Pan-lyssavirus Real Time RT-PCR for Rabies Diagnosis
Summary
This real-time RT-PCR using dsDNA intercalating dye is suitable to diagnose lyssavirus infections. The method begins with RNA extracted from rabies suspected ante-mortem or post-mortem samples, detailing master mix preparation, RNA addition, setup of the real-time machine and correct interpretation of results.
Abstract
Molecular assays are rapid, sensitive and specific, and have become central to diagnosing rabies. PCR based assays have been utilized for decades to confirm rabies diagnosis but have only recently been accepted by the OIE (World Organisation for Animal Health) as a primary method to detect rabies infection. Real-time RT-PCR assays provide real-time data, and are closed-tube systems, minimizing the risk of contamination during setup. DNA intercalating fluorochrome real-time RT-PCR assays do not require expensive probes, minimizing the cost per sample, and when the primers are designed in conserved regions, assays that are specific across virus genera rather than specific to just one virus species are possible. Here we describe a pan-lyssavirus SYBR real-time RT-PCR assay that detects lyssaviruses across the Lyssavirus genus, including the most divergent viruses IKOV, WCBV and LLEBV. In conjunction with dissociation curve analysis, this assay is sensitive and specific, with the advantage of detecting all lyssavirus species. The assay has been adopted in many diagnostic laboratories with quality assured environments, enabling robust, rapid, sensitive diagnosis of animal and human rabies cases.
Introduction
Diagnosis of rabies using molecular methodologies was accepted by the OIE in 20181, recognizing the advantages of these techniques in confirming rabies cases, particularly in situations when the samples are sub-optimal, or for ante-mortem diagnosis, as there is no requirement for live virus or fresh samples. PCR assays for lyssaviruses require a reverse transcription (RT) before PCR can commence as the genome is RNA. RT-PCR assays that detect the 3’ proximal region of the genome are considered the most sensitive, as transcriptional gradients occur during lyssavirus replication. Commonly used RT-PCR assays can be divided broadly into two categories, end-point (or gel-based) and real-time. Both approaches are sensitive and specific; however, the real-time assay has some additional benefits such as obtaining results in ‘real-time’ and being performed in an entirely closed tube system, thereby reducing the potential for operator contamination. There are two main approaches to detect lyssavirus-specific amplicons obtained using real-time assays. The first utilizes hydrolysis probes (such as TaqMan probes) that contain a fluorophore and a quencher. When the probe binds to the target region during amplification, the exonuclease activity of the polymerase results in dissociation of fluorophore and quencher, enabling the resulting fluorescence to be measured. The second utilizes a DNA intercalating dye (fluorochrome such as SYBR Green) that binds to double stranded DNA during amplification. The bound fluorochromes emit fluorescence that is detected at each cycle, allowing real time detection and quantification of the product. Due to the non-specific nature of binding to any dsDNA, a dissociation curve analysis is undertaken to confirm the specificity of the reaction. Real-time RT-PCRs are rapid due to the small amplicon sizes, typically less than 200 bp in length; however, identifying suitable regions to design primers and probes in conserved regions, can prove challenging, therefore removing the requirement for a probe is a distinct advantage.
A number of real-time RT-PCRs have been designed for specifically detecting individual strains or lineages of RABV2 and also to detect lyssaviruses across the genus3,4,5,6,7,8,9. All assays will have a limit of detection dependent on how conserved the primer (and if necessary, the probe) sequences are across the genus. Indeed, emerging or novel virus strains may render the highly specific probe-based assays ineffective. The choice of the real-time detection (dye vs. probe) will depend on the intended application. For a laboratory conducting surveillance on locally sourced brain material and expecting high numbers of negative samples, the use of the cheaper intercalating dye is a sensible choice. The SYBR Green approach would also be optimal when conducting scanning surveillance where the presence of novel or divergent lyssaviruses would remain undetected by more restricted probe-based assays.
All members of the genus Lyssavirus cause the disease rabies, which is fatal once symptoms appear. The vast majority of human and animal rabies cases are due to rabies virus (RABV), the dominant reservoir for which is the domestic dog10. Bats are important host reservoirs for lyssaviruses and all but two lyssavirus species characterized have been identified directly in bats — Ikoma lyssavirus (IKOV) and Mokola virus (MOKV) — and of these two, IKOV has been speculated to have a bat host reservoir11. In addition to the 16 recognized lyssavirus species12, there are two lyssaviruses that have been recently described: Taiwan bat lyssavirus (TWBLV)13 and Kotalahti bat lyssavirus (KBLV)14. Lyssaviruses can be genetically divided into three phylogroups, with the majority of lyssaviruses, including RABV, belonging to phylogroup I. However, the most divergent lyssaviruses belong to phylogroup III and are unlikely to be detected by RT-PCRs designed to target RABV or phylogroup I virus sequences.
