nm dichroic filter) by clicking the Load… button in the upper right of the Multi-Dimensional Acquisition tool window and navigating to the excitation settings saved previously (in step 2.1.4.10). See discussion for information regarding appropriate spectral ranges.
- Acquire a background/blank image that contains no fluorescence data to use for background and noise subtraction. This can be performed using a blank sample (step 4.1) or by navigating to a blank region of an experimental sample. As described in step 5.6, this is most easily achieved by finding the edge of the sample then positioning it so that the edge is centered within the field of view.
- Once the acquisition settings are confirmed and a background region is visible, click the Acquire! button to acquire a spectral image stack containing background and noise to use for subtraction later. It should be noted that samples are likely to have more intense fluorescence away from the edge of the sample. It may be advisable to move about the sample to find the "brightest" regions and perform several test image stacks to determine appropriate acquisition times and avoid overexposure.
- Take a single image stack on a region of the sample that appears to have intense fluorescence to ensure that no combination of wavelengths and that exposure times result in overexposure.
- Use ImageJ to confirm that no wavelengths contain overexposed pixels.
- In ImageJ, click File > Import > Image Sequence and navigate to the folder containing the spectral images taken in step 5.10. This will open a new window. Click the box next to Number of images and enter the number of wavelengths included in the spectral scan (e.g., 26). Leave Starting image and Increment as "1". Click the OK button to continue.
- Press M on the keyboard to utilize ImageJ's measure function. Ensure that the number listed under Max is not the upper detection limit of the camera (e.g., 65,535). Repeat for each image in the spectral scan. Note that the detection limit of the camera depends on the camera itself. The upper limit is displayed on the top right portion of the histogram in the main window of MicroManager. Alternatively, the upper limit can be determined by opening an image in ImageJ and navigating to Image > Adjust > Brightness/Contrast. Click the Set button in the resulting pop-up window, enter an excessively large value (e.g., 999,999) in the blank next to Maximum displayed value, and click OK to continue. The maximum value displayed in the new histogram should be the upper detection limit of the camera.
- If any images contained the upper detection limit of the camera (e.g., 65,535), adjust the exposure time of the spectral scan to ensure that the maximum signal throughout the spectral range does not exceed the dynamic range of the camera.
- Acquire spectral image data from unlabeled samples to determine any autofluorescence. Acquire the scan for the unlabeled samples using an identical wavelength range and camera settings as the remainder of the experiment (e.g., 360-485 nm at 100 ms per wavelength).
- Acquire spectral image data from single-labeled samples to use as spectral controls to build the spectral library. Perform the scan for each label using an identical wavelength range, exposure time, and camera settings. Note that a longer acquisition time may be needed for some samples to allow accurate label spectrum detection with minimum noise contribution.
- Perform measurements on an experimental sample (e.g., human airway smooth muscle cells with GCaMP probe and mitochondrial label).
- Place a slide or coverslip containing the experimental sample on the microscope.
- Select a field of view with appropriately labeled cells of tissues.
- Acquire the spectral image data using the desired acquisition settings, as described above.
6. Image analysis
- Correct images to a flat spectral response.
- Subtract the background spectrum obtained in section 5.9 and multiply by the correction coefficient determined in section 3.1.5. This can be done with a simple MATLAB script or ImageJ routine. A subtraction/correction MATLAB code (used in this example) is available on the University of South Alabama Bioimaging Resources website.
- Load the subtraction/correction code in MATLAB.
- In the EDITOR tab, click Run. This will open a new window containing a Bsq Correction button. Click the Bsq Correction button to open another window containing choices for working directory, background file, correction factor file, and uncorrected tiff folder.
- Use the Browse… button beside Working Directory to choose the file path where subsequent windows will open. Navigate to the folder containing the saved background spectrum (in .dat format).
- Use the Browse… button beside Background File (.dat) to open the directory chosen in step 6.1.1.2.1 and choose the desired background file (in .dat format). Click the Open button to continue.
- Use the Browse… button beside Correction Factor (.dat) to choose a correction factor. The directory for the correction factor defaults to the location of the parent MATLAB code. If necessary, navigate to the subfolder containing the correction factor file (in .dat format). Highlight the proper correction factor file and click the Open button to continue.
- Use the Browse… button beside Uncorrected Tiff Folder to open the directory chosen in step 6.1.1.2.1 and choose the folder for background subtraction and image correction (this is usually the Pos0 folder that immediately contains the raw spectral images). Click the Open button to continue.
- Click the Click for Spectral Correction button. After processing completes, a window will open with the message "Bsq Correction Complete". Click the OK button to continue. The correction images are stored in a new folder named with the original raw spectral image folder and an additional "_Corrected" (e.g., Pos0_Corrected).
- Subtract the background spectrum obtained in section 5.9 and multiply by the correction coefficient determined in section 3.1.5. This can be done with a simple MATLAB script or ImageJ routine. A subtraction/correction MATLAB code (used in this example) is available on the University of South Alabama Bioimaging Resources website.
- Generate the spectral library. Several software packages can be used to extract spectral data from images, including ImageJ. For best results, normalize each endmember to the wavelength band of greatest intensity by determining which wavelength band has the most intense value and dividing the measurement at each wavelength band by that maximum value. It should also be noted that single-label controls generated within autofluorescent samples will need additional modifications28,29,30,31,32. See step 6.2.4 and discussion for details.
- In ImageJ, click File > Import > Image Sequence. This will open a new window. Navigate to the folder containing the corrected spectral images. Double-click any image file in the folder to open a new window. Click the box next to Number of images and enter the number of wavelengths included in the spectral scan (e.g., 26). Leave Starting image and Increment as "1". Click the OK button to continue.
- In the ImageJ main window, click the Polygon selections icon, then use the mouse pointer by clicking on the image to create a polygon around a region containing fluorescence data. Note that there may be little to no fluorescence data in the initial wavelength. Navigate to another wavelength image and/or use the brightness/contrast tool (Image > Adjust > Brightness/Contrast > Auto) to better visualize regions containing fluorescence data of interest.
