Use of Alu Element Containing Minigenes to Analyze Circular RNAs
Summary
We clone and analyze reporter genes generating circular RNAs. These reporter genes are larger than constructs to analyze linear splicing and contain Alu elements. To investigate the circular RNAs, the constructs are transfected into cells and resulting RNA is analyzed using RT-PCR after removal of linear RNA.
Abstract
In addition to linear mRNAs, many eukaryotic genes generate circular RNAs. Most circular RNAs are generated by joining a 5' splice site with an upstream 3' splice site within a pre-mRNA, a process called back-splicing. This circularization is likely aided by secondary structures in the pre-mRNA that bring the splice sites into close proximity. In human genes, Alu elements are thought to promote these secondary RNA structures, as Alu elements are abundant and exhibit base complementarities with each other when present in opposite directions in the pre-mRNA. Here, we describe the generation and analysis of large, Alu element containing reporter genes that form circular RNAs. Through optimization of cloning protocols, reporter genes with up to 20 kb insert length can be generated. Their analysis in co-transfection experiments allows the identification of regulatory factors. Thus, this method can identify RNA sequences and cellular components involved in circular RNA formation.
Introduction
Circular RNAs
Circular RNAs (circRNAs) are covalently closed single stranded RNAs that are expressed in most organisms. They are generated by joining a downstream 5' splice site to an upstream 3' splice site, a process called back-splicing (Figure 1A)1. Sequences in the pre-mRNA that exhibit base complementary as short as 30-40 nt bring back-splice sites into proper alignment for circRNA formation2. In humans, Alu elements1, representing about 11% of the genome3, form extensive double stranded RNA structures in pre-mRNA due to their self-complementarity4,5 and thus promote the formation of circRNAs1.
Currently, three major functions of circRNAs have been described. Some circRNAs bind microRNAs (miRNAs) and through sequestration act like miRNA sponges6. CircRNAs have been implicated in transcriptional and post transcriptional regulation, through competition with linear splicing7 or modulation of transcription factor activity8. Finally, circRNAs contain short open reading frames and proof of principle studies show that they can be translated9,10. However, the function of most circRNAs remains enigmatic. The majority of circular RNAs have been detected using next-generation sequencing methods11. Detailed analyses of individual genes using targeted RT-PCR approaches reveal that a large number of circular RNAs remains to be discovered12.
Use of reporter genes to analyze pre-mRNA processing
The analysis of mRNA derived from DNA reporter constructs transfected into cells is a well-established method to study alternative pre-mRNA splicing, which can be applied to circular RNAs. In general, the alternative exon, its surrounding introns, and constitutive exons are amplified and cloned into a eukaryotic expression vector. Frequently, the introns are shortened. The constructs are transfected into eukaryotic cells and usually analyzed by RT-PCR13,14. This approach has been extensively used to map regulatory splice sites and trans-acting factors in co-transfection experiments13,15,16,17,18. In addition, the generation of protein-expressing minigenes allowed for screening of substances that change alternative splicing19,20.
The method has been applied to circular RNAs. Currently, at least 12 minigene backbones have been described in the literature and are summarized in Table 1. With the exception of the tRNA based expression system21,22, they are all dependent on polymerase II promoters. Here, we describe a method to generate human reporter minigenes to determine cis and trans-acting factors involved in the generation of circular RNAs. An overview of the method using sequences of a published reporter gene23 is shown in Figure 1.
Protocol
1. Design of the constructs
- Use the UCSC genome browser24 to identify repetitive elements necessary for circular RNA formation and incorporate them in the constructs. Importantly, primers for amplification need to be outside the repetitive elements.
- Paste the circular RNA sequence (Supplemental Figure 1 is a test sequence) into https://genome.ucsc.edu/cgi-bin/hgBlat?command=start and select the right organism. Submit the sequence and go to browser view, zoom out 1.5x or as appropriate (Figure 2A).
NOTE: The search sequence appears in the top line (Figure 2A, 1). Depending on the order of the exons in the circular RNA, BLAT will not connect all the exons. In this example, exon 12 (Figure 2A, 4) is not connected to 11 (Figure 2A, 2, 3), because exon 12 is upstream of exon 11 in the circular RNA sequence (Supplemental Figure 1). The repetitive elements are in the 'repeat masker' track, indicated by boxes, where black to gray color indicates the evolutionary conservation (Figure 2A, 5). - Mouse over the repetitive elements to identify their subtype in a floating window. Alu elements are in the SINE (short interspersed nuclear element) line. Use the 'default tracks' button under the window to reset the browser if a different picture than Figure 2 is obtained.
NOTE: Mousing over the exons in the gene display generates a window with exon numbers that are computer generated. These numbers do not correspond to the exon numbering established in the literature and also change between isoforms.
2. Select the sequence to be cloned in an expression vector
- Download the DNA sequence shown in the window by going to View → DNA on the top line of the UCSC genome browser. In the Sequencing Formatting Option, select Extended Case/Color Options.
- Select the default case as Lower and select toggle case for NCBI refseq. Select underline, and bold, and italic for Repeat Masker. Click Invia. There will be exons as capital letters and introns as small letters. Check the exon/intron borders.
- In this example there is a 'ccctttacCTTTTT' sequence, indicating that the browser shows the reverse complement. If this is the case, go back and select the reverse complement box until seeing the correct exon intron borders (agEXONgt), in this example AAAAAGgtaaaggg.