The assay described here utilizes the real-time primer pair JW12-N165, first described in 20053. The primers were designed to be pan-lyssavirus albeit the original application was as a TaqMan assay with probes to differentiate lyssavirus species. Subsequent confirmation that the primer pair was pan-lyssavirus in specificity was achieved utilizing a 2-step SYBR real-time assay on all lyssavirus species available including WCBV15. The primer pair is described here in a one-step RT-PCR real-time assay utilizing an intercalating fluorochrome, validated using representatives from all 16 recognized lyssavirus species. This one-step real-time assay is a rapid, sensitive, lyssavirus-specific assay and demonstrates that the robustness of the primer set to identify even highly divergent lyssavirus species.
Protocol
Samples from diagnostic material received at APHA after natural infection, or obtained by inoculating mice using protocols assessed by the APHA ethics and statistical committee under UK Home Office regulations under licence 70/7394.
1. Quantification of RNA Using a Micro-volume Spectrophotometer
- Ensure the settings of the spectrophotometer are set to RNA.
- Use 1-2 µL of molecular grade water to initialize the machine and set a baseline.
- Use 1-2 µL of each test RNA sample to assess the RNA quantity.
- Save the readings and document.
- Adjust the RNA to 1 µg/µL, if required.
NOTE: RNA must be kept on ice (or in a cool block) at all times. If RNA is obtained using a column, or bead-based method the RNA is usually less than 1 µg/µL. In this situation use the RNA neat.
2. Preparation of RNA Dilution Series to Determine End Point Sensitivity
- Make a 10-fold serial dilution of the RNA.
- Label tubes with the dilution series (e.g., 10-1, 10-2, etc.) and the RNA details.
- Add 45 µL of molecular grade water to each tube.
- Add 5 µL of the RNA (previously diluted to 1 µg/µL) and mix well.
- Dispose of pipette tip in appropriate disinfectant and replace with a fresh tip.
- Take 5 µL from the 10-1 and add to 10-2 tube and mix well.
- Repeat 2.1.3-2.1.5 with the remaining dilutions.
NOTE: RNA must be kept on ice (or in a cool block) at all times.
3. Preparation of Real-time RT-PCR Reactions
- Using a spreadsheet, plan the plate layout according to the number of test samples and control samples, for both the lyssavirus and ß-actin assays.
NOTE: If 4 samples are to be tested in duplicate with a positive and negative control, this equates to 10 reactions for both assays. - In a ‘clean room’ or area separate from the RNA template, wipe down the surfaces with an appropriate disinfectant prior to use or prepare a PCR workstation (if using). To prepare the workstation, wipe the cabinet surface with an appropriate disinfect and place the items required into the workstation and close the doors. Switch on the UV light for 10 minutes
- Remove regents and primers from the freezer and thaw (reagents listed in Table 1 and primers in Table 2).
NOTE: The enzyme mix is stored in glycerol so does not require thawing and must be kept on ice (or in a cool block) at all times. All other reagents can be thawed at room temperature. - Once thawed, mix the reagents and centrifuge briefly to collect liquid.
NOTE: Do not vortex the enzyme mix, just centrifuge briefly. - Prepare separate master mixes for lyssavirus and ß-actin. For each reaction, add 7.55 µL of molecular grade water, 10 µL of 2x Universal SYBR green reaction mix, 0.6 µL of forward primer, 0.6 µL of reverse primer, 0.25 µL of iTaq RT enzyme mix.
- Use a spreadsheet to calculate correct volumes to avoid errors in manual calculating. Ensure enough master-mix is prepared to compensate for pipetting error. Therefore if 10 reactions are required (see NOTE in 3.1), prepare 12 reactions
- Prepare the master-mix on ice (or in a cool block) and remain on ice until placed into the real-time machine.
- Mix the prepared master-mixes, centrifuge briefly and dispense 19 µL into the relevant wells of strip tubes or a 96 well plate compatible with the real-time machine in use.
NOTE: Minimize the production of bubbles in the wells whilst pipetting. - In a separate room, or in a UV cabinet prepared as described in 3.2, carefully add 1 µL of the RNA previously adjusted to 1 µg/µL (see step 1.5) below the surface of the appropriate master-mix well and mix gently. Discard pipette tip into disinfectant directly after use (under the surface).
- Add the controls after the test samples, with the positive control added next and the no template control (NTC – molecular grade water) added last. The amount of RNA used can be altered depending on the sample type, and RNA extraction used. The amount used must be validated to ensure the reaction is optimized.
- Seal plate using strip lids or sealer taking care to ensure all the lids are firmly closed and labelled sufficiently to orientate the samples. Label the edge of the plate/strip tubes.
- Spin down the samples using a centrifuge to collect all the liquid at the bottom of the wells.
- Transfer plate to the real-time PCR machine, open the door and place in the holder ensuring the correct location/orientation of the samples according to the plate layout.
NOTE: If tube strips are used, ensure the holder is in place. - Open up the real-time PCR machine program and choose the option for SYBR real-time experiment with dissociation curve. Program the real-time PCR machine using the thermal cycling conditions specified in Table 3, including the data collection points.
- Select SYBR as the fluorescent dye and select unknown as the sample type and insert a name into the correct sample name box.
NOTE: Differentiate between the replicates and also between the lyssavirus and ß-actin wells. - Choose a file location to save the experimental data, ensure the lamp will be switched off at the end of the run, then start the run.