- With the polygon drawn, generate the Z-axis profile by clicking Image > Stacks > Plot Z-axis Profile. This will open a new window. Click Save to save the spectral data as a .csv file.
- Perform a subtraction to ensure a proper spectral library with pure labels devoid of autofluorescence contamination. Use the following equation29 to isolate a pure spectrum from the mixture of a single-label and fluorescence:
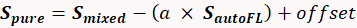 (1)
(1)
Where: Spure is the pure spectrum of the label, Smixed is the measured spectrum of the sample contaminated with autofluorescence, S autoFL is the measured autofluorescence spectrum, "a" is a scalar multiplier of the autofluorescence spectrum (between 0 and 1; e.g., 0.4), and "offset" is an offset (usually 0). Vary the "a" term until a near-zero value is achieved for the apparent contribution of autofluorescence. Additional information can be found in several publications28,29,30,31,32.
- Unmix the spectral image data. The unmixing step will generate an abundance image for each fluorescent label, where abundance is the amount of relative fluorescence signal in the image from the respective label. Several unmixing algorithms are available for use with ImageJ33,34,35. Additionally, a spectral unmixing MATLAB code (used in this example) is available on the University of South Alabama Bioimaging Resources website. A more comprehensive overview of spectral unmixing is available36.
- Load the spectral unmixing code in MATLAB.
- Edit the Wavelength.mat and Library.mat files to correspond to the experimental conditions (e.g., "Wavelength" as 360-485 in increments of five).
NOTE: The corresponding values for those wavelengths should be loaded into "Library" for each fluorescent label (and autofluorescence). Endmember_Name should list the labels in the same order as the column values of "Library". Note that the Library.mat file should contain both "Library" and "Endmember_Name" variables. - In the EDITOR tab, click Run. This will open a new window allowing the user to choose a directory. Navigate to the folder containing the spectral images to be unmixed. Once selected, this will open a new window.
- In the resultant blanks, enter the image name (e.g., HASMC with GCaMP and Mito FOV1), number of wavelength bands (e.g., 26), number of timepoints (e.g., 1) and whether a FRET measurement is desired (e.g., n) in their respective blanks. If "n" is selected for FRET, leave both End-Member blanks empty. Press OK to continue, which will open a new window.
- Naviagate to the folder containing the Wavelength.mat file. Once selected, click Open to continue, which will open a new window.
- Navigate to the folder containing the Library.mat file. Once selected, click Open to continue, which will save the unmixed images in a new "Unmixed" folder inside the directory containing the spectral images.
- Inspect the unmixed spectral images for quality.
- Open each unmixed image to visually inspect the distribution of pure components.
- Compare the magnitude of the error image to those of the unmixed images (similar to step 5.11, this can be done through ImageJ's measure function). If the magnitude of the error image is similar in magnitude to the unmixed abundance images, it is likely that there are signals in the spectral image data that are not accurately accounted for by the spectral library and the linear spectral unmixing process.
- Compare the error image to the unmixed images to see if there are unidentified residual structures.
- Open each unmixed image to visually inspect the distribution of pure components.
Representative Results
Several important steps from this protocol are necessary to ensure the collection of data that is both accurate and devoid of imaging and spectral artifacts. Skipping these steps may result in data that appear significant but cannot be verified or reproduced with any other spectral imaging system, thereby effectively nullifying any conclusions made with said data. Chief among these important steps is proper spectral output correction (section 3). The correction factor compensates for wavelength-dependent variations in the spectral output of the tunable excitation system. This is accomplished by scaling wavelengths with high power excitation such that the optical illumination power is comparable to wavelengths with a low power excitation, achieving a flat spectral excitation profile. An example of an inappropriate correction factor is one that contains very low values (e.g., <0.001) at one or more wavelengths, indicating that the intensity values measured at those wavelengths must be greatly attenuated to achieve a flat spectral response.
Calibration of the system to a NIST-traceable standard ensures that data collected with the excitation-scanning spectral imaging system is comparable to other systems also calibrated to NIST-traceable standards. Therefore, it is imperative to ensure that any correction factor adjusts the collected data appropriately. Accuracy of a correction factor can be verified with the use of a fluorescence standard, such as NIST-traceable fluorescein. Figure 1 illustrates an appropriate and inappropriate correction factor, visualized through graphical and image data. In this case, the spectral output at 340 nm was virtually nonexistent compared to the rest of the spectral range, resulting in a near-zero (<0.001) value for virtually every wavelength. Applied to the image stack, this results in near-zero values through most of the image stack for most pixels.
As stated in section 5, the excitation range, selected fluorophores, and acquisition settings may create the potential that one or many excitation wavelength image bands contain oversaturated pixels. Figure 2 illustrates an example in which the exposure time was set too long for several of the excitation wavelengths, causing the subsequent images to contain oversaturated pixels. This is important to note because, as the figure shows, both the individual images and the false-colored images may visually appear to be within acceptable intensity ranges.
Appropriate selection of a background region for subtraction is also important for data comparison among systems, as it removes elements of camera noise or stray light prior to NIST-traceable spectral correction (Figure 3). It is often helpful for subsequent image processing and data analysis steps to generate an RGB false-colored image of the spectral image stack in order to visualize spectral features within the image. Figure 4 shows an RGB false-colored image generated by merging three selected wavelength bands (370 nm = blue, 420 nm = green, 470 nm = red).
Spectral unmixing requires knowledge of the spectral profile of each individual fluorescence source. In the case of excitation-scanning hyperspectral imaging, this is done by acquiring spectral image stacks for each fluorophore (and autofluorescence). Figure 5 is included as an example for choosing regions from single-label controls to generate a spectral library. It should be noted that each measurement has been normalized to its peak wavelength.