- Copy the file with the correct orientation (internal exons are surrounded by intronic ag…gt) into a word processing document and highlight the exons (Supplemental Figure 2).
NOTE: In this example, the genomic fragment encompassing exons 9-12 is around 24 kb and thus too large to be amplified from genomic DNA. Therefore, each exon surrounded by about 1 kb intronic region is individually amplified and these four fragments are assembled in a cloning vector. - Select fragments to be amplified (exon ± 500 nt intron). Make sure that the intron does not begin or end in a repetitive region, as primers in these regions will not amplify specific sequences. The selected regions are shown in Figure 2B, and their sequences are shown in Supplemental Figure 3.
NOTE: In general, the larger the constructs, the more difficult the cloning will be. The fragments can be assembled either step wise (i.e., exon 9 is combined with the vector and in the next step exon 10 is introduced into this construct via cloning until all exons are in place), or alternatively, all fragments are assembled simultaneously. A stepwise approach always works but requires more time. A simultaneous assembly does not always work and depends on how well the individual fragments can be amplified from genomic DNA and on the overall size of the construct. We therefore usually start with both approaches simultaneously.
3. Design primers for cloning
- Use a web tool (https://nebuilder.neb.com/#!/) to design the primers for cloning. For example, enter fragments 9, 10, 11 and 12 and the vector sequence (Supplemental Figure 3) to this tool.
- For the vector sequence, add the insertion site as the last nucleotide and subsequently add the fragments. Since the vector numbering does not start with a given insertion site, the site of insertion in the vector is located and the downstream part is put in in front of the upstream sequence. In this example the inserts start directly after the HindIII [AAGCTT] site and ends directly after the PmeI [GTTTAAAC] site of pcDNA3.1. The sequence from cccgctgatcag … ccgtaaaaaggccgc is pasted in front of position 1 in the pcDNA3.1 sequence (gttgctggcgtttttcc …).
- Adjust primers if their melting points are more than 4 °C apart, as they will not work in amplification. The assembly of the fragments and primer sequences designed are shown in Supplemental Figure 4. Primers for cloning can also be designed manually25.
4. PCR and amplicon detection
- Standard PCR Reaction: Make a reaction mix for total volume of 50 µL per reaction. Below is the recipe for one reaction using polymerase 1:
10 µL of 5x Reaction Buffer
1 µL of 10 mM dNTPs
2.5 µL of 10 µM Forward Primer
2.5 µL of 10 µM Reverse Primer
0.5 µL of Polymerase 1
32.5 µL of Nuclease free H2O- Optionally, add 10 µL of 5x GC Enhancer if the product has high GC content; make a separate mix containing GC Enhancer to test PCR reaction.
- Aliquot 49 µL of the mix into PCR tubes per reaction sample.
- Add 1 µL of DNA to the PCR, the amount ranges from 10 pg to 1 ng.
- Spin down the samples to remove residue off the sides and place them in a PCR machine. Use the same machine as well as same spots in the machine when optimizing the PCR conditions.
5. Optimization for longer DNA fragments for use of different polymerases
- Temperature
- Optimize the long-range Polymerase 2.
- Use a lower denaturation temperature (Polymerase 1: 98 °C, Polymerase 2: 94 °C).
- Use a longer denaturation time (Polymerase 1: 10 s, Polymerase 2: 30 s).
- Use a longer annealing time (Polymerase 1: 30 s, Polymerase 2: 60 s).
- Use a longer extension time (Polymerase 1: 30 s/kb, Polymerase 2: 50 s/kb).
- Use a lower extension temperature (Polymerase 1: 72 °C, Polymerase 2: 65 °C).
- Use an annealing temperature 5 °C below the Tm of the primers.
- Optimize the primer concentrations (5x less to 5x more than the original primer concentration). Optimize DNA concentrations from 10 pg to 50 pg, 100 pg, 1 ng, 5 ng, 10 ng.
- As an example of a PCR program to amplify a 15 kb product size using Polymerase 2, perform an initial denaturation at 94 °C for 30 s, DNA denaturation at 94 °C for 30 s followed by annealing at 58 °C for 30 s, and DNA extension at 65 °C for 12 min 30 s. Perform a final extension at 65 °C for 10 min with a final 4 °C hold.
NOTE: The annealing temperature (Ta) specific for the primers determined by web-based temperature calculations that are specific for each polymerase. Extension times may vary and need to be optimized if fragments are larger than 10 kb.
- Optimize the long-range Polymerase 2.
- Extension times and DNA concentrations
- Use longer extension times for DNA fragments over 6 kb: 1 min per 1 kb. Longer fragments should use less DNA.
- Perform dilutions of DNA to find the optimum DNA concentration for amplification, usually 1 pg to 1 ng for plasmids or 1 ng to 1 µg for genomic DNA.
NOTE: For most cloning use polymerase 1, engineered by fusing the Sso7d DNA binding domain to a proprietary thermostable DNA polymerase26. The polymerase has a low error rate and due to the Sso7d domain a high processivity, needed to amplify large (10-15 kb) genomic fragments. Due to this large fragment size, enzymatic assembly of DNA molecules27 is used for insertion into vectors, as this method does not require restriction enzymes. The amplification of fragments longer than 15 kb gets increasingly difficult with polymerase 1. For large fragment amplification use polymerase 2 made from pyrococcus-like proofreading polymerase fused to Sso7d or the long range PCR kit that gives, however, higher error rates.
