Multi-Fiber Photometry to Record Neural Activity in Freely-Moving Animals
Summary
This protocol details how to implement and perform multi-fiber photometry recordings, how to correct for calcium-independent artifacts, and important considerations for dual-color photometry imaging.
Abstract
Recording the activity of a group of neurons in a freely-moving animal is a challenging undertaking. Moreover, as the brain is dissected into smaller and smaller functional subgroups, it becomes paramount to record from projections and/or genetically-defined subpopulations of neurons. Fiber photometry is an accessible and powerful approach that can overcome these challenges. By combining optical and genetic methodologies, neural activity can be measured in deep brain structures by expressing genetically-encoded calcium indicators, which translate neural activity into an optical signal that can be easily measured. The current protocol details the components of a multi-fiber photometry system, how to access deep brain structures to deliver and collect light, a method to account for motion artifacts, and how to process and analyze fluorescent signals. The protocol details experimental considerations when performing single and dual color imaging, from either single or multiple implanted optic fibers.
Introduction
The ability to correlate neural responses with specific aspects of an animal’s behavior is critical to understand the role a particular group of neurons plays in directing or responding to an action or stimulus. Given the complexity of animal behavior, with the myriad of internal states and external stimuli that can affect even the simplest of actions, recording a signal with single-trial resolution equips researchers with the necessary tools to overcome these limitations.
Fiber photometry has become the technique of choice for many researchers in the field of systems neuroscience because of its relative simplicity compared to other in vivo recording techniques, its high signal-to-noise ratio, and the ability to record in a variety of behavioral paradigms1,2,3,4,5,6,7,8. Unlike traditional electrophysiological methods, photometry is the optical approach most commonly used in conjunction with genetically-encoded calcium indicators (GECIs, the GCaMP series)9. GECIs change their ability to fluoresce based on whether or not they are bound to calcium. Because the internal concentration of calcium in neurons is very tightly regulated and voltage-gated calcium channels open when a neuron fires an action potential, transient increases in internal calcium concentration, which result in transient increases in the ability of a GECI to fluorescence, can be a good proxy for neuronal firing9.
With fiber photometry, excitation light is directed down a thin, multimode optic fiber into the brain, and an emission signal is collected back up through the same fiber. Because these optic fibers are lightweight and bendable, an animal can move largely unhindered, making this technique compatible with a wide array of behavioral tests and conditions. Some conditions, such as rapid movements or bending of the fiber-optic patch cord beyond the radius at which it can maintain total internal reflection, can introduce signal artifacts. To disambiguate signal from noise, we can exploit a property of GCaMP known as the “isosbestic point.” Briefly, with GCaMP, as the wavelength of the excitation light is shifted to the left, its emission in the calcium-bound state decreases and the emission in the calcium-unbound state marginally increases. The point at which the relative intensity of these two emissions are equal is termed the isosbestic point. When GCaMP is excited at this point, its emission is unaffected by changes in internal calcium concentrations, and variance in the signal is most often due to attenuation of the signal from overbending of the fiber-optic patch cord or movement of the neural tissue relative to the implanted fiber.
Single unit electrophysiology is still the gold standard for freely-moving in vivo recordings due to its single-cell and single-spike level resolution. However, it can be difficult to pinpoint the molecular identity of the cells being recorded, and the post-hoc analysis can be quite laborious. While fiber photometry does not have single-cell resolution, it does allow researchers to ask questions impossible to address with traditional techniques. Combining viral strategies with transgenic animals, the expression of GECIs can be directed to genetically-defined neuronal types to record population- or projection-defined neural activity, which can be performed by monitoring calcium signal directly at axon terminals10,11. Moreover, by implanting multiple fiber-optic cannulas, it is possible to simultaneously monitor neural activity from several brain regions and pathways in the same animal12,13.
In this manuscript, we describe a technique for single and multi-fiber photometry, how to correct for calcium-independent artifacts, and detail how to perform mono- and dual-color recordings. We also provide examples of the types of questions it enables one to ask and their increasing levels of complexity (see Figure 1). The fiber photometry setup for multi-fiber recordings detailed in this protocol can be built using a list of materials found at https://sites.google.com/view/multifp/hardware (Figure 2).
It is essential that the system be equipped for both 410 nm and 470 nm excitation wavelengths for calcium-independent and calcium-dependent fluorescence emission from GCaMP6 or its variants. For custom-built setups or if there is no available software to run the system, the free, open source program Bonsai (http://www.open-ephys.org/bonsai/) can be used. Alternatively, fiber photometry can be run through MATLAB (e.g., https://github.com/deisseroth-lab/multifiber)12 or other programming language14. The software and hardware of the system should allow manipulation of both the 410 nm and 470 nm LEDs and the camera, extraction of images (Figure 2), and calculation of the mean fluorescent intensity in the regions of interest (ROIs) drawn around the fibers on the images. The output should be a table of mean intensity values recorded with the 470 nm and 410 nm LEDs from each fiber in the patch cord. When performing multi-fiber experiments, 400 µm bundled fibers may limit the movement of mice. In such cases, we recommend using 200 µm patch cords, which provide more flexibility. It may also be possible to use smaller dummy cables during training of mice.