When performed correctly, the spectral unmixing process allows separation of a spectral image stack into respective contributions from each fluorescence label. Figure 6 shows an example unmixed spectral image set, with individual images for each of the following signals: airway smooth muscle cell autofluorescence, a GCaMP probe, and a mitochondrial label. The error associated with spectral unmixing is also shown and can be examined to compare intensity levels of the error to those of the unmixed signals. Calculation and interpretation of this error has been discussed previously37. As shown in Figure 6, there is high error associated with the nuclear and perinuclear regions of the cells, indicating that the measured spectra of those regions are not well accounted for by the spectra in the spectral library. One potential source of error may be that the single-label controls for GCaMP and the mitochondrial label were prepared using airway smooth muscle cells, which have a high native autofluorescence. Hence, the GCaMP and the mitochondrial label may not represent pure endmember spectra. In addition, the autofluorescence signal may influence the library spectra for the two other labels in a way that was not properly accounted for, resulting in a less accurate fitting than if the labels had been acquired using a cell line with little to no autofluorescence. Additionally, examples are included for when too few or too many components of a spectral library are available, resulting in underfitting and overfitting of the spectral image data, respectively (Figure 6, Figure 7, Figure 8, Figure 9, and Table 1).
As noted in section 6.2 and specifically step 6.2.4, "pure" spectra collected from labeled cells that are also autofluorescent will likely contaminate the spectral profile for the pure component. As such, care should be taken to separate the pure spectra of the labels of interest from their autofluorescent hosts. Figure 10 shows the difference of unmixing with "pure" spectra contaminated with autofluorescence (mixed) versus pure spectra calculated by the method described in step 6.2.4. The difference occurs mainly in unmixing the autofluorescence signal. Without proper signal separation (e.g., subtraction of autofluorescence contamination from the GCaMP spectrum), the autofluorescence and GCaMP library components compete for the same spectral data in each pixel, resulting in characteristic holes or dark spots in the autofluorescence image.
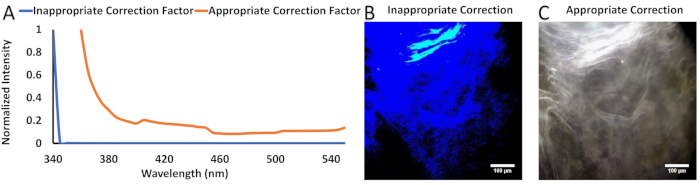
Figure 1: Example applications of inappropriate and appropriate correction factors used to correct images to a flat spectral response. (A) Plotted inappropriate and appropriate correction factors. (B) An RGB image generated with use of the inappropriate correction factor in (A). (C) An RGB image generated with the same field of view as (B), except with the use of an appropriate correction factor. Please click here to view a larger version of this figure.
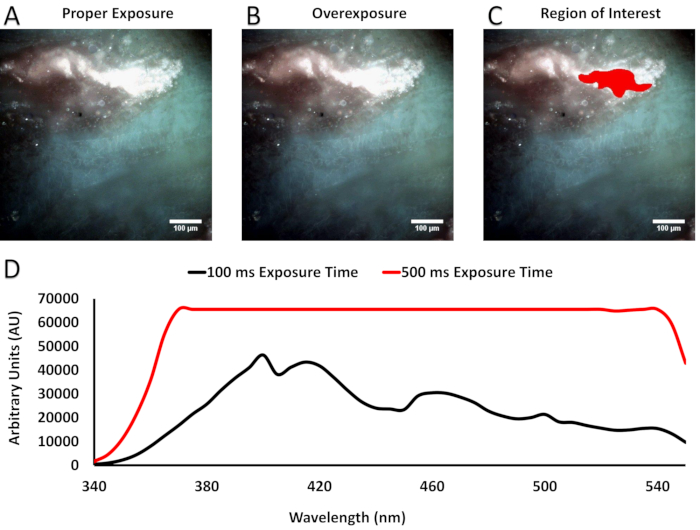
Figure 2: Examples that illustrate the importance of appropriate acquisition settings. (A,B) RGB images generated by an appropriate acquisition time (A, 100 ms) vs. the same field of view with a saturating acquisition time (B, 500 ms). It should be noted that the RGB images appear identical when false-colored. (C) The region of interest selected to survey intensity values from the most intense regions of (A) and (B). (D) Plotted intensity values per wavelength from the 100 ms exposure time image (A, black line) and 500 ms exposure time (B, red line). It should be noted that pixel intensities for the 500 ms exposure time have reached the limit of the dynamic range of the detector (65,535 AU) at 370 nm and do not decrease until 525 nm, resulting in spectral artifact. Please click here to view a larger version of this figure.
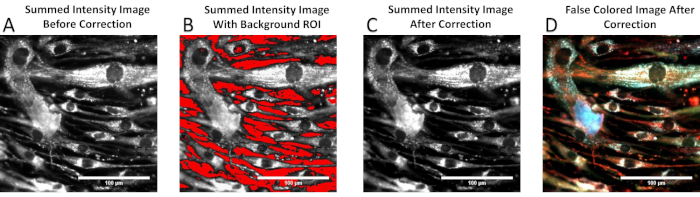
Figure 3: Selection of a region of interest for background subtraction. (A) A raw, wavelength-summed intensity image. (B) Regions of the image selected to determine the pixel-averaged background spectrum for background subtraction shown in red. (C) The background-subtracted, corrected, and summed intensity image. (D) An RGB coloring of the corrected image (C). The process of generating an RGB false-colored image is shown in Figure 4. Please click here to view a larger version of this figure.
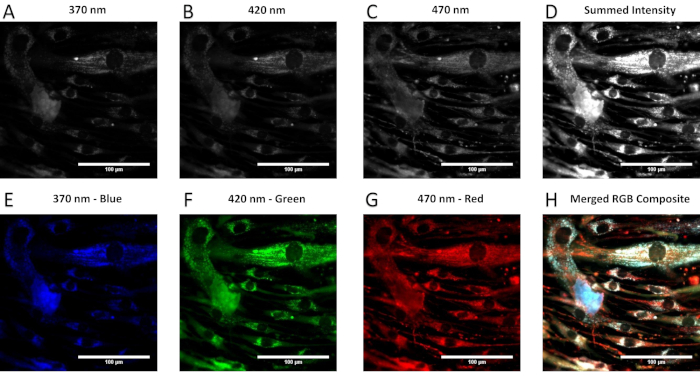
Figure 4: Process of generating an RGB false-colored image. (A–C) Three wavelength bands spaced evenly throughout the spectral acquisition range were selected for false-coloring (blue = 370 nm, green = 420 nm, red = 470 nm). (D) The summed intensity image generated by adding pixel intensities from all wavelength bands in the image cube. (E–G) The images in panels (A-C) with their respective false-color look-up tables applied. (H) The resultant merged image of (E-G). Please click here to view a larger version of this figure.