6. Purification of PCR products for cloning
- Run half of the PCR products on 1% agarose gels containing an intercalating nucleic acid stain (e.g., 1x GelGreen). Visualize the stained gels on a dark reader transilluminator. This stained DNA does not run true to size.
NOTE: GelGreen intercalates into DNA, similar to ethidium bromide, but it is excited by light around 500 nm (cyan), a wavelength that does not damage DNA, which highly improves cloning. - In parallel, run half of the PCR products on a 1% gel that is stained post-run with ethidium bromide (Figure 3A,B).
- Excise bands of the right size from the stained gel (step 6.1) and isolate DNA using a gel and PCR cleanup kit. In deviation from its standard protocol, elute the DNA in only 20 µL of double distilled water, as usually the DNA concentrations are low.
- Prior to cloning, check the isolated DNA on an agarose gel for concentration and integrity (Figure 3B). Aim for a 2:1 molar insert to vector ratio. When using several inserts, the ratio is 2:2:2:1.
7. DNA assembly and clone detection
NOTE: Cloning is done using an enzymatic DNA assembly kit, with minor modifications. The assembly is performed for 60 min at 50 °C and generally the lower range of DNA is used for assembly (20-100 fmol/20 µL reaction). The whole reaction mix is added to chemical competent cells and the whole cells plated out on a 6 cm agar plate.
- Combine the vector and insert in a 1:2 molar ratio (20-500 fmol each) in 10 µL of water.
- Add 10 µL of DNA Assembly Master Mix.
- Incubate samples for 60 minutes at 50 °C.
- Next, transform competent cells with the total assembly reaction. Here, use E. coli strain 1 cells for shorter constructs or E. coli strain 2 cells for longer or unstable constructs. Cells should be in a 50 µL volume.
- Thaw cells on ice and add 2 µL of the chilled assembled product to the competent cells. Mix by gently flicking the tube 4-5 times. Do not vortex.
- Let the mixture sit on ice for 30 min.
- Heat shock at 42 °C for 30 s. Do not mix.
- Transfer the tube back to ice for 2 min.
- Add 950 µL of room-temperature SOC Media to the tube.
- Incubate the reaction tube at 37 °C for 60 min. Shake vigorously (300 rpm).
- Warm selection plates with the appropriate antibiotic during incubation at 37 °C.
- Pellet the cells through centrifugation (10,000 x g, 30 s) and plate out ¼ and ¾ of cells on two selection plates and incubate at 37 °C overnight.
8. Validation of clones
- Use colony PCR, employing PCR primers spanning the assembly sites (Figure 1C) for detection.
- Take a sterile toothpick and touch a single bacterial colony.
- Touch the bottom of a PCR tube with the toothpick that has the colony and then streak the toothpick on an antibiotic agar plate, incubate the streaked plate at 37 °C or room temperature for sensitive constructs overnight.
- Overlay the PCR tube touched with the colony with a PCR mix containing the detection primers. These are designed using primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/). The program has the option to force primer design across a defined sequence using the "[ccc]" signs to select primers across the assembly junction. DNA from positive strains is isolated and verified by sequencing.
9. Analysis of circular RNA expressing reporter genes
- For analysis, transfect reporter genes into eukaryotic cells. Here, use HEK293 cells (ATTC #CRL-1573) as they give high transfection efficiency. To save costs, use PEI (polyethyleneimine) solutions.
- For the PEI solution, dissolve linear polyethyleneimine hydrochloride (PEI) at 1 mg/mL in water at low pH, pH 2. Bring up the pH to 7 with NaOH. Sterile filter with 0.22 µm filters and store at 4 °C.
- Split cells into six wells (approximately 150,000 cells per well) and let them grow overnight in 10% FBS in DMEM media.
- Aliquot 1 µg of the reporter gene in a sterile tube and add 200 µL of sterile filtered 150 mM NaCl. Mix with the DNA by vortexing.
- Add PEI solution to this mix and vortex, briefly centrifuge to collect samples at bottom of tube. Use a ratio of 1 µg of DNA per 3 µL of PEI.
- Incubate at room temperature for 10 min and then add directly to HEK 293 cells.
- Incubate HEK293 cells at 37 °C, 5% CO2, overnight.
- Isolate RNA for RT-PCR via an RNA isolation kit.
10. RNase R treatment to remove linear RNAs
- Use 10 µg of total RNA in an RNase-free tube.
- Add 10 µL of 10x RNase R buffer (0.2 M Tris-HCl (pH 8.0), 1 M KCl, 1 mM MgCl2) to RNA.
- Add RNase R to RNA (1 µl = 20U).
- Add 1 µL of glycol blue to the RNA and bring the volume up to 100 µL with sterile water.
- Incubate samples at 37 °C for 30 min.
- Add 100 µL of phenol/chloroform and vortex for 1 min.
- Centrifuge at 21,000 x g for 1 min to separate phases.
- Take the supernatant (aqueous phase) and add 1 volume (around 80 µL of chloroform).
- Vortex for 1 min, and then centrifuge for 1 min to separate phases.
- Take supernatant, add 1:10 vol KAc and 2.5 vol ethanol, and precipitate at -20 °C for 1-4 h. Centrifuge at 4 °C for 30 min at full speed (21,000 x g). There will be a small blue pellet at the bottom.
- Remove the supernatant, and wash with 80% ethanol. Let air dry for 5 min at room temperature, and dissolve in 10 µL water.
11. RT-PCR analysis
- Use 1 µg of RNA per RT reaction.