It is crucial to be able to extract time points for events of interest during fiber photometry acquisition. If the system does not readily provide a built-in system to integrate TTLs for specific events, an alternative strategy is to assign a time stamp to individual time points recorded to align with specific times and events during the experiment. Time stamping can be done using the computer clock.
Protocol
All experiments were done in accordance with the Institutional Animal Care and Use Committees of the University of California, San Diego, and the Canadian Guide for the Care and Use of Laboratory Animals and were approved by the Université Laval Animal Protection Committee.
1. Alignment of the optical path between the CMOS (complementary metal oxide semiconductor) camera and the individual or branching patch cord
- Loosen all screws on the 5-axis translator (11, Figure 2B).
- Screw in the patch cord (12, Figure 2B) to the adaptor [SMA (sub-miniature A) or FC (fiber optic connector)] that is affixed to the 5-axis translator.
- Turn on the 470 nm excitation light (1, Figure 2B) at low power (100 µW), and place the tip of the patchcord pointing to an autofluorescent plastic slide. This does not have any bearing on future recordings but is solely for visualizing the alignment process.
- Record from the CMOS camera (13, Figure 2B) in live mode. Increase the gain or adjust the lookup table (LUT) until the image is not entirely black. The point is to be able to see an image at the focal point of the objective (10, Figure 2B).
- Advance the 5-axis translator towards the objective, ensuring that the 470 nm light is centered on the fiber at the SMA or FC end of the patch cord, until an image can be resolved on the camera.
- Adjust the X and Y axes until the image is centered and well-resolved.
- Visualize the light emitted from the ferrule-end of the patch cord. It should appear as an isotropic circle. If a branching patch cord is used, the amount of light emitted at the ferrule-ends of each patch cord should be similar. If the circle is not isotropic or the emitted light is unequal, adjust the 5-axis translator in the X-Y axis.
2. Setup of ROIs around fibers for measurement of mean fluorescent intensity
- Turn on all the excitation lights to better visualize the fibers. Adjust the camera gain such that no pixels are saturated and a clear image of the fibers are present.
- Live record or take a preliminary image.
- Draw ROIs around the fibers and keep them for the measurement of the mean intensity values during recordings (Figure 2A).
- For multiple fiber recordings, test for independence in signals.
- Live record from all fibers.
- Point one fiber towards a light source and tap with a finger. Very large fluctuations should occur solely in that channel (acceptable leakage 1:1000).
- If the signals are not independent, redraw more conservative ROIs and repeat the independence test.
- To label and keep track of which ROI corresponds to which fiber, colored tape or nail polish can be applied to the end of the fibers. Take a picture prior to the start of any experiment as a secondary reminder.
3. Setup of recording arena
- Hang the patch cord above the arena using stands, clamps, or holders.
- Make sure that the animal can freely move throughout the entire arena, uninhibited by the length of the fiber.
- Whether an operant box or open field is used, ensure that the patch cord will be able to reach the animal with minimal bending. If this requires a nose poke, ensure that there is enough room overhead to prevent bending of the fiber. Avoid any excessive bending or twisting of the patch cord.
4. In vivo recordings
NOTE: The procedure of optic fiber cannula implantation for fiber photometry experiments is identical to the procedure for optogenetics as described in Sparta et al15. We recommend using dental cement (see Table of Materials), which provides robust anchoring of the headcap to the skull bone. Dental cement will be particularly useful in cases where anchoring screws cannot be used.
- Visually inspect the distal end of the fibers of the patch cord by eye and with a minifiber microscope. If the surface of the fibers is scratched, repolish the fibers using fiber polishing/lapping film with fine grit (1 µm and 0.3 µm).
- Clean the distal ends of the patch cord with 70% ethanol and a cotton tip applicator.
- Clean the fiber-optic cannulas using 70% ethanol and a cotton tip applicator.
- Connect the ferrule end of the patch cord to the implanted fiber using a ceramic split-sleeve covered with a black shrink tube. During the connection, make sure that the sleeve is tight, otherwise use a new sleeve.
NOTE: There will be a large amount of signal loss if there is any space between the patch cord ferrule and the implant, and the recordings will not work. - Allow the animal to recover for a few minutes prior to the start of behavioral testing.
- Start recording the optical signal and run the experiment.
- While recording, keep a careful eye on the live-trace to ensure quality recordings. The signal is expected to rapidly decrease as a function of time in the first 2 min of recording. This effect is caused by heat-mediated LED decay, whereby the increase in heat increases the resistance of the optical element.
- If a jump in the signal that exceeds the on/off kinetics of GCaMP occurs, this is often an indication that the sleeve is not tight enough and the space between the patch cord and the implant is changing. In this case, stop the experiment and reconnect the animal using a new sleeve.