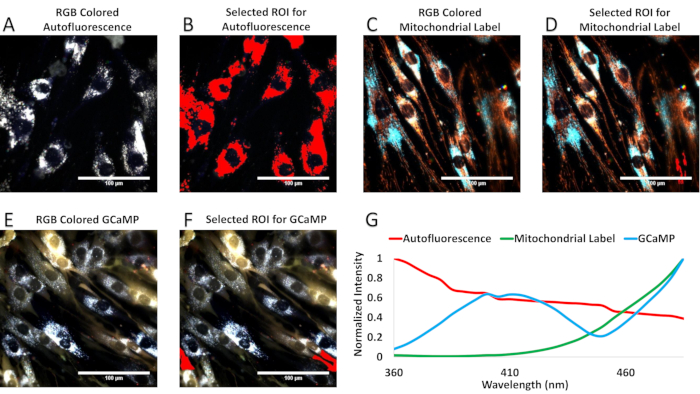
Figure 5: Region selection of single-label controls for spectral library generation. (A) RGB false-colored image of airway smooth muscle (ASM) cell autofluorescence. (B) Shown in red, regions of (A) chosen for the autofluorescence component of the spectral library. (C) RGB false-colored ASM cells labeled with the mitochondrial label. (D) Shown in red, regions of (C) chosen for the mitochondiral label component of the spectral library. Due to the autofluorescence of the ASM cells localized near the nucleus, small regions were selected far from the nuclei to identify the mitochondrial label spectrum. (E) RGB false-colored ASM cells transfected with the GCaMP probe. (F) Regions of (E) selected for the GCaMP component of the spectral library are shown in red. Similar to (D), regions away from the nuclei were selected. (G) The spectral library obtained from A-F, normalized to a value of unity at the wavelength with the strongest signal. Please click here to view a larger version of this figure.
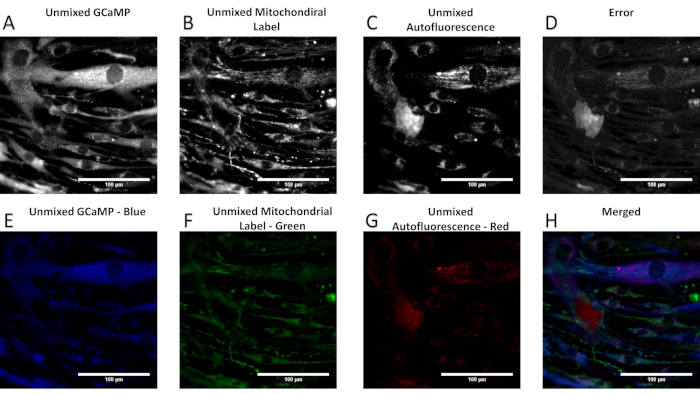
Figure 6: Example of unmixed image data in which unmixed relative signal contributions from each library component can be visualized. (A–D) The unmixed abundance of GCaMP, mitochondrial label, autofluorescence, and error term. (E–G) The images in panels (A-C) with their respective false-color look-up tables applied. (H) The composite, merged, false-colored image. Please click here to view a larger version of this figure.
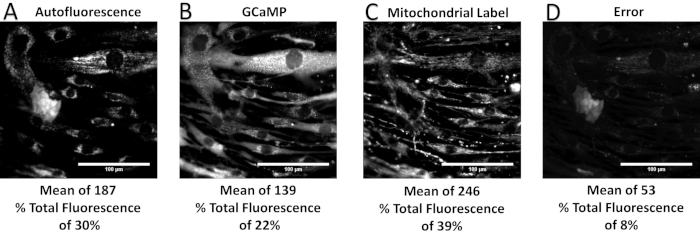
Figure 7: Unmixed relative signal contributions, including mean intensity and percent of total fluorescence, from a properly defined spectral library. (A–D) The unmixed abundance for autofluorescence, GCaMP, mitochondrial label, and error term. It should be noted that the error term comprises less than 10% of the total fluorescence signal measured. Please click here to view a larger version of this figure.
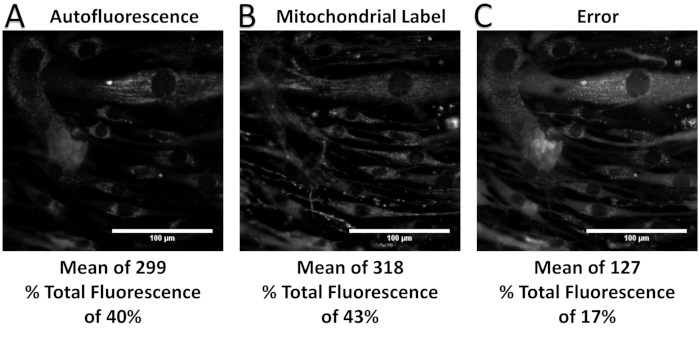
Figure 8: Unmixed relative signal contributions, including mean intensity and percent of total fluorescence, from a spectral library missing a component known to be included in the sample (i.e., an underdefined spectral library). (A–C) The unmixed abundance of autofluorescence, mitochondrial label, and error term. Note that the omission of GCaMP from the spectral library has increased the calculated relative signal contributions from the library components as well as the error term, when compared to Figure 7. Please click here to view a larger version of this figure.