- Make reaction mix for a final total volume of 20 µL per reaction. For one reaction, use 1 µL of 10 mM dNTPs, 1 µL of 0.1 M Dithiothreitol (DTT), 4 µL of 5x First-Strand Buffer, and 0.5 µL of Reverse Transcriptase.
- Aliquot 6.5 µL of reaction mix into new PCR tubes.
- Mix up to 5 primers in one mix. However, some primers may mis-pair with other primers in the PCR (Figure 5). Primer sequences are in Table 2. We routinely use gene-specific exon-junction primers but priming with random hexamers is also possible.
- Add 1 µL of 10 µM Reverse primer to desired RT reaction tube.
- Add 1 µg of RNA to PCR reaction tube.
- Add RNase-free H2O up to a total volume of 20 µL.
- Spin tubes down to remove residue on side of tubes and place in thermocycler.
- Run the RT reaction in thermocycler at 50 °C for 50 minutes.
- Store RT cDNA at -20 °C or proceed to the PCR reaction.
Representative Results
Reporter genes allow determination of regulatory factors that influence circular RNA formation. However, these reporter genes are large and contain repetitive elements that often make DNA constructs unstable. Due to their large size, it is often necessary to delete parts of the introns, which is achieved by amplifying genomic pieces containing the exons and smaller flanking intronic parts. These DNA pieces are enzymatically assembled, allowing construction without restriction enzymes.
The example of a circular RNA generated from the microtubule associated protein tau (MAPT) shows an application of the minigene approach to analyze circular RNAs. The tau 9→12 minigene used in this example was co-transfected with different splicing factors and the effect of these splicing factors was detected by RT-PCR (Figure 6). Different trans-acting factors influence both circular RNA and linear pre-mRNA formation. The experiment also shows that all the sequence elements necessary for circular RNA formation are localized in the cloned fragment.
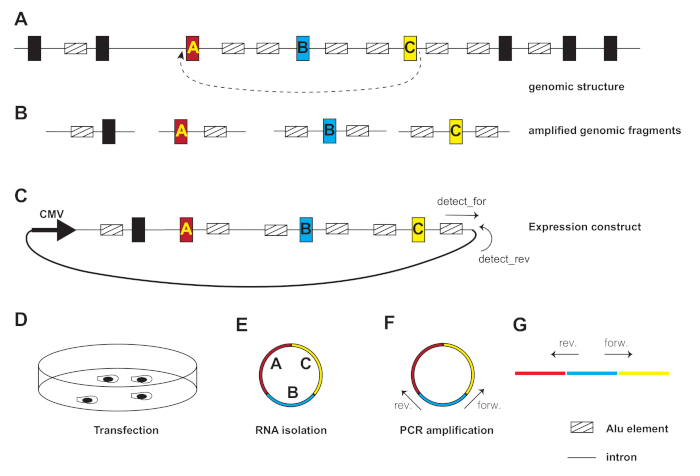
Figure 1: Overview of the technique. (A) A hypothetical gene is shown. Introns are lines, exons are boxes, Alu elements are smaller striped boxes. Backsplicing from exon C to A creates a circular RNA. The structure of this circular RNA is shown in panel (E). (B) To create a reporter gene, exons and surrounding introns (at least 500 nt on each side) are amplified. The constructs should contain repetitive elements, which are usually Alu elements in humans. An exon upstream of exon A was included to provide an additional Alu element. The genomic fragments will overlap with their flanking 25 nts. (C) Fragments are cloned into an expression vector, driven by a CMV promoter. The successful recombination is detected by detection primers and validated by sequencing. (D) Cells are transfected with this construct. (E) Circular RNA is isolated and (F) amplified using circular RNA specific primers, preferable exon junction primers. During PCR amplification, linear RNA can also be amplified (G). (G) Orientation of the primers used to detect circular RNAs. The forward primer is in sense orientation (i.e., has the same sequence as the RNA) and the reverse primer is in antisense orientation (i.e., is the reverse complement of the RNA). Note that different from RT-PCR for linear mRNAs, the reverse primer is upstream of the forward primer. Please click here to view a larger version of this figure.
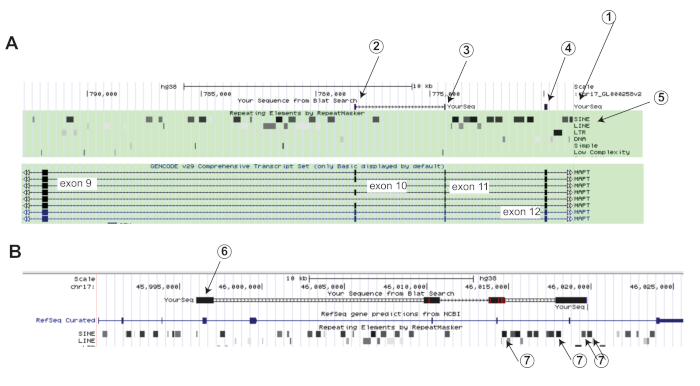
Figure 2: Selection of the sequence for minigene construction. (A) Browser display after the sequence shown in Supplemental Figure 1 is run against the human genomic database using BLAT. Literature exon numbers36 are indicated in the gene display, they are different from the numbers given by the browser.
1. The aligned sequences are shown under 'YourSeq'
2-4. Note that due to the circularity of the RNA, BLAT does not connect all exons with lines as it does in linear RNA. Exons 10 and 11 (corresponding to 2 and 3) are connected, but exon 12 (corresponding to 4) is not connected to exon 11.