NOTE: As the first step is an RT stage, no data is collected during this time, therefore, if the lamp requires a warm up period this can occur during the RT stage. The real-time machine and software will display the amplification curves in real-time, while the melting curve will be generated at the end of the cycle.
4. Data Analysis
- Once the run has been completed perform the data analysis as follows.
- First analyze the amplification plot results of the test samples alongside the control samples. Positive samples display exponential ramps, usually followed by plateau and a Ct value. Negative samples display flat amplification plots with no Ct values (Figure 1A). The Ct value is automatically calculated by the software, although this should be checked and manually altered if required.
- Second, analyze the dissociation curve results of the test samples alongside the control samples. A positive sample will have a melting temperature (Tm) 77 – 80 °C, and overlap with the positive control Figure 1B).
- Obtain the overall diagnostic result by ensuring the controls are valid. Use Table 4 to interpret the results in relation to the internal ß-actin control. If the positive control samples are negative and/or the negative samples are positive, the run should be disregarded.
- Record the Ct and Tm values obtained for the control RNA in a ‘control card’ to enable trend analysis and help identify drifts in the assay sensitivity.
Representative Results
Following the protocol describe above, the sensitivity of the pan-lyssavirus RT-PCR was demonstrated on a dilution series of the control standard virus (CVS) (Figure 1) and a range of other lyssaviruses (Figure 2 and Figure 3). SYBR Green I dye was utilized as a universal one-step RT-PCR, where cDNA synthesis and PCR amplification are carried out in a single tube. As the amplification of the specific target occurs, more dye is bound, resulting in real-time increased levels of fluorescence. All dye intercalating real-time assays must be interpreted in two phases: amplification and dissociation. The amplification phase is identical to any real-time amplification (Figure 1A). There is a linear ‘early phase’ during the early cycles where DNA amplification cannot be calculated due to insufficient signal in relation to the background. The length of this is directly related to the amount of target in the sample. Subsequently, there is an exponential phase where the doubling of DNA molecules is detected and recorded. Finally, the plateau phase is reached (apart from the highly diluted samples which may not reach this phase before the end of the program). In this phase, the intensity of fluorescence levels out, due to the exhaustion of reagents. The amplification plots observed using a 10-fold serial dilution of CVS, conformed to the expected plots (Figure 1A,C) where the lyssavirus assay demonstrated a higher sensitivity than the ß-actin assay. The dissociation curve was calculated after amplification, where the dsDNA was dissociated into ssDNA by an incremental increase in temperature and the fluorescence monitored as a function of temperature (Figure 1B, D-F). The threshold temperature at which the specific amplicon dissociates into ssDNA, caused a release of fluorescence, which was measured by the thermocycler software (Tm). This dissociation phase provided data on the amplicon size, enabling the user to interpret the result in comparison to a positive control, resulting in a negligible likelihood of false-positive results. The Tm observed for CVS using the pan-lyssavirus assay (Figure 1B) and ß-actin assay (Figure 1D) are distinct, and aided the user to confirm the correct assay analysis by noting the Tm obtained (Figure 1E). Furthermore, the Ct and Tm values between runs and operators was assessed and shown to be reproducible (Table 5). The threshold used to calculate the Ct value was calculated automatically by the software and dependent on many factors, including the reaction mix or instrument used. Over the 12 independent runs the mean Ct was 20.66 (SD 0.63) for the lyssavirus assay and 27.5 (SD 1.13) for the ß-actin assay. In contrast the variation observed in the Tm values was markedly lower, due to the lack of external influences on this measurement. For example, the mean Tm for the CVS lyssavirus assay was 78.92 (SD 0.16) (Table 5), when compared to the mean of all lyssaviruses 78.81 °C (SD 0.531) (Table 6 and Figure 1F). This lack of variation in the Tm across the Lyssavirus genus is advantageous as the same control RNA can be used irrespective of the lyssavirus in the sample, however differentiating between the lyssavirus species using the Tm is not possible, particularly because different RABV sub-lineages spanned the range of Tm values observed (Table 6). Non-specific amplification plots are rarely observed with this assay; however, specific parameters to define a positive result vs. a non-specific negative result are required. The SD observed across all lyssaviruses (0.531) was applied to the lowest (77.34 – LBVa) and highest (79.67 – IKOV) observed Tm values to set a range 76.8 °C – 80.2 °C for positive specific bands. Therefore, Tm values outside this range were considered non-specific and therefore a negative result. Occasionally multiple peaks are observed for a sample. If the dominant peak is at the correct Tm (for each replicate) then the sample is considered positive. The most common reason a non-specific peak is observed is due to primer-dimers, the assay has been optimized to minimize primer dimers. Primer dimers typically result in an amplicon smaller than that of the target sequence, therefore would have a Tm lower than the specific product.