5. Fiber photometry data analysis
NOTE: This is a method for data analysis that works well for most recordings. However, alternative approaches can be implemented. Example code for data analysis can be found here: https://github.com/katemartian/Photometry_data_processing.
- Extract mean fluorescence intensity values recorded from 470 nm (Int470) and 410 nm (Int410) LEDs, corresponding to each individual fiber.
- Smooth each signal using a moving mean algorithm (Figure 3A).
- Perform baseline correction of each signal (Figure 3A and 3B) using the adaptive iteratively reweighted Penalized Least Squares (airPLS) algorithm (https://github.com/zmzhang/airPLS) to remove the slope and low frequency fluctuations in signals.
- Standardize each signal using the mean value and standard deviation (Figure 3C):
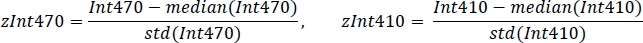
- Using non-negative robust linear regression, fit standardized zInt410 to zInt470 signals (Figure 3D) to the regression function:
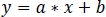
- Use the parameters of the linear regression (a, b) to find new values of zInt410 fitted to zInt470 (fitInt410, Figure 3D,E):
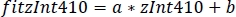
- Use the parameters of the linear regression (a, b) to find new values of zInt410 fitted to zInt470 (fitInt410, Figure 3D,E):
- Calculate the normalized dF/F (z dF/F) (Figure 3F):
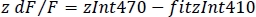
6. Simultaneous dual-color recordings
- Add to the photometry system a 560 nm LED to excite the red fluorescent calcium sensor and appropriate dichroic mirrors and filters (see Kim et al., 2016 for detailed description)12.
- Add an image splitter between the objective and the CMOS camera to separate the green and red emission wavelengths (see Figure 5). The image splitter will form two mirrored images on the camera sensor, corresponding to the red and green signals (e.g., a patch cord with 3 branches will create an image with 6 fibers).
- Draw ROIs around all fibers in both colors as detailed above. Make sure to clearly identify each ROI with the corresponding fiber and channel (green and red) (Figure 4A).
- Trigger simultaneous excitation with 470 nm and 560 nm LEDs and alternate them with 410 nm LED (Figure 5A).
7. Dual color data analysis
- Follow the steps in Section 5 to find fitInt410 for the Int470 signal and calculate z dF/F.
- Because the isosbestic point for red-shifted GECIs is generally unknown, the signal recorded with 410 nm LED in the green channel can be used for movement correction across both channels. Follow the steps in Section 5 to find fitInt410 for the Int560 signal and calculate z dF/F.
Representative Results
Neural correlates of behavioral responses can vary depending on a variety of factors. In this example, we used in vivo fiber photometry to measure the activity of axon terminals from the lateral hypothalamic area (LHA) that terminate in the lateral habenula (LHb). Wild type mice were injected with an adeno-associated virus (AAV) encoding GCaMP6s (AAV-hSyn-GCaMP6s) in the LHA and an optic fiber was implanted with the tip immediately above the LHb (Figure 4A). GCaMP6s expression is found in the cell bodies of the LHA and their axon terminals projecting to the LHb, where calcium signal can be recorded. Activation of the LHA-LHb pathway promotes passive avoidance in the real-time preference test suggesting that this pathway transmits aversive signals16. Mice were then connected to the fiber photometry system, placed in an open arena for 6 min, and exposed to 1 sec aversive airpuffs every 60 sec. The measured fluorescence significantly increased concurrently with the administration of airpuffs (Figure 4B-C). In mice expressing green fluorescent protein (GFP), no change in the signal was detected during the administration of airpuffs (Figure 4C). Following behavioral testing, the site of injection in the LHA and fiber placement above the LHb were confirmed histologically (Figure 4A).