5. Alu elements are shown in the repetitive element track.
(B) Sequence alignment between the planned construct and genomic DNA.
6. The planned construct was run against the database using BLAT.
7. Note the inclusion of several Alu elements in the construct. Please click here to view a larger version of this figure.
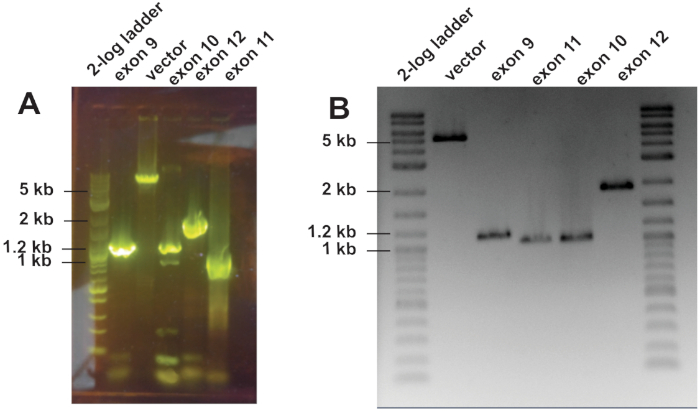
Figure 3: Example of the amplicons prior to cloning. (A) Optimized PCR products separated on a 1% agarose gel containing 1x GelGreen. The individual bands represent the PCR products that will be used in enzymatic DNA assembly. (B) The bands from (A) were cut out from the gel and purified. The purified PCR products were separated on a 1% agarose gel, which was subsequently stained with ethidium bromide. Please click here to view a larger version of this figure.
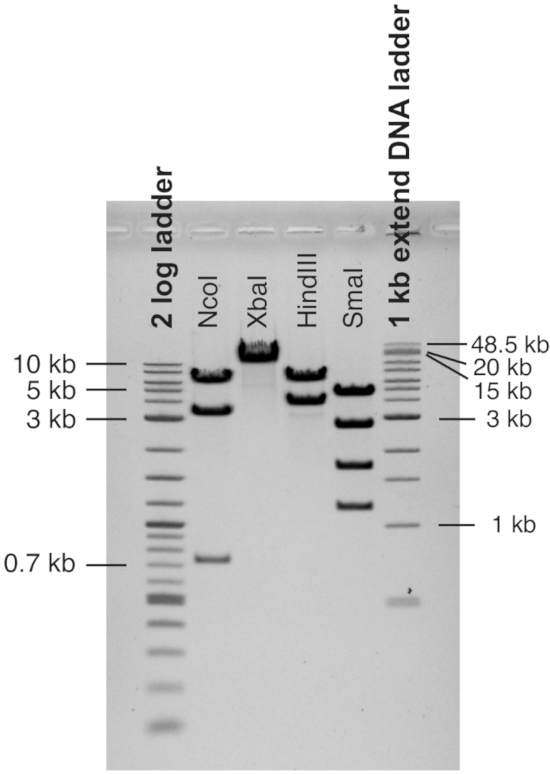
Figure 4: Restriction analysis of reporter genes. The tau 9-12 minigene used as an example was cut with restriction enzymes indicated to rule out major recombinations. Lane 1: cut with NcoI expected sizes 735 bp, 3345 bp, 6266 bp, lane 2 cut with XbaI expected size 10346 bp, lane 3 cut with HindIII expected sizes 3951 bp, 6395 bp, lane 4 cut with SmaI expected sizes 1168 bp, 1688 bp, 2708 bp, 4782 bp. Please click here to view a larger version of this figure.
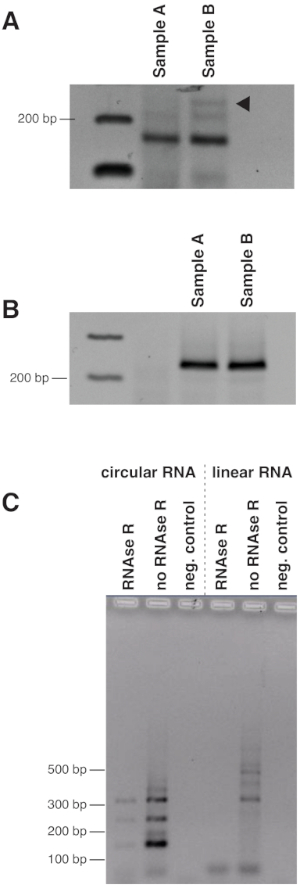
Figure 5: Effect of primer multiplexing and RNase R treatment on circular RNA detection. (A) cDNA from samples A and B derived from human brain tissues was amplified with circular RNA primers circTau exon12_10 Reverse and circTau exon10_11 Forward. The reverse transcription for the cDNA was performed with the primers for linear and circular tau RNA. The expected band corresponding to tau circular RNA is shown by a triangle. The other strong bands are artifacts that did not match the human genome. (B) The experiment was repeated with identical PCR conditions, but the reverse transcription was performed only with the circTau exon12_10 Reverse primer. Only the expected band was amplified and validated through sequencing. (C) The RNA was treated with RNase R that removes linear RNA. The circular RNA is detectable after the treatment (left), whereas linear RNA gives no longer a detectable signal (right) Please click here to view a larger version of this figure.