A 10-fold serial dilution of three lyssavirus positive brain sample RNAs extracted using TRIzol, were run in parallel and plotted (Figure 2). The limit of detection for the three lyssaviruses varied, but none exceeded Ct 36. The R2 coefficient values for the viruses plotted in Figure 2 ranged from 0.9637 and 0.996. For all viruses analyzed (Table 6) the range did not exceed this, furthermore, 7 of the 29 lyssavirus had R2 >0.99 (data not shown). Taking into account that the preparation of the dilution series is from total RNA extractions, the linearity observed provides evidence that the assay is robust. Finally, detection of all lyssavirus species (particularly the most diverse phylogroup III viruses) was investigated using a panel of RNAs spanning all three phylogroups in the Lyssavirus genus. RNA extracted from either original, or experimentally infected mice, brain material, was utilized using the protocols described above. The results confirm that the primers amplify all lyssavirus species, including the divergent phylogroup III lyssaviruses IKOV, WBCV and LLEBV (Table 6 and Figure 3). A diverse panel of non-lyssavirus rhabdoviruses, originally collected and analyzed antigenically16, and more recently genetically17 were screened and no-cross reactivity was detected, indicating that the primers are specific for members of the Lyssavirus genus only (data not shown). The pan-lyssavirus real-time assay has been included in the EURL (EU Reference Laboratory) inter-laboratory proficiency schemes since 2013, demonstrating 100% concordance with other molecular assays such as the pan-lyssavirus TaqMan assay and the conventional RT-PCR assay in addition to the FAT (Fluorescent Antibody test).
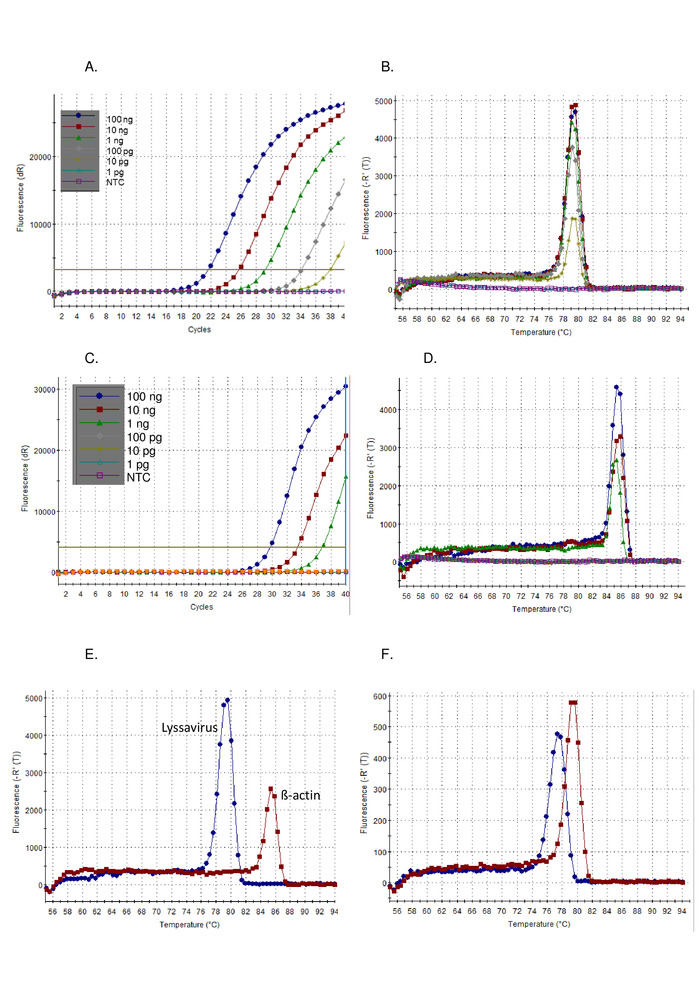
Figure 1: 10-fold serial dilution of CVS positive control RNA, run on the pan-lyssavirus RT-PCR assay, visualized as the amplification plot (A), and dissociation curve (B), and run on the ß-actin RT-PCR assay visualized as the amplification plot (C), and dissociation curve (D). NTC = no template control. Comparison of the CVS control RNA run on both the pan-lyssavirus RT-PCR assay (blue) and the ß-actin RT-PCR assay (red) demonstrating the difference in dissociation curves (E) — see Table 5 for mean values; and finally dissociation curves for LBVa (blue) and IKOV (red) demonstrating the range of Tm values observed across the Lyssavirus genus (F). Please click here to view a larger version of this figure.