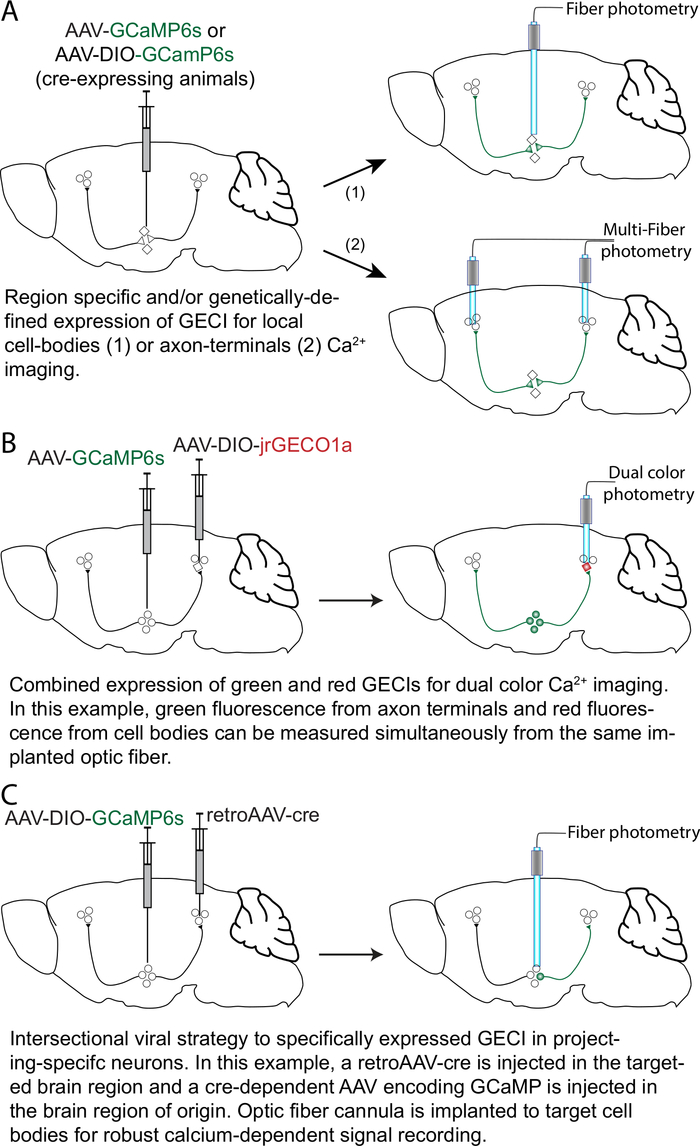
Figure 1: Strategies and approaches for GECI expression with anatomical and cell-type specificity. (A) A viral vector adeno-associated virus (AAV) encoding GCaMP6 (AAV-GCaMP6) is injected in a brain region of interest. The optic fiber can be chronically implanted with the tip placed over the cell bodies (1) or over the axon terminals (2). For selective expression in a genetically-defined neuronal population, a cre-dependent AAV (e.g., AAV-DIO-GCaMP6) can be injected in transgenic mice expressing the cre recombinase in a specific neuronal population. (B) For dual-color calcium imaging, in this example an AAV-GCaMP6s is injected in a brain region of interest, and a cre-dependent AAV encoding a red-shifted GECI (e.g., jrGECO1a; AAV-DIO-jrGECO1a) is injected in a genetically-defined neuronal population in a target brain region. The optic fiber is implanted for simultaneous calcium signal imaging of the axon terminals (green fluorescence) and cell bodies (red fluorescence). (C) For an intersectional viral strategy, a viral vector with retrograde transport properties (like retroAAV17) encoding the cre recombinase (retroAAV-cre) is injected in the target brain region together with an AAV-DIO-GCaMP6s injected in the projecting brain region in the same mouse. Optic fiber cannulas are implanted over the cell bodies for robust calcium-dependent signal recording. Please click here to view a larger version of this figure.
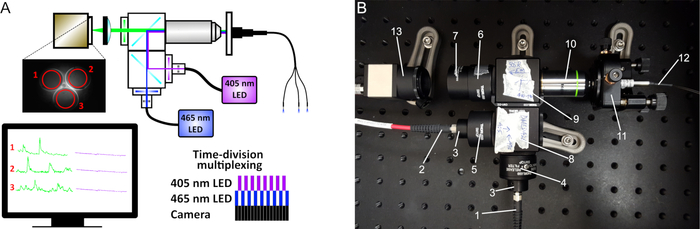
Figure 2: Fiber photometry schematic. (A) The excitation light from two LEDs (410 nm and 470 nm) passes through a series of filters and dichroic mirrors and produces an excitation spot at the working distance of the 20x objective. The light passes through either a single patch cord or bundled fibers (for multiple site recordings) that are connected to the implanted cannulas. The emitted fluorescence is collected by the same fibers, filtered, and projected on a CMOS camera sensor. On the captured images, the mean fluorescence intensity is recorded at the ROIs of each fiber. To simultaneously acquire signals from both 410 nm and 470 nm LEDs, a time-division multiplexing is implemented (lower right diagram). (B) Image of our custom-made photometry system and its components: (1) Fiber to the 465 nm LED, (2) Fiber to the 405 nm LED, (3) Collimators, (4) 470 nm bandpass filter, (5) 410 nm bandpass filter, (6) 535 nm bandpass filter, (7) Tube lens, (8) Cube with longpass 425 dichroic mirror, (9) Cube with longpass 495 dichroic mirror, (10) 20x objective, (11) 5-axis translator, (12) Mono- or bundled-fiber patch cord, (13) CMOS camera. Please click here to view a larger version of this figure.