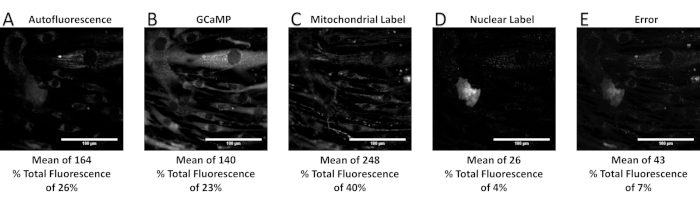
Figure 9: Unmixed relative signal contributions, including mean intensity and percent of total fluorescence, from a spectral library containing an additional component known to be missing in the sample (i.e., an overdefined spectral library). (A–E) The unmixed abundance of autofluorescence, GCaMP, mitochondrial label, nuclear label, and error term. Note that the addition of a nuclear label to the spectral library has decreased the calculated relative signal contributions from autofluorescence, when compared to Figure 7. Furthermore, the error term is decreased below the percent error specified by the properly defined spectral library. This is because an overdefined library will almost always allow a better fit to the experimental data than a properly defined library, even though abundance signals for components known to be absent from the sample are (in reality) artifacts of overdefining the spectral library. Please click here to view a larger version of this figure.
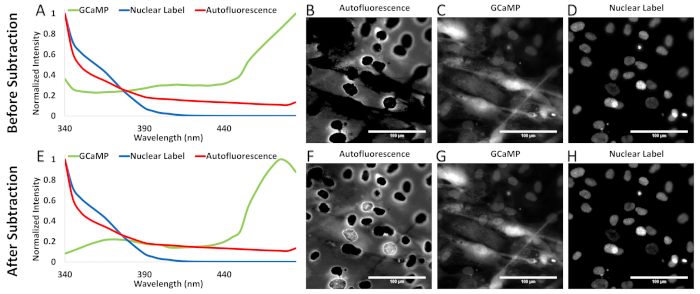
Figure 10: Comparison of unmixed images when using a library before and after proper autofluorescence contamination signal subtraction. (A) The original "pure" spectra derived from the single-label controls in highly autofluorescent airway smooth muscle cells before the scaled subtraction detailed in step 6.2.4. (B–D) The unmixed autofluorescence, GCaMP, and nuclear label images generated using (A) as the library. (E) The corrected pure spectra derived from single-label controls in highly autofluorescent airway smooth muscle cells after scaled subtraction. (F–H) The unmixed autofluorescence, GCaMP, and nuclear label images generated using (E) as the library. Please click here to view a larger version of this figure.
| Mean Intensity (AU) | |||
| Proper Fitting | Underfitting | Overfitting | |
| Autofluorescence | 187 | 299 | 164 |
| GCaMP | 139 | – | 140 |
| Mitochondrial Label | 246 | 318 | 248 |
| Nuclear Label | – | – | 26 |
| RMS Error | 53 | 126 | 43 |
| Standard Deviation (AU) | |||
| Proper Fitting | Underfitting | Overfitting | |
| Autofluorescence | 362 | 442 | 315 |
| GCaMP | 168 | 168 | |
| Mitochondrial Label | 344 | 388 | 345 |
| Nuclear Label | – | – | 93 |
| RMS Error | 62 | 126 | 44 |
| Maximum Intensity (AU) | |||
| Proper Fitting | Underfitting | Overfitting | |
| Autofluorescence | 6738 | 7409 | 6738 |
| GCaMP | 1336 | – | 1336 |
| Mitochondrial Label | 5098 | 5194 | 5098 |
| Nuclear Label | – | – | 1257 |
| RMS Error | 1050 | 1286 | 910 |
| % of Total Fluorescence | |||
| Proper Fitting | Underfitting | Overfitting | |
| Autofluorescence | 30% | 40% | 26% |
| GCaMP | 22% | 43% | 23% |
| Mitochondrial Label | 39% | – | 40% |
| Nuclear Label | – | – | 4% |
| RMS Error | 8% | 17% | 7% |
Table 1: A table comparing the average, standard deviation, and maximum unmixed abundance intensity values per unmixed abundance image from a proper spectral library and from underdefined or overdefined spectral libraries, as well as the percent of total fluorescence per unmixed abundance image (the minimum unmixed abundance intensity for each image was always zero). Data taken from Figure 7, Figure 8, and Figure 9.
Discussion
The optimal use of an excitation-scanning hyperspectral imaging set-up begins with construction of the light path. In particular, choice of light source, filters (tunable and dichroic), filter switching method, and camera determine the available spectral range, possible scan speed, detector sensitivity, and spatial sampling. Mercury arc lamps offer many excitation wavelength peaks but do not provide a flat spectral output and will require significant signal reduction at the output peaks to correct the spectral image data back to a NIST-traceable response38. Alternative light sources, such as Xe arc lamps and white light supercontinuum lasers, may provide a more uniform spectral output that is better suited for excitation-scanning hyperspectral imaging38,39,40. Choice of light source, tunable filters, and dichroic filters determine the available spectral range. This range should be chosen with careful consideration for the desired spectral information of the experimental sample.
Additionally, consideration should be given to the switching mechanism used for the tunable filters, as well as various camera factors such as quantum efficiency, pixel size, and available frame rates, as these factors will affect potential sampling rates41,42,43. However, all other factors being constant, utilization of the excitation-scanning approach should provide increased sensitivity and the ability for faster imaging, compared to most emission-scanning spectral imaging approaches21.
As noted in the introduction, hyperspectral imaging here refers to the contiguous and spectrally overlapping nature of the acquired data. As such, the capabilities of the system need to be able to collect data using spacing of excitation center wavelengths less than half the distance of the FWHM of the filters. As reported previously, a carefully chosen array of thin-film tunable filters allows data acquisition with center wavelengths spaced 5 nm apart, a distance sufficient to oversample the excitation spectrum given filters with FWHM of between 14-20 nm. Such a spacing gives slight redundancy in spectral data collection with likely increases the accuracy of the unmixing process. Consideration of both spectral ranges and necessary minimum number of wavelength channels for accurate unmixing have been discussed previously44,45,46. To this end, given the bandwidth of these filters, the excitation range should be selected to end at a wavelength that is somewhat lower (5-10 nm lower) than the cut-off wavelength of the dichroic beamsplitter. This will ensure that the entire bandwidth of the excitation illumination is below the cutoff wavelength of the dichroic beamsplitter (e.g., 360-485 nm for the 495 dichroic filter) to avoid excitation-emission cross-talk.