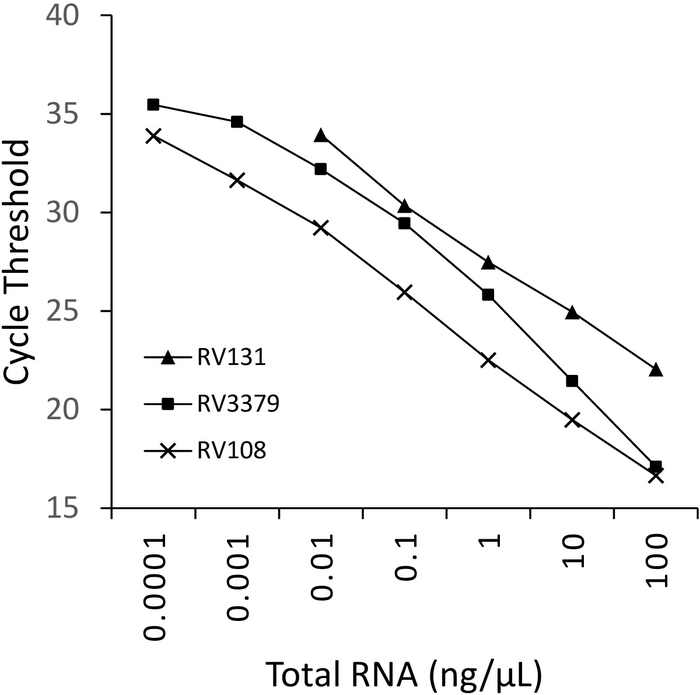
Figure 2: 10-fold serial dilutions for three lyssavirus species: RABV (RV108), DUVV (RV131) and ARAV (RV3379). R2 = 0.996, 0.9962 and 0.9637 respectively. Data points at 10-7 (0.0001 ng/µL) reached the limit of detection (where a value was obtained). Please click here to view a larger version of this figure.
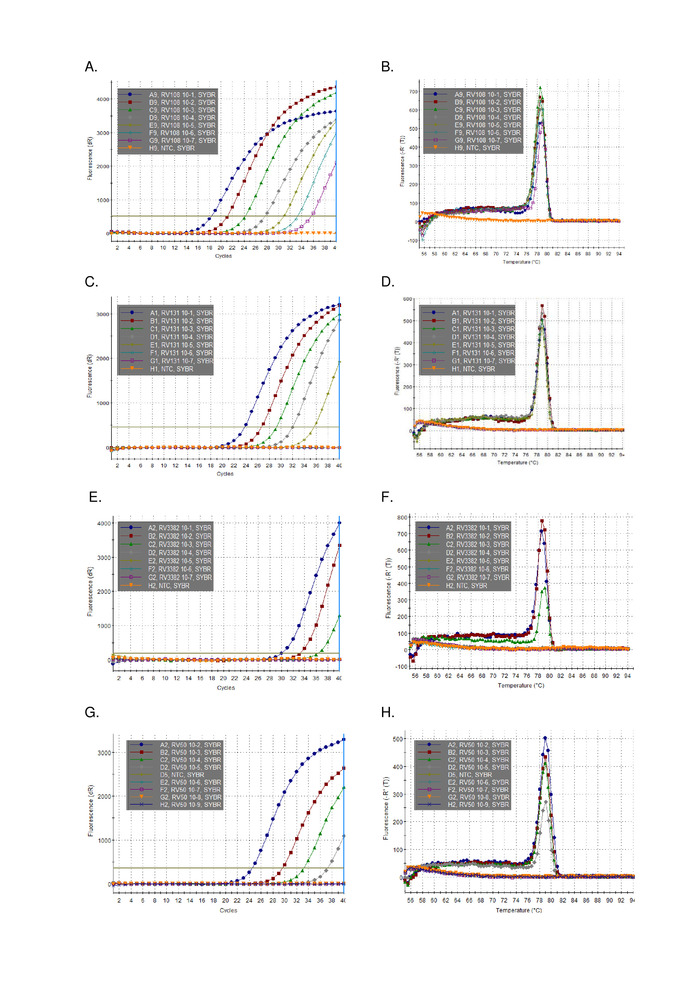
Figure 3: Representative 10-fold serial dilution data from across the Lyssavirus genus (see individual legends for lyssavirus identity and Table 6 for tabulated results in comparison to other lyssaviruses). Panels A, C, E, G amplification plots and B, D, F, H dissociation curves for A, C, E, and G respectively. Please click here to view a larger version of this figure.
| Reagent | μL/Reaction |
| Molecular grade water | 7.55 |
| 2x Universal RT PCR reaction mix | 10 |
| Primer Forward [20 μM] | 0.6 |
| Primer Reverse [20 μM] | 0.6 |
| RT enzyme mix | 0.25 |
| Total per reaction | 19 |
Table 1: Pan-lyssavirus real-time RT-PCR master mix reagents.
| Assay | Primer name | Primer role | Sequence 5’-3’ | Posizione1 |
| Lyssavirus | JW12 | RT-PCR | ATG TAA CAC CYC TAC AAT G | 53-73 |
| N165 | PCR | GCA GGG TAY TTR TAC TCA TA | 165-146 | |
| ß-actin | ß-actin intronic | PCR | CGA TGA AGA TCA AGA TCA TTG | 1051-1072 |
| ß-actin reverse | RT-PCR | AAG CAT TTG CGG TGG AC | 1204-1188 | |
| Primer positions are given in relation to Pasteur virus sequence (M13215) and mouse ß-actin gene sequence (NM_007393) | ||||
Table 2: Pan-lyssavirus real-time RT-PCR primer details.
| Stage | Cycles | Temperature | Time | Data Collection |
| Reverse Transcription | 1 | 50 °C | 10 min | |
| RT inactivation/initial denaturation | 1 | 95 °C | 5 min | |
| Amplification | 40 | 95 °C | 10 s | |
| 60 °C | 30 s | end point | ||
| Dissociation curve analysis | 1 | 9 °C | 1 min | |
| 55 °C | 1 min | |||
| 55 – 95 °C | 10 s | all points |
Table 3: Pan-lyssavirus real-time RT-PCR cycling conditions.