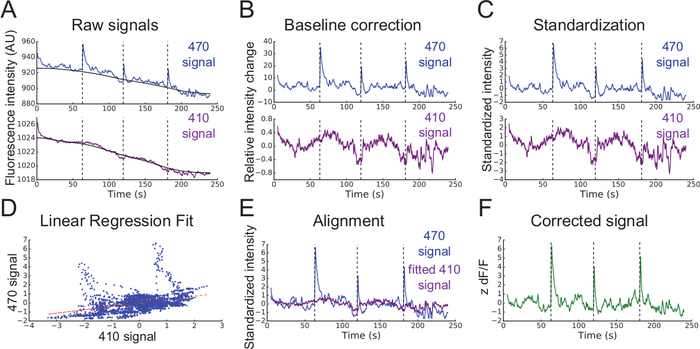
Figure 3: Analysis of fiber photometry data. (A) Smoothed mean fluorescent intensities (Int) recorded from 470 nm (top blue line) and 410 nm (bottom purple line) excitation wavelengths. Black lines are baselines found using the airPLS algorithm. (B) Relative intensity changes in signals after baseline correction. (C) Standardized 470 nm and 410 nm signals (zInt470, top; zInt410, bottom). (D) Non-negative robust linear fit of 470 nm and 410 nm signals. (E) Alignment of the trace Int410 to Int470 based on the fit. (F) Corrected and normalized calcium-dependent change in fluorescence (z dF/F). Please click here to view a larger version of this figure.
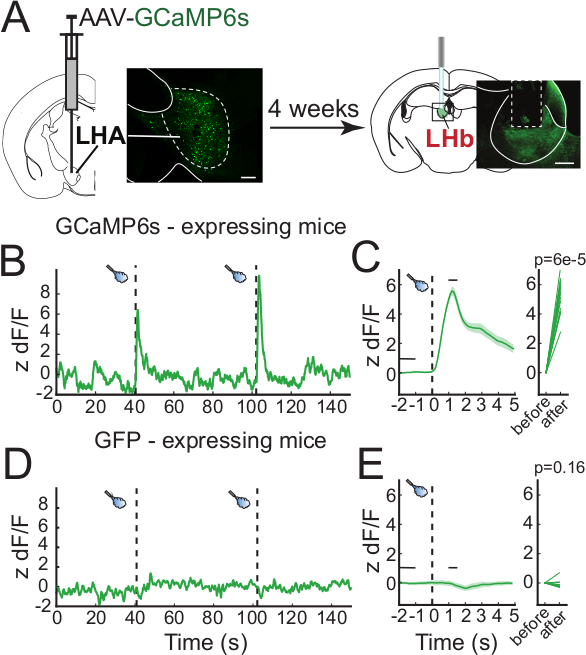
Figure 4: Representative results. (A) Diagram of the experimental procedure. An AAV-GCaMP6s is injected in the LHA of mice and 4 weeks later, an optic fiber cannula is implanted over the LHb for axon terminal signal recording. Inset are representative confocal images of GCaMP6s expression in the cell bodies of LHA neurons (left) and their axon terminals projecting to the LHb (right). (B) Representative calcium signal trace to airpuffs (dashed vertical bars) measured from LHb-projecting LHA axon terminals measured from a GCaMP6s-expressing mouse. (C) Peri-event plot of the average calcium response to airpuff events. The thick green line represents the average and the green-shaded regions represent the standard error of the mean (SEM, left panel), and the signal measured before and after an airpuff (3 mice, 15 events). (D,E) Same measurements as (B,C) for GFP-expressing mice (2 mice, 10 events). Scale bars are 200 µm. Please click here to view a larger version of this figure.
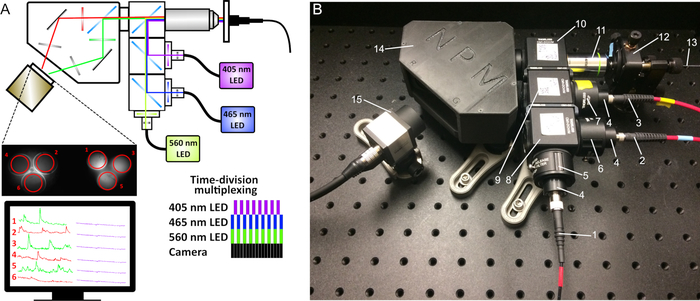
Figure 5: Schematic of dual-color fiber photometry. (A) An additional 560 nm LED, appropriate filters and dichroic mirrors, and an image splitter before the camera sensor were added to the original setup. (B) Photometry system components: (1) Fiber to the 560 nm LED, (2) Fiber to the 465 nm LED, (3) Fiber to the 405 nm LED, (4) Collimators, (5) 560 nm bandpass filter, (6) 470 nm bandpass filter, (7) 410 nm bandpass filter, (8) Cube with longpass 495 dichroic mirror, (9) Cube with longpass 425 dichroic mirror, (10) Cube with 493/574 dichroic mirror, (11) 20x objective, (12) 5-axis translator, (13) Mono- or bundled-fiber patch cord, (14) Image splitter, (15) CMOS camera. Please click here to view a larger version of this figure.
Discussion
Fiber photometry is an accessible approach that allows researchers to record bulk-calcium dynamics from defined neuronal populations in freely-moving animals. This method can be combined with a wide range of behavioral tests, including “movement heavy” tasks such as forced swim tests2, fear-conditioning18, social interactions1,4, and others7,8,19,20. This allows researchers to observe what behaviors or stimuli drive activity in a particular neural population or vice versa.