Software to independently control each component of the hardware to achieve high-speed spectral imaging scans is required. The software should be able to operate the shutter, select excitation wavelengths, and acquire images at sufficiently high speeds to meet experimental conditions (exact sample rate requirements will vary experiment, but an example goal might be to acquire four complete spectral image stacks per minute). More complex experiments may make use of multiple dichroic mirrors, objectives, or XYZ locations. There are a number of software packages available for data acquisition47,48. Micro-Manager is a free open-source software for microscope automation that offers a variety of customization options. Furthermore, Micro-Manager includes a scripting panel for additional customization not available within the main user interface. For example, it is possible to use a custom script to reduce the 250 ms delay of the tunable filter switcher to 10 ms at all excitation wavelengths except the wavelength transitions in which the filter wheel rotates to a new tunable filter, reducing the effective imaging time by 240 ms per wavelength for most wavelengths. This customization has allowed acquisition of up to 30 wavelengths in under 4 s. Finally, Micro-Manager can be operated alongside other environments, such as MATLAB, to further customize microscope device control.
Determining the proper spectral excitation range, acquisition time, and initial wavelength for sample viewing and subsequent data acquisition is very important. However, incorrect operation of individual components of the system may require further troubleshooting. Each component of the optical path contributes to the image data acquired. Hence, it is important to verify the spectral response and optical transmission of optical components within the light path, especially if trying to optimize overall system response. Light sources often have variable intensity settings and may have reduction in power over the lifetime of the bulb40. If a sample appears dim, the cause may be reduced output from the light source. The autoshutter function in Micro-Manager, in our experience, does not operate 100% of the time. Lack of signal may indicate a closed shutter. The initial excitation wavelength saved within Micro-Manager's configuration file may default to a wavelength with little or no spectral output, such as the 340 nm example shown in Figure 1.
Additionally, it is not uncommon to mistakenly choose an excitation wavelength that is above the cutoff wavelength of the long-pass dichroic beamsplitter, resulting in additional cross-talk and/or excitation light being shunted directly to the camera which may potentially damage the camera sensor. Similarly, adjusting the light path for transmission imaging may result in loss of signal to the camera, depending on the configuration of the microscope. Furthermore, as indicated in Figure 2, choice of initial excitation wavelength and exposure time for sample viewing may appear appropriate but actually result in oversaturated pixels at other excitation wavelengths. This fact will not be apparent unless pixel saturation is checked for each image, so it is often prudent to perform a test image stack on an area of the sample that appears to contain the most intense fluorescence.
It is also worth noting that variable intensities may be present in images chosen for background regions. Care should be taken to ensure that these regions do not actually contain relevant image data, as subsequent data correction steps will then subtract this signal from the image data acquired at other regions in the sample. It is sometimes necessary to use the transmission settings and/or drastically increase the exposure time to check that the region selected for measuring sample background does indeed contain no sample. Similarly, unmixed images may present with dark spots or holes in the autofluorescence image. This may be due to an improper library caused by autofluorescence "contamination" of the "pure" spectral signals from single-labeled cells. This is because any "pure" spectral signal collected from a sample containing autofluorescence will invariably contain some of the autofluorescence spectra itself, even if in trace amounts. Care should be taken to subtract out autofluorescence signals from the single-labeled controls to ensure a proper spectral library, as described on several occasions by Mansfield et al.28,29,30,31,32
Though not demonstrated in this article, the excitation-scanning spectral imaging system can also be used for time-lapse imaging and imaging over multiple XY locations (or a large field of view due to image stitching) in multiple focal planes. If these options are desired for a single experiment, careful thought should be given to the importance of acquisition order and potential effects of photobleaching. It is also worth noting that if the actual time taken exceeds the estimated time (e.g., an image stack takes 11 s to acquire rather than the estimated 10 s), Micro-Manager will continue to acquire the selected number of timepoints and/or positions. This miscalculation can compound with multiple timepoints and alter calculated temporal sampling, potentially skewing any conclusions made from the data.
This example demonstrates the ability to separate three sources of fluorescence within a 145 nm excitation scanning range. Previous experiments using this system have been able to separate up to five sources of fluorescence within the same range. The time required for this image acquisition is less than 1 min, which is significantly faster than most emission-scanning spectral imaging systems. Improvements in the light path, such as higher speed excitation wavelength tuning, may further advance this imaging technology to speeds that are sufficient for video-rate spectral imaging.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge support from NSF 1725937, NIH P01HL066299, NIH R01HL058506, NIH S10OD020149, NIH UL1 TR001417, NIH R01HL137030, AHA 18PRE34060163, and the Abraham Mitchell Cancer Research Fund.