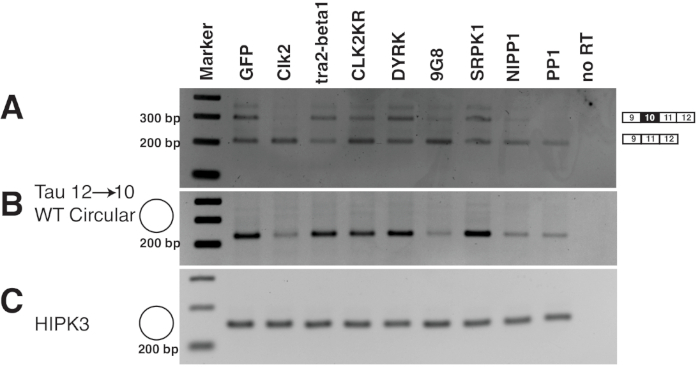
Figure 6: Example of an analysis of a circRNA reporter gene. 1 µg of the tau 9→1237 reporter gene was transfected with 1 µg of splicing factors indicated. RNA was isolated 24 h post transfection and analyzed by RT-PCR. (A) Amplification of the linear tau mRNA. Due to alternative splicing of exon 10 two bands are observed. Their ratio changes due to the overexpression of splicing factors38,39. (B) Amplification of the circular 12→10 tau RNA23. Note the dependency of tau circRNA expression on expression of some splicing factors, especially the cdc2 like kinase clk2 and the SR protein 9G8. (C) The circular RNA of HIPK3 was used as a positive control indicating equal loading. Please click here to view a larger version of this figure.
Table 1: List of current minigenes expressing circular RNAs. Please click here to download this file.
| Circular HIPK3 Control Primers | |
| HIPK3 Reverse HIPK3 Forward |
TGCTTGGCTCTACTTTGAGTTTC TCGGCCAGTCATGTATCAAA |
| Linear Primers | |
| Tau Exon 12 Reverse Tau Exon 9 Forward |
CCCAATCTTCGACTGGACTC TGTCAAGTCCAAGATCGGCT |
| Circular Primers | |
| circTau exon12_10 Reverse circTau exon10_11 Forward |
CAGCTTCTTATTAATTATCTGCACCTTTT GAGGCGGCAGTGTGCAA |
Table 2: List of Primers.

Supplemental Figure 1: Tau circular RNA test sequence. Test sequence corresponding to a circular RNA from the MAPT locus. Different exons are indicated by underline, small caps and large caps. Please click here to view a larger version of this figure.

Supplemental Figure 2: Genomic sequence containing the planned minigene. Exons are highlighted in color and repetitive elements are underlined, italic and bold. Gray shading indicates flanking regions of low complexity that can be used to generate primers. Please click here to download this file.

Supplemental Figure 3: Sequences of the planned reporter gene. The vector sequence and the planned genomic fragments are shown. Please click here to download this file.
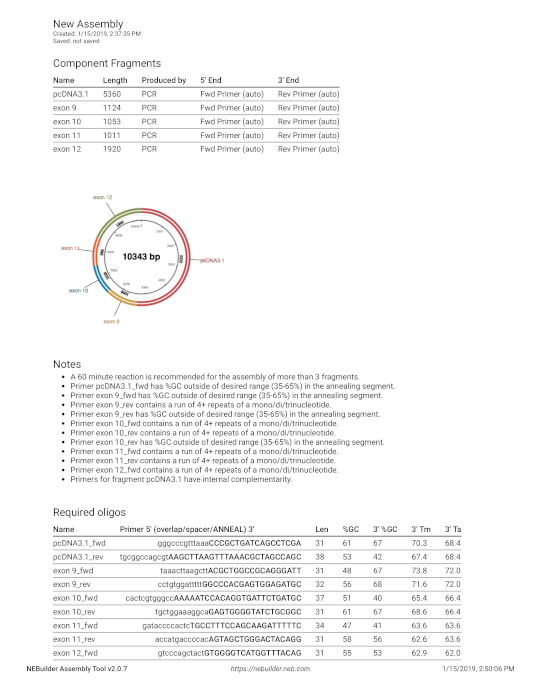
Supplemental Figure 4: Primers design for assembly. The sequence from Supplemental Figure 3 was entered into the builder tool. Please click here to view a larger version of this figure.

Supplemental Figure 5: Sequence of the tau 9->12 reporter gene used as an example. Please click here to download this file.
Discussion
In general, circular RNAs are low abundant1, which complicates the study of their function and formation. Similar to linear RNAs13, the use of reporter minigenes allows the identification of cis and trans-acting factors that regulate the formation of circular RNAs. Thus, this approach generates hypotheses that can be further tested using the endogenous genes.
The most critical step is the design of the reporter gene. The enzymatic assembly of DNA fragments ("Gibson cloning"27) facilitates this design, as it allows construction of large reporter genes independent of restriction sites.
The back-splicing sites are brought together through flanking inverted repeats, which should be taken into account in reporter gene construction. The repeats are annotated in the genome browser 'repeat track' and selecting them shows their orientation. Keep in mind that proteins can also force the back-splicing sites into a secondary structure needed for circular RNA expression28 and for an unbiased analysis 1-2 kb of flanking intronic regions should be investigated.