Despite its prima facie simplicity, there are important considerations when implementing fiber photometry, and the system detailed in this protocol offers several advantages circumventing common pitfalls. First, it takes advantage of the fact that GCaMP variants have a calcium-independent isosbestic excitation point around 410 nm21 and time-division multiplexing to correct calcium-dependent changes in signals almost simultaneously12. When neurons expressing GCaMP are excited with 410 nm wavelength, the emission intensity from GCaMP is independent from its binding to calcium and behaves essentially like a low-efficiency GFP. Time-division multiplexing uses both 410 nm calcium-independent and 470 nm calcium-dependent excitation wavelengths in the same recording. The signal recorded with the 410 nm excitation wavelength is used as a control for calcium-independent movement and artifact changes in fluorescence intensity. Alternative strategies that do not include simultaneous uses of isosbestic excitation of GCaMP can be envisioned. For example, it is possible to prepare two animals cohorts, one expressing GFP and the other expressing GCaMP2. However, this is not a perfect control, as it is across animals and it is more difficult to identify if a given response in a given animal was an artifact. Second, the described fiber photometry setup allows researchers to measure activity in several pathways simultaneously, opening the door to questions regarding signal propagation throughout the brain. Third, dual-color recording allows additional flexibility to dissect different pathways in the same brain region combining green and red-shifted GECIs (Figure 1).
With fiber photometry recordings, very low emission fluorescence needs to be recorded through background noise. Therefore, it is crucial to ensure the maximum collection of the fluorescence emitted from GECIs. First, when developing a viral strategy one must consider that recording from terminals can be challenging, because axon terminals in the recording field can be sparse. To resolve this problem, an alternative intersectional viral strategy can be envisioned allowing expression of GCaMP in a target-projecting region of interest and optic fiber cannulas implanted above the cell bodies, where more robust fluorescence can be measured22,23,24. Another factor that may negatively impact the signal-to-noise ratio is the quality of the optic fiber cannula and patch cord. For implantations, stainless steel ferrule is preferable, because zirconia is highly autofluorescent and will increase the background signal. It is essential to use the highest-quality optic fiber cannulas with well-polished terminal connectors and high transmission rates (>85% transmission). Any defects in the fibers will lead to a decrease in the signal-to-noise ratio. The optic fiber patch cord should be of high quality as well. Providers produce fibers specifically designed for fiber photometry, in which autofluorescence is minimized as much as possible. Custom-made patch cords often do not reach requisite efficiency. It is also important to optimize the alignment of the optical path to get a picture with all fibers well-resolved at the focal point of the objective. Moreover, the numerical aperture (NA) of the fiber must match that of the patch cord as well as the objective used with the system. Any NA mismatch will result in either excitation or emission light loss. All previous steps will provide clear calcium-dependent signals. However, a signal correction is still required. Most recordings have an exponential decrease in fluorescence because of autofluorescence in the fibers, heat-mediated LED decay, and photobleaching. This decrease varies with different fibers and channels, and it is important to remove it in each channel separately using the airPLS algorithm described in the protocol section. Second, movement correction must be taken into account. Even if GCaMP signals seem high where any artifact changes seem meaningless, it is important to correct it with calcium independent signal. The graph with aligned 410 nm and 470 nm signals is a reference showing which changes in intensity are caused by GCaMP and not artifacts.
Adding a 560 nm excitation LED, proper dichroic lenses, and a beamsplitter enables simultaneous recording of green and red fluorescence. This opens the possibility to monitor neural activity from two genetically distinct neuronal populations or to monitor presynaptic activity from axon terminals (e.g., expressing GCaMP6), and postsynaptic neuronal activity (e.g., expressing jrGECO1a). When implementing dual-color recordings, important considerations need to be kept in mind. Significant photoconversion and photoactivation has been reported for many red-shifted GECIs, where illumination with 405 nm, 488 nm, and 560 nm can increase calcium-independent fluorescence25. Use of jrGECO1a and RCaMP1b may minimize this problem26. Another concern during dual color imaging is green signal that leaks into the red channel. Many strategies can be envisioned to avoid this problem. For example, it is possible to interweave the three excitation wavelengths to excite the isosbestic point of the green calcium sensor (410 nm), the calcium-dependent signal from the green calcium sensor (470 nm), and the red calcium sensor (560 nm) in sequence. In that case, it is possible to rely on the isosbestic point of the green sensor for movement correction. Finally, when performing dual color imaging, it is best to acquire stronger signal from the red calcium sensor (e.g., from cell somas) because these have weaker fluorescence emissions compared to green sensors.
New genetically encoded fluorescent sensors have been developed for rapid and specific in vivo detection of neurotransmitter release22,23,24. These sensors emit green fluorescence when bound with their respective endogenous ligand and can be combined with red-shifted GECIs, such as jrGECO1a, for simultaneous detection of neurotransmitter release concurrently with changes in neuronal activity from single multiple optic fibers5,27.