Materials
| Airway Smooth Muscle Cells | National Disease Research Interchange (NDRI) | Isolated from human lung tissues obtained from NDRI | Highly autofluorescent, calcium sensitive cells |
| Automated Shutter | Thorlabs Inc. | SHB1 | Remote-controllable shutter to minimize photobleaching |
| Automated Stage | Prior Scientific | H177P1T4 | Remote-controllable stage for automated multiple field of view or stitched image collection. |
| Automated Stage Controller (XY) | Prior Scientific | Proscan III (H31XYZE-US) | For interfacing automated stage with computer and joystick |
| Buffer | Made in-house | Made in-house | 145 mM NaCl, 4 mM KCl, 20 mM HEPES, 10 mM D-glucose, 1 mM MgCl2, and 1mM CaCl2, at pH 7.3 |
| Cell Chamber | ThermoFisher Scientific | Attofluor Cell Chamber, A7816 | Coverslip holder composed of surgical stainless steel and a rubber O-ring to seal in media and prevent sample and/or objective contamination |
| Excitation Filters | Semrock Inc. | TBP01-378/16 | Center wavelength range (340-378 nm), Bandwidth (Minimum 16 nm, nominal FWHM 20 nm), Refractive index (1.88) |
| Semrock Inc. | TBP01-402/16 | Center wavelength range (360-400 nm), Bandwidth (Minimum 16 nm, nominal FWHM 20 nm), Refractive index (1.8) | |
| Semrock Inc. | TBP01-449/15 | Center wavelength range (400-448.8 nm), Bandwidth (Minimum 15 nm, nominal FWHM 20 nm), Refractive index (1.8) | |
| Semrock Inc. | TBP01-501/15 | Center wavelength range (448.8-501.5 nm), Bandwidth (Minimum 15 nm, nominal FWHM 20 nm), Refractive index (1.84) | |
| Semrock Inc. | TBP01-561/14 | Center wavelength range (501.5-561 nm), Bandwidth (Minimum 14 nm, nominal FWHM 20 nm), Refractive index (1.83) | |
| Fluorescence Filter Cube Dichroic Beamsplitter | Semrock Inc. | FF495-Di03 | Separates excitation and emission light at 495 nm (>98% reflection between 350-488 nm, >93% transmission between 502-950 nm), Filter effective index (1.78) |
| Fluorescence Filter Cube Longpass Filter | Semrock Inc. | FF01 496/LP-25 | Allows passage of light longer than 496 nm ( >93% average transmission between 503.2-1100 nm), Refractive index (1.86) |
| GCaMP Probe | Addgene | G-CaMP3; Plasmid #22692 | A single-wavelength GCaMP2-based genetically encoded calcium indicator |
| Integrating Sphere | Ocean Optics | FOIS-1 | Used for accurate measurement of wide-angle illumination |
| Inverted Fluorescence Microscope | Nikon Instruments | TE2000 | Inverted microscopes allow direct excitation of sample without the need to penetrate layers of media and/or tissue. |
| Mitotracker Green FM | ThermoFisher Scientific | M7514 | Labels mitochondria |
| NIST-Traceable Calibration Lamp | Ocean Optics | LS-1-CAL-INT | A lamp with a known spectrum for use as a standard |
| NIST-Traceable Fluorescein | ThermoFisher Scientific | F36915 | For verifying appropriate spectral response of the system |
| NucBlue | ThermoFisher Scientific | R37605 | Labels cell nuclei |
| Objective (10X) | Nikon Instruments | Plan Apo λ 10X/0.45 ∞/0.17 MRD00105 | Useful for large fields of view |
| Objective (20X) | Nikon Instruments | Plan Apo λ 20X/0.75 ∞/0.17 MRD00205 | Most often used for tissue samples |
| Objective (60X) | Nikon Instruments | Plan Apo VC 60X/1.2 WI ∞/0.15-0.18 WD 0.27 | Most often used for cell samples |
| sCMOS Camera | Photometrics | Prime 95B (Rev A8-062802018) | For acquiring high-sensitivity digital images |
| Spectrometer | Ocean Optics | QE65000 | Used to measure spectral output of excitation-scanning spectral system |
| Tunable Filter Changer | Sutter Instrument | Lambda VF-5 | Motorized unit for automated excitation filter tuning/switching |
| Xenon Arc Lamp | Sunoptic Technologies | Titan 300HP Lightsource | Light source with relatively uniform spectral output |
Riferimenti
- Hagen, N. A., Kudenov, M. W. Review of snapshot spectral imaging technologies. Optical Engineering. 52 (9), 90901 (2013).
- Li, Q., He, X., Wang, Y., Liu, H., Xu, D., Guo, F. Review of spectral imaging technology in biomedical engineering: achievements and challenges. Journal of Biomedical Optics. 18 (10), 100901 (2013).
- Lu, G., Fei, B. Medical hyperspectral imaging: a review. Journal of Biomedical Optics. 19 (1), 10901 (2014).
- Mehta, N., Shaik, S., Devireddy, R., Gartia, M. R. Single-Cell Analysis Using Hyperspectral Imaging Modalities. Journal of Biomechanical Engineering. 140 (2), 20802 (2018).
- Goetz, A. F. H. Three decades of hyperspectral remote sensing of the Earth: A personal view. Remote Sensing of Environment. 113, S5-S6 (2009).
- Goetz, A. F. Measuring the Earth from Above: 30 Years(and Counting) of Hyperspectral Imaging. Photonics Spectra. 45 (6), 42-47 (2011).
- Schröck, E. Multicolor spectral karyotyping of human chromosomes. Science. 273 (5274), 494-497 (1996).
- Tsurui, H. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. Journal of Histochemistry & Cytochemistry. 48 (5), 653-662 (2000).
- Lu, R., Chen, Y. R. Hyperspectral imaging for safety inspection of food and agricultural products. SPIE. 3544, 121-134 (1999).
- Hege, E. K., O’Connell, D., Johnson, W., Basty, S., Dereniak, E. L. Hyperspectral imaging for astronomy and space surviellance. SPIE. 5159, 380-392 (2004).
- Vo-Dinh, T. A hyperspectral imaging system for in vivo optical diagnostics. IEEE Engineering in Medicine and Biology Magazine. 23 (5), 40-49 (2004).
- Dorrepaal, R. M., Gowen, A. A. Identification of Magnesium Oxychloride Cement Biomaterial Heterogeneity using Raman Chemical Mapping and NIR Hyperspectral Chemical Imaging. Scientific Reports. 8 (1), 13034 (2018).
- Swayze, G. A. Using imaging spectroscopy to map acidic mine waste. Environmental Science & Technology. 34 (1), 47-54 (2000).
- Khoobehi, B., Beach, J. M., Kawano, H. Hyperspectral imaging for measurement of oxygen saturation in the optic nerve head. Investigative Ophthalmology & Visual Science. 45 (5), 1464-1472 (2004).
- Gowen, A., O’Donnell, C., Cullen, P., Downey, G., Frias, J. Hyperspectral imaging-an emerging process analytical tool for food quality and safety control. Trends in Food Science & Technology. 18 (12), 590-598 (2007).