To ensure stability of the constructs, an important consideration is the type of bacterial strains and their growth conditions. For shorter, simple constructs standard cloning bacteria are used, which are almost identical to DH5-alpha (huA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17). For longer fragments, containing more than 6 Alu elements, "stable" competent cells are used that lack a recombinase (recA) and endonuclease (endA1) (F' proA+B+ lacIq ∆(lacZ)M15 zzf::Tn10 (TetR) ∆(ara-leu) 7697 araD139 fhuA ∆lacX74 galK16 galE15 e14- Φ80dlacZ∆M15 recA1 relA1 endA1 nupG rpsL (StrR) rph spoT1 ∆(mrr-hsdRMS-mcrBC). If problems appear with recombination, indicated by low transformation counts, plate the transformed bacteria on two plates and let them grow at 30 ºC and 37 ºC, respectively. Due to the presence of numerous repetitive elements in the minigenes, they need to be fully sequenced using next generation sequencing, which is commercially available for around $150 per plasmid at 2019 rates. The sequence of the example is shown in Supplemental Figure 5. In addition, restriction fragment length polymorphism analysis for new larger preparation of the constructs is routinely performed. For example, using sites that cut 1-4 times results in a characteristic band pattern that rules out recombinations (Figure 4). Enzymes should be selected that give a characteristic band pattern of fragments that can be separated on an agarose gel.
Circular RNAs are analyzed with RT-PCR using exon junction primers that overlap with the backsplicing event (Figure 1F). Due to the circular nature of the RNA, the reverse (i.e. antisense) primer is upstream of the forward (i.e. sense) primer (Figure 1G). Primers detecting the abundantly expressed homeodomain-interacting protein kinase 3 (HIPK3) circular RNA1 are used as a positive control. HIPK3 and minigene specific reverse primers are reverse transcribed in the same tube, which allows their comparison. PCR reactions are performed with primers amplifying the linear mRNAs to compare processing patterns of circular and linear pre-mRNAs. We frequently observed aberrant bands when primers for linear and circular RNAs were mixed (Figure 5), and thus keep the reverse transcription of these samples separate.
RT-PCR analysis of circular RNAs is challenging and needs to be carefully controlled. While sensitive and convenient, it can produce artifacts unique for circular RNAs29. The reverse transcriptase can move several times around the RNA circle, which generates concatemers. Most circular RNA reporter genes generate both circular and linear RNA, which can cross-hybridize, leading to more PCR artifacts21,30,31. It is thus imperative to sequence the PCR products and validate findings using different techniques using Northern blots32 or RNase protections23.
Unexplained bands can also originate from aberrant amplification of linear RNA. Linear RNA can be removed using the exonuclease RNase R, which enriches circular RNAs33 (Figure 5C). RNase R treatment helps in the initial optimization of detection primers and can often be omitted once primers are optimized.
Alternative back-splicing can also contribute to unexplained bands as multiple circular RNAs can be formed from a genomic locus34. This alternative back-splicing is often the result of competing pre-mRNA structures formed by more than two inverted repeat elements. In addition, cryptic back-splice sites can occur32,35. Depending on the experimental goal, Alu elements can be repeated or added to the constructs. The complementary regions flanking back-splicing sites can be as short as 30-40 nt35 and replacement of Alu elements with shorter complementary regions can increase circular RNA formation2, which can be tested to improve circular RNA formation. Once the pre-mRNA sequences that cause back-splicing have been identified, it is thus possible to shorten circular RNA expressing constructs, which can improve transfection efficiency in some cases.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Department of Defense DoD grant AZ180075. Stefan Stamm thanks Jacqueline Noonan Endowment. Anna Pawluchin was supported by the DAAD, German academic exchange program, Justin R. Welden was a recipient of the University of Kentucky Max Steckler Award.
Materials
| (PEI) Hydrochloride | Polysciences | 24765-1 | |
| Builder tool | NEB | https://nebuilder.neb.com/#!/ | |
| Dark Reader Transilluminator. | Clare Chemical Research | ||
| Enzymatic DNA assembly kit | NEB | E2621S | |
| Gel and PCR cleanup kit | Promega | A9282 | |
| Glyco Blue | Thermo Fisher | AM9516 | |
| pcDNA3.1 cloning site | Polycloning site | https://assets.thermofisher.com/TFS-Assets/LSG/manuals/pcdna3_1_man.pdf | |
| Polymerase 1 | NEB | M0491L | Q5 DNA polymerase |
| Polymerase 2 | Biorad | 1725310 | Long range polymerase (NEB), iproof (BioRad) |
| Polymerase 2 | Qiagen | 206402 | Qiagen long range polymerase kit |
| Reverse Transcriptase | Thermo Fisher | 18080044 | |
| RNA isolation kit | Life Technologies | 12183025 | Ambion by Life Technologies |
| RNAse R | Lucigen | RNR07250 | Epicenter/Lucigen |
| Stable competent cells | NEB | C3040H | NEB stable cells |
| Standard cloning bacteria | NEB | C2988J | NEB5-alpha competent |
| Web tool to design primers | NEB | https://nebuilder.neb.com/#!/ | |
| Web-based temperature calculations | NEB | https://tmcalculator.neb.com/#!/main |
Riferimenti
- Jeck, W. R., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19 (2), 141-157 (2013).
- Zhang, X. O., et al. Complementary sequence-mediated exon circularization. Cell. 159 (1), 134-147 (2014).
- Deininger, P. Alu elements: know the SINEs. Genome Biology. 12 (12), 236 (2011).
- Bazak, L., Levanon, E. Y., Eisenberg, E. Genome-wide analysis of Alu editability. Nucleic Acids Research. 42 (11), 6876-6884 (2014).
- Levanon, E. Y., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature Biotechnology. 22 (8), 1001-1005 (2004).
- Hansen, T. B., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 495 (7441), 384-388 (2013).