| Test Result | Internal ß-actin control | Overall result |
| Negative | Negative1 | Invalid. Repeat extraction and assay2 |
| Negative | Positive | Negative result reported |
| Positive | Positive | Positive result reported |
| Positive | Negative1 | Repeat extraction and assay3 |
| 1Use of heterologous external control would be beneficial during repeat extraction. | ||
| 2 If a second negative result is obtained for the internal control, the sample will be reported as untestable by this assay. | ||
| 3 If a second negative result is obtained for the internal control, alongside a positive test result a secondary rabies | ||
| diagnostic test should be undertaken to confirm this result. | ||
Table 4: Summary of outcomes and overall results for pan-lyssavirus real-time RT-PCR. Negative is designated to a sample with no Ct value (amplification) and no melt temperature (dissociation), or a melt temperature which is outside of the Tm range for positive lyssaviruses (76.8 °C – 80.2 °C). Positive is designated to a sample with a Ct value (amplification) and a melt temperature (dissociation) which is inside of the Tm range for positive lyssaviruses.
| Lyssavirus assay | ß-actin assay | |||
| Ct | Tm | Ct | Tm | |
| Mean | 20.66 | 78.92 | 27.5 | 85.26 |
| SD | 0.63 | 0.16 | 1.13 | 0.35 |
| LCL (95%) | 19.39 | 78.59 | 25.23 | 84.56 |
| UCL (95%) | 21.93 | 79.25 | 29.76 | 85.96 |
Table 5: Inter-run analysis of CVS positive control across 12 independent runs including multiple operators.
| Phylogroup | Species | Virus ID | Lineage | Limit of detection | Tm |
| I | RABV | RV50 | US bat | 10-5 | 79.5 |
| I | RABV | RV51 | US Fox | 10-7 | 77.6 |
| I | RABV | RV108 | Chile bat | 10-7 | 78.63 |
| I | RABV | RV313 | European Fox | 10-9 | 78.53 |
| I | RABV | RV437 | European RacDog | 10-7 | 78.09 |
| I | RABV | RV1237 | European Deer | 10-8 | 78.76 |
| I | RABV | RV334 | Chinese Vaccine | 10-8 | 79.03 |
| I | RABV | RV102 | Africa 2 | 10-7 | 78.58 |
| I | RABV | RV995 | Africa 3a | 10-8 | 79.66 |
| I | RABV | RV410 | Africa 3b | 10-7 | 79.03 |
| I | RABV | RV2324 | Africa 4 | 10-7 | 79.17 |
| I | RABV | RV2417 | Sri Lanka Dog | 10-9 | 78.71 |
| I | RABV | CVS-11 | 10-7 | 79.17 | |
| I | EBLV-1 | RV20 | Germany | 10-6 | 79.05 |
| I | EBLV-2 | RV1787 | UK | 10-7 | 78.76 |
| I | BBLV | RV2507 | Germany | 10-9 | 78.71 |
| I | ABLV | RV634 | 10-8 | 78.25 | |
| I | DUVV | RV131 | 10-5 | 79.03 | |
| I | GBLV | RV3269 | 10-7 | 79.15 | |
| I | ARAV | RV3379 | 10-7 | 79.46 | |
| I | KHUV | RV3380 | 10-7 | 78.97 | |
| I | SHIBV | RV3381 | 10-7 | 78.59 | |
| I | IRKV | RV3382 | 10-3 | 78.59 | |
| II | LBVa | RV767 | 10-5 | 77.34 | |
| II | LBVd | RV3383 | 10-7 | 78.59 | |
| II | MOKV | RV4 | 10-3 | 78.81 | |
| III | IKOV | RV2508 | 10-5 | 79.67 | |
| III | LLEBV | RV3208 | 10-4 | 79.15 | |
| III | WCBV | RV3384 | 10-3 | 79 |
Table 6: Summary of pan-lyssavirus real-time RT-PCR specificity, sensitivity and Tm for representative lyssaviruses across all three phylogroups. The mean Tm across lyssaviruses was 78.81 (SD 0.531).