- Edelman, G., van Leeuwen, T. G., Aalders, M. C. Hyperspectral imaging for the age estimation of blood stains at the crime scene. Forensic Science International. 223 (1), 72-77 (2012).
- Edelman, G., Gaston, E., Van Leeuwen, T., Cullen, P., Aalders, M. Hyperspectral imaging for non-contact analysis of forensic traces. Forensic Science International. 223 (1), 28-39 (2012).
- Markgraf, W., Feistel, P., Thiele, C., Malberg, H. Algorithms for mapping kidney tissue oxygenation during normothermic machine perfusion using hyperspectral imaging. Biomedical Engineering/Biomedizinische Technik. 63 (5), 557-566 (2018).
- Boubanga-Tombet, S. Thermal Infrared Hyperspectral Imaging for Mineralogy Mapping of a Mine Face. Remote sensing. 10 (10), 1518 (2018).
- Yuen, P. W., Richardson, M. An introduction to hyperspectral imaging and its application for security, surveillance and target acquisition. The Imaging Science Journal. 58 (5), 241-253 (2010).
- Favreau, P. F. Excitation-scanning hyperspectral imaging microscope. Journal of Biomedical Optics. 19 (4), 046010-046010 (2014).
- Favreau, P. Thin-film tunable filters for hyperspectral fluorescence microscopy. Journal of biomedical optics. 19 (1), 011017-011017 (2014).
- Leavesley, S. J. Hyperspectral imaging microscopy for identification and quantitative analysis of fluorescently-labeled cells in highly autofluorescent tissue. Journal of Biophotonics. 5 (1), 67-84 (2012).
- Annamdevula, N. S. Spectral imaging of FRET-based sensors reveals sustained cAMP gradients in three spatial dimensions. Cytometry Part A. 93 (10), 1029-1038 (2018).
- Deshpande, D. A., Walseth, T. F., Panettieri, R. A., Kannan, M. S. CD38/cyclic ADP-ribose-mediated Ca2+ signaling contributes to airway smooth muscle hyper-responsiveness. The FASEB Journal. 17 (3), 452-454 (2003).
- Deshpande, D. A. Modulation of calcium signaling by interleukin-13 in human airway smooth muscle: role of CD38/cyclic adenosine diphosphate ribose pathway. American Journal of Respiratory Cell and Molecular Biology. 31 (1), 36-42 (2004).
- Guo, M. Cytokines regulate β-2-adrenergic receptor responsiveness in airway smooth muscle via multiple PKA-and EP2 receptor-dependent mechanisms. Biochimica. 44 (42), 13771-13782 (2005).
- Mansfield, J. R., Gossage, K. W., Hoyt, C. C., Levenson, R. M. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. Journal of Biomedical Optics. 10 (4), 41207 (2005).
- Mansfield, J. R., Hoyt, C., Levenson, R. M. Visualization of microscopy-based spectral imaging data from multi-label tissue sections. Current Protocols in Molecular Biology. 84 (1), 14-19 (2008).
- Bouchard, M. B. Recent advances in catheter-based optical coherence tomography (OCT) have provided the necessary resolution and acquisition speed for high-quality intravascular imaging. Complications associated with clearing blood from the vessel of a living patient have. Journal of Biomedical Optics. 12 (5), 51601 (2007).
- Mansfield, J. R. Distinguished photons: a review of in vivo spectral fluorescence imaging in small animals. Current Pharmaceutical Biotechnology. 11 (6), 628-638 (2010).
- Levenson, R. M., Mansfield, J. R. Multispectral imaging in biology and medicine: slices of life. Cytometry Part A. 69 (8), 748-758 (2006).
- Gammon, S. T., Leevy, W. M., Gross, S., Gokel, G. W., Piwnica-Worms, D. Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Analytical Chemistry. 78 (5), 1520-1527 (2006).
- . . Spectral Unmixing Plugins. , (2006).
- . . Spectral Unmixing of Bioluminescence Signals. , (2006).
- Keshava, N., Mustard, J. F. Spectral unmixing. IEEE Signal Processing Magazine. 19 (1), 44-57 (2002).
- Deal, J. Identifying molecular contributors to autofluorescence of neoplastic and normal colon sections using excitation-scanning hyperspectral imaging. Journal of Biomedical Optics. 23 (12), (2018).
- Microscopy Key, . Microscopy: Key Considerations for Nonlaser Light Sources | Features. BioPhotonics. , (2016).
- Chiu, L., Su, L., Reichelt, S., Amos, W. Use of a white light supercontinuum laser for confocal interference-reflection microscopy. Journal of Microscopy. 246 (2), 153-159 (2012).
- . . Choosing the best light source for your fluorescence experiment. , (2019).
- Beier, H. T., Ibey, B. L. Experimental comparison of the high-speed imaging performance of an EM-CCD and sCMOS camera in a dynamic live-cell imaging test case. PLoS ONE. 9 (1), e84614 (2014).
- Tutt, J. Comparison of EM-CCD and scientific CMOS based camera systems for high resolution X-ray imaging and tomography applications. Journal of Instrumentation. 9 (6), P06017 (2014).
- Coates, C. New sCMOS vs. current microscopy cameras. Biophotonics International. 18 (5), 24-27 (2011).
- Neher, R., Neher, E. Optimizing imaging parameters for the separation of multiple labels in a fluorescence image. Journal of Microscopy. 213 (1), 46-62 (2004).
- Deal, J. Hyperspectral imaging fluorescence excitation scanning spectral characteristics of remodeled mouse arteries. SPIE. 10890, 108902M (2019).
- Deal, J., Rich, T. C., Leavesley, S. J. Optimizing channel selection for excitation-scanning hyperspectral imaging. SPIE. , 108811B (2019).
- Biehlmaier, O., Hehl, J., Csucs, G. Acquisition speed comparison of microscope software programs. Microscopy Research and Technique. 74 (6), 539-545 (2011).
- . . Comparison with other microscopy software – Micro-manager. , (2012).