- Kelly, S., Greenman, C., Cook, P. R., Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. Journal of Molecular Biology. 427 (15), 2414-2417 (2015).
- Li, Z., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology. 22 (3), 256-264 (2015).
- Yang, Y., et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Research. 27 (6), 626-641 (2017).
- Abe, N., et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Scientific Reports. 5, 16435 (2015).
- Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L., Brown, P. O. Cell-type specific features of circular RNA expression. PLOS Genetics. 9 (9), 1003777 (2013).
- Ottesen, E. W., Luo, D., Seo, J., Singh, N. N., Singh, R. N. Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs. Nucleic Acids Research. 47 (6), 2884-2905 (2019).
- Stoss, O., Stoilov, P., Hartmann, A. M., Nayler, O., Stamm, S. The in vivo minigene approach to analyze tissue-specific splicing. Brain Research Protocols. 4, 383-394 (1999).
- Mardon, H. J., Sebastio, G., Baralle, F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Research. 15, 7725-7733 (1987).
- Gaildrat, P., et al. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods in Molecular Biology. 653, 249-257 (2010).
- Cooper, T. A. Use of minigene systems to dissect alternative splicing elements. Methods. 37 (4), 331-340 (2005).
- Baralle, D., Baralle, M. Splicing in action: assessing disease causing sequence changes. Journal of Medical Genetics. 42 (10), 737-748 (2005).
- Percifield, R., Murphy, D., Stoilov, P. Medium throughput analysis of alternative splicing by fluorescently labeled RT-PCR. Methods in Molecular Biology. 1126, 299-313 (2014).
- Stoilov, P., Lin, C. H., Damoiseaux, R., Nikolic, J., Black, D. L. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proceedings of the National Academy of Sciences of the United States of America. 105 (32), 11218-11223 (2008).
- Shen, M., et al. Pyrvinium pamoate changes alternative splicing of the serotonin receptor 2C by influencing its RNA structure. Nucleic Acids Research. 41 (6), 3819-3832 (2013).
- Noto, J. J., Schmidt, C. A., Matera, A. G. Engineering and expressing circular RNAs via tRNA splicing. RNA Biology. , 1-7 (2017).
- Schmidt, C. A., Noto, J. J., Filonov, G. S., Matera, A. G. A Method for Expressing and Imaging Abundant, Stable, Circular RNAs In Vivo Using tRNA Splicing. Methods in Enzymology. 572, 215-236 (2016).
- Welden, J. R., van Doorn, J., Nelson, P. T., Stamm, S. The human MAPT locus generates circular RNAs. Biochimica et Biophysica Acta – Molecular Basis of Disease. , 2753-2760 (2018).
- Casper, J., et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Research. 46, 762-769 (2018).
- Grozdanov, P. N., MacDonald, C. C. Generation of plasmid vectors expressing FLAG-tagged proteins under the regulation of human elongation factor-1alpha promoter using Gibson assembly. Journal of Visualized Experiments. (96), (2015).
- Wang, Y., et al. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Research. 32 (3), 1197-1207 (2004).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 343-345 (2009).
- Conn, S. J., et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 160 (6), 1125-1134 (2015).
- Jeck, W. R., Sharpless, N. E. Detecting and characterizing circular RNAs. Nature Biotechnology. 32 (5), 453-461 (2014).
- Pamudurti, N. R., et al. Translation of CircRNAs. Molecular Cell. 66 (1), 9-21 (2017).
- Kramer, M. C., et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes & Development. 29 (20), 2168-2182 (2015).
- Starke, S., et al. Exon circularization requires canonical splice signals. Cell Reports. 10 (1), 103-111 (2015).
- Suzuki, H., Tsukahara, T. A view of pre-mRNA splicing from RNase R resistant RNAs. International Journal of Molecular Sciences. 15 (6), 9331-9342 (2014).
- Zhang, X. O., et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Research. 26 (9), 1277-1287 (2016).
- Liang, D., Wilusz, J. E. Short intronic repeat sequences facilitate circular RNA production. Genes & Development. 28 (20), 2233-2247 (2014).
- Wang, Y., Mandelkow, E. Tau in physiology and pathology. Nature Reviews Neuroscience. 17 (1), 5-21 (2016).
- Fejes-Toth, K., et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 457 (7232), 1028-1032 (2009).
- Gao, Q. S., et al. Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. Journal of Neurochemistry. 74 (2), 490-500 (2000).
- Hartmann, A. M., et al. Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Molecular and Cellular Neuroscience. 18 (1), 80-90 (2001).
- Wang, Y., Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 21 (2), 172-179 (2015).
- Yang, Y., Wang, Z. Constructing GFP-Based Reporter to Study Back Splicing and Translation of Circular RNA. Methods in Molecular Biology. 1724, 107-118 (2018).
- Zheng, Q., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications. 7, 11215 (2016).
- Jia, W., Xu, B., Wu, J. Circular RNA expression profiles of mouse ovaries during postnatal development and the function of circular RNA epidermal growth factor receptor in granulosa cells. Metabolism. 85, 192-204 (2018).
- Liang, D., et al. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Molecular Cell. 68 (5), 940-954 (2017).
- Legnini, I., et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Molecular Cell. 66 (1), 22-37 (2017).
- Li, X., et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Molecular Cell. 67 (2), 214-227 (2017).
- Zhang, Y., et al. The Biogenesis of Nascent Circular RNAs. Cell Reports. 15 (3), 611-624 (2016).