Discussion
The pan-lyssavirus real-time RT-PCR assay described is a closed-tube, one-step assay which detects lyssaviruses across all three phylogroups. The assay has been validated for both animal and human rabies diagnosis, including post-mortem brain tissue (optimally brainstem), and ante-mortem samples such as skin biopsy, serially collected saliva, or cerebral spinal fluid (CSF). The primers utilized in this assay were first designed and utilized for a probe-based assay to differentiate between RABV, EBLV-1 and EBLV-23, which has been used in many OIE rabies laboratories and performs consistently in the EURL proficiency schemes. Subsequently the ‘pan-lyssavirus’ nature of these primers has been confirmed using a 2-step real-time assay15. The assay described here has utilized the primers to further optimize the RT-PCR in a one-step SYBR assay enabling a closed-tube, rapid system. Furthermore, training in rabies endemic countries using this assay has confirmed suitability to implement in any laboratory with basic PPE and quality systems to reduce cross contamination and trace samples, facilities to store the reagents and a real-time machine with SYBR detection. The assay is extremely robust and has 100% correlation with the FAT, with improved sensitivity for decomposed samples3. One of the main advantages for a DNA intercalating dye based assay in comparison to a probe based assay is the relative cost. An additional benefit is that the assay only utilizes two primers, therefore there is less risk of failed detection due to sequence divergence, which has been a weakness in previously published probe-based assays. Indeed, representatives of all lyssavirus species (apart from TWBLV and KBLV) are detected using this assay, and sequence analysis of TWBLV and KBLV across the primer sites reveals no significant divergence strongly suggesting that they will also be detected using this method. The range of limit of detection observed across the viruses analyzed, can be considered to be due to two main factors. The first is that the RNA was isolated from clinical brain material, therefore the amount of genome copies in each undiluted sample is not directly comparable. The RNA was ‘normalized’ by adjusting the total RNA to 1 µg/µL; however, the proportion of viral genome RNA within that sample will vary. The second is the diversity of lyssavirus sequences, despite the primer sites being located in conserved regions, there remain positions of variation. Therefore, it is not surprising that the majority of lyssaviruses with lower sensitivity for the assay are phylogroup II and III viruses. The dissociation curve analysis represents an essential parameter, minimizing a false positive result which could otherwise occur due to the formation of primer dimers, or amplification of a non-specific region in the host genome. In reality, this is a rare occurrence and the dissociation curve analysis is equivalent to running an agarose gel to visualize correctly sized conventional RT-PCR amplicons. The range of acceptable Tm values has been provided (77-80 °C), based on the data collected in our laboratory. It is strongly recommended that individual laboratories collate in-house data to ensure the range is transferrable and amend accordingly. Interpretation of the results from both the amplification and dissociation plots, alongside the positive and negative controls and the ß-actin results, enables robust and reproducible diagnostic outcomes.
Outside the scope of this protocol is the RNA extraction method used to obtain high quality RNA. All RNA analyzed in this protocol was prepared using TRIzol; however, there are many suitable guanidium-based extractions RNA extraction protocols available, including column and bead-based extraction kits. Handling of lyssavirus positive (or suspected positive) samples must be within licenced biocontainment facilities approved within country. However, the total RNA extracted is non-infectious, therefore handled within low containment laboratories. Depending on the extraction method used, the requirement to quantify and dilute the RNA would need to be assessed. For phenol-based extractions, including TRIzol, this step is required and prevents inhibition of the assay from contaminating gDNA; however, column and bead-based extractions (particularly those with a DNA depletion stage) do not require dilution prior to testing.
Throughout the protocol, it is essential that care and diligence are used to prevent cross-contamination and accurate addition of sample to the correct wells. A spreadsheet with the reagent calculations and plate layout is available for download as a supplemental file. Good laboratory practice, including clean work surfaces, regular changes of gloves, use of barrier tips and different rooms/UV cabinets to separate each stage will minimize the chance of contamination. To ensure the test is performing with expected parameters, positive and negative controls must be included and all test samples run in duplicate (or triplicate).
The inclusion of controls is an essential feature of any PCR, particularly for diagnostics. Positive control RNA was prepared from CVS (challenge virus standard) infected mouse brains in batches and validated and calibrated to ensure consistency between batches. The control RNA was quantified and diluted to 1 µg/µL. RNA for which a positive result was obtained in a serial dilution down to at least 10-4 (equal to 100 pg/µL) was considered fit for purpose. The positive control RNA was stored at -80 °C in 10-1 aliquots. When required, an aliquot was diluted 1:100 to provide a working stock at 1 ng/µL and stored at -80 °C in 5 µL single use aliquots. The diluted positive control RNA was used to represent low level positive samples, and to ensure that any reduction in sensitivity of the assay was detected. A ‘control card’ was kept for each control to monitor the Ct values and identify trends (Table 5). Table 5 demonstrated good inter-run comparability for the CVS positive control sample Ct and Tm values across multiple days and operators, providing reassurance that the assay is robust and reproducible. Results that deviate from these measurements should be investigated and test samples repeated if necessary. Molecular grade water was included in every run as an NTC to confirm the reagents were free from contamination with lyssavirus RNA and confirm a negative sample. Furthermore, to ensure RNA extraction efficacy, ß-actin was tested alongside the test samples in a separate tube. The lyssavirus positive control RNA was also used for the ß-actin positive control. Other endogenous genes or heterologous internal control systems can be utilized. Use of these controls ensured that all steps were analyzed under the same conditions as the test samples. Occasionally, if the sample was highly degraded or did not contain sufficient host RNA (such as saliva or CSF), the endogenous gene PCR can fail. In this instance, where the lyssavirus real-time RT-PCR result was positive, confirmation on an independent RNA extraction – to rule out contamination during RNA extraction on the original test or by a secondary test (either molecular, such as conventional RT-PCR), or FAT. Use of Table 4 during analysis of a diagnostic sample ensured the correct interpretation.