Fiber photometry provides exquisite flexibility to monitor neural activity in freely-moving animals through flexible and lightweight optic fiber patch cords. However, it is not well-suited for situations requiring movement between compartments, such as light-dark chamber tests. This will be possible with further developments in the wireless photometry system28. Finally, while it provides great advantages when monitoring neural dynamics from multiple brain regions, or from distinct neural populations, the signal obtained in fiber photometry is an aggregate of many neurons that may not fire with the same temporal dynamics and will not be resolved. However, it provides a complementary approach to in vivo microendoscopic calcium imaging resolving somatic calcium signal from tens to hundreds of individual neurons29. Finally, when recording from multiple fibers in one single animal, it may be difficult to differentiate whether the signal is coming from the terminals or from cell somas. Carefully designing experiments can overcome most of these limitations. However, the development of new GECIs exclusively localized at cell somas will provide more resolution during multi-fiber calcium imaging experiments.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC: RGPIN-2017-06131) to C.P. C. P. is a FRSQ Chercheur-Boursier. We also thank the Plateforme d’Outils Moléculaires (https://www.neurophotonics.ca/fr/pom) for the production of the viral vectors used in this study.
Materials
| 1/4"-20 Stainless Steel Cap Screw, 1" Long | Thorlabs | SH25S100 | |
| 1/4"-20 Stainless Steel Cap Screw, 1/2" Long | Thorlabs | SH25S050 | |
| 1/4"-20 Stainless Steel Cap Screw, 3/8" Long | Thorlabs | SH25S038 | |
| 1000 µm, 0.50 NA, SMA-SMA Fiber Patch Cable | Thorlabs | M59L01 | |
| 12.7 mm Optical Post | Thorlabs | TR30/M | |
| 12.7 mm Pedestal Post Holder | Thorlabs | PH20EM | |
| 15 V, 2.4 A Power Supply Unit with 3.5 mm Jack Connector for T-Cube | Thorlabs | KPS101 | |
| 20x objective | Thorlabs | RMS20X | #10 in Figure 2, #11 in Figure 5 |
| 30 mm Cage Cube with Dichroic Filter Mount | Thorlabs | CM1-DCH/M | #8-9 in Figure 2, #8-10 in Figure 5 |
| 405 nm LED | Doric Lenses | CLED_405 | #2 in Figure 2 |
| 410 nm bandpass filter | Thorlabs | FB410-10 | #5 in Figure 2; #7 in Figure 5 |
| 465 nm. LED | Doric Lenses | CLED_465 | #1 in Figure 2 |
| 470 nm bandpass filter | Thorlabs | FB470-10 | #4 in Figure 2; #6 in Figure 5 |
| 560 nm bandpass filter | Semrock | FF01-560/14-25 | #5 in Figure 5 |
| 560 nm LED | Doric Lenses | CLED_560 | #1 in Figure 3 |
| 5-axis kinematic Mount | Thorlabs | K5X1 | #11 in Figure 2, #12 in Figure 5 |
| Achromatic Doublet | Thorlabs | AC254-035-A-ML | #7 in Figure 2 |
| Adaptor for 405 collimator | Thorlabs | AD11F | #3 in Figure 2; #4 in Figure 5 |
| Adaptor for ajustable collimator | Thorlabs | AD127-F | #3 in Figure 2; #4 in Figure 5 |
| Aluminum Breadboard | Thorlabs | MB1824 | |
| Clamping Fork | Thorlabs | CF125 | |
| Cube connector | Thorlabs | CM1-CC | |
| Dual 493/574 dichroic | Semrock | FF493/574-Di01-25×36 | #10 in Figure 5 |
| Emission filter for GCaMP6 | Semrock | FF01-535/22-25 | #6 in Figure 2 |
| Enclosure with Black Hardboard Panels | Thorlabs | XE25C9 | |
| Externally SM1-Threaded End Cap for Machining | Thorlabs | SM1CP2M | |
| Fast-change SM1 Lens Tube Filter Holder | Thorlabs | SM1QP | #4-6 in Figure 2, #5-7 in Figure 5 |
| Fixed Collimator for 405 nm light | Thorlabs | F671SMA-405 | #3 in Figure 2; #4 in Figure 5 |
| Fixed collimator for 470 and 560 nm light | Thorlabs | F240SMA-532 | #3 in Figure 2; #4 in Figure 5 |
| Green emission filter | Semrock | FF01-520/35-25 | In light beam splitter |
| High-Resolution USB 3.0 CMOS Camera | Thorlabs | DCC3260M | #13 in Figure 2, #15 in Figure 5 |
| Light beam splitter | Neurophotometrics | SPLIT | #14 in Figure 5 |
| Longpass Dichroic Mirror, 425 nm Cutoff | Thorlabs | DMLP425R | #8 in Figure 2, #9 in Figure 5 |
| Longpass Dichroic Mirror, 495 nm Cutoff | Semrock | FF495-Di03 | #9 in Figure 2, #8 in Figure 5 |
| Metabond dental cement | C&B | ||
| M8 – M8 cable | Doric Lenses | Cable_M8-M8 | |
| Optic fiber cannulas | Doric Lenses | Need to specify that these will be used to photometry experiments requiring low autofluorescence | |
| Optic fiber Patchcords | Doric Lenses | Need to specify that these will be used to photometry experiments requiring low autofluorescence | |
| Red emission filter | Semrock | FF01-600/37-25 | In light beam splitter |
| T7 LabJack | LabJack | ||
| T-cube LED Driver | Thorlabs | LEDD1B | |
| USB 3.0 I/O Cable, Hirose 25, for DCC3240 | Thorlabs | CAB-DCU-T3 |
Riferimenti
- Gunaydin, L. A., et al. Natural Neural Projection Dynamics Underlying Social Behavior. Cell. 157 (7), 1535-1551 (2014).