Regardless of the molecular assay used to confirm rabies infection, follow up investigations using Sanger sequencing to determine the lyssavirus species and classical techniques in virology such as FAT or virus isolation should also be undertaken to allow for further virus characterisation and support the notification of positive cases to OIE and WHO.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors wish to thank Miss Emma Wise and Miss Megan Golding for assistance in completing the experiments. The development of this protocol was financially supported by the UK Department for Environment, Food and Rural Affairs (Defra), Scottish Government and Welsh Government by grants [SV3500, and SE0431] and by European Virus Archive global (EVAg) project that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 653316.
Materials
| Art Barrier pipette tips (various sizes) | Thermofisher | various | |
| Centrifuge | Beckman | Allegra 21R | Rotor capable of holding 96 well plates required. Step 3.8. |
| Centrifuge (mico) | Sigma | ||
| Finnpipettes (to dispense 0.5-1000 µL) | Thermofisher | various | |
| iTaq Universal SYBR Green One-Step RT-PCR kit | Bio-Rad | 172-5150 | Equivalent kits can be used if validated |
| MX3000P or MX3005P real-tme PCR system | Stratagene | N/A | Eqivalent machines can be used if validated |
| MicroAmp reaction plate base | Any suitable | Used to hold tube strip and plates securely. | |
| Optically clear flat clear strips (8) | ABgene | AB-0866 | |
| Perfect fit frame (if using tube strips) | Stratagene | N/A | Specific to machine |
| Primers: for primer details see Table 2. | Ordered at 0.05 µmole scale HPLC purified. | ||
| Thermo-Fast 96 well plares, non skirted | ABgene | AB-600 | |
| Thermo-Fast strips (8) Thermo-tubes | ABgene | AB-0452 | |
| Vortex machine / Whirlimixer | Fisons Scientific equipment | SGP-202-010J | |
| Unless stated, alternative equipment can be used |
Riferimenti
- . OIE. OIE Terrestrial Manual 2018. , 8-14 (2018).
- Panning, M., et al. Comparative analysis of rabies virus reverse transcription-PCR and virus isolation using samples from a patient infected with rabies virus. Journal of Clinical Microbiology. 48 (8), 2960-2962 (2010).
- Wakeley, P. R., et al. Development of a real-time, TaqMan reverse transcription-PCR assay for detection and differentiation of lyssavirus genotypes 1, 5, and 6. Journal of Clinical Microbiology. 43 (6), 2786-2792 (2005).
- Wang, L., et al. A SYBR-green I quantitative real-time reverse transcription-PCR assay for rabies viruses with different virulence. Virologica Sinica. 29 (2), 131-132 (2014).
- Wadhwa, A., et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR Assay for the Detection of Highly Variable Rabies virus and Other Lyssaviruses. PLOS Neglected Tropical Diseases. 11 (1), e0005258 (2017).
- Nadin-Davis, S. A., Sheen, M., Wandeler, A. I. Development of real-time reverse transcriptase polymerase chain reaction methods for human rabies diagnosis. Journal of Medical Virology. 81 (8), 1484-1497 (2009).
- Hoffmann, B., et al. Improved safety for molecular diagnosis of classical rabies viruses by use of a TaqMan real-time reverse transcription-PCR "double check" strategy. Journal of Clinical Microbiology. 48 (11), 3970-3978 (2010).
- Faye, M., et al. Development and validation of sensitive real-time RT-PCR assay for broad detection of rabies virus. Journal of Virological Methods. 243, 120-130 (2017).
- Dacheux, L., et al. Dual Combined Real-Time Reverse Transcription Polymerase Chain Reaction Assay for the Diagnosis of Lyssavirus Infection. PLOS Neglected Tropical Diseases. 10 (7), e0004812 (2016).
- Fooks, A. R., et al. Rabies. Nature Reviews Disease Primers. 3, 17091 (2017).
- Marston, D. A., et al. Ikoma lyssavirus, highly divergent novel lyssavirus in an african civet. Emerging Infectious Diseases. 18 (4), 664-667 (2012).
- . Virus Taxonomy: 2018 Release: Email ratification October 2018 (MSL #33) Available from: https://talk.ictvonline.org/taxonomy/ (2018)
- Hu, S. C., et al. Lyssavirus in Japanese Pipistrelle, Taiwan. Emerging Infectious Disease. 24 (4), 782-785 (2018).
- Nokireki, T., Tammiranta, N., Kokkonen, U. M., Kantala, T., Gadd, T. Tentative novel lyssavirus in a bat in Finland. Transboundary and Emerging Diseases. 65 (3), 593-596 (2018).
- Hayman, D. T., et al. A universal real-time assay for the detection of Lyssaviruses. Journal of Virological Methods. 177 (1), 87-93 (2011).
- Calisher, C. H., et al. Antigenic relationships among rhabdoviruses from vertebrates and hematophagous arthropods. Intervirology. 30 (5), 241-257 (1989).
- Aznar-Lopez, C., et al. Detection of rhabdovirus viral RNA in oropharyngeal swabs and ectoparasites of Spanish bats. Journal of General Virology. 94 (1), 69-75 (2013).