- Proulx, C. D., et al. A neural pathway controlling motivation to exert effort. Proceedings of the National Academy of Sciences of the United States of America. 115 (22), 5792-5797 (2018).
- Muir, J., et al. In Vivo Fiber Photometry Reveals Signature of Future Stress Susceptibility in Nucleus Accumbens. Neuropsychopharmacology. 43 (2), 255-263 (2017).
- Wang, D., et al. Learning shapes the aversion and reward responses of lateral habenula neurons. eLife. 6, (2017).
- de Jong, J. W., et al. A Neural Circuit Mechanism for Encoding Aversive Stimuli in the Mesolimbic Dopamine System. Neuron. 101 (1), 133-151 (2018).
- Lerner, T. N., et al. Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell. 162 (3), 635-647 (2015).
- Calipari, E. S., et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proceedings of the National Academy of Sciences of the United States of America. 113 (10), 2726-2731 (2016).
- González, A. J., et al. Inhibitory Interplay between Orexin Neurons and Eating. Current Biology. 26 (18), 2486-2491 (2016).
- Chen, T. -. W., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 499 (7458), 295-300 (2013).
- Barker, D. J., et al. Lateral Preoptic Control of the Lateral Habenula through Convergent Glutamate and GABA Transmission. Cell Reports. 21 (7), 1757-1769 (2017).
- Siciliano, C. A., Tye, K. M. Leveraging calcium imaging to illuminate circuit dysfunction in addiction. Alcohol. 74, 47-63 (2018).
- Kim, C. K., et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nature Methods. 13 (4), 325-328 (2016).
- Sych, Y., Chernysheva, M., Sumanovski, L. T., Helmchen, F. High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nature Methods. 16 (6), 553-560 (2019).
- Akam, T., Walton, M. E. pyPhotometry: Open source Python based hardware and software for fiber photometry data acquisition. Scientific Reports. 9 (1), 3521 (2019).
- Sparta, D. R., et al. Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nature Protocol. 7 (1), 12-23 (2011).
- Stamatakis, A. M., et al. Lateral Hypothalamic Area Glutamatergic Neurons and Their Projections to the Lateral Habenula Regulate Feeding and Reward. The Journal of Neuroscience. 36 (2), 302-311 (2016).
- Tervo, G. D., et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 92 (2), 372-382 (2016).
- Yu, K., da Silva, P., Albeanu, D. F., Li, B. Central Amygdala Somatostatin Neurons Gate Passive and Active Defensive Behaviors. The Journal of Neuroscience. 36 (24), 6488-6496 (2016).
- Falkner, A. L., Grosenick, L., Davidson, T. J., Deisseroth, K., Lin, D. Hypothalamic control of male aggression-seeking behavior. Nature Neuroscience. 19 (4), 596-604 (2016).
- Ren, J., et al. Anatomically Defined and Functionally Distinct Dorsal Raphe Serotonin Sub-systems. Cell. 175 (2), 472-487 (2018).
- Barnett, L. M., Hughes, T. E., Drobizhev, M. Deciphering the molecular mechanism responsible for GCaMP6m’s Ca2+-dependent change in fluorescence. PLOS ONE. 12 (2), 0170934 (2017).
- Sun, F., et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell. 174 (2), 481-496 (2018).
- Patriarchi, T., et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science. 360 (6396), (2018).
- Feng, J., et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Detection of Norepinephrine. Neuron. 102 (4), 745-761 (2019).
- Akerboom, J., et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in Molecular Neuroscience. 6, 1-29 (2013).
- Dana, H., et al. Sensitive red protein calcium indicators for imaging neural activity. eLife. 5, (2016).
- Wang, H., Jing, M., Li, Y. Lighting up the brain: genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Current Opinion in Neurobiology. 50, 171-178 (2018).
- Lu, L., et al. Wireless optoelectronic photometers for monitoring neuronal dynamics in the deep brain. Proceedings of the National Academy of Sciences. 115 (7), 1374-1383 (2018).
- Jennings, J. H., et al. Visualizing Hypothalamic Network Dynamics for Appetitive and Consummatory Behaviors. Cell. 160 (3), 516-527 (2014).

