High-Resolution Studies of Proteins in Natural Membranes by Solid-State NMR
Summary
This work details robust basic routines on how to prepare isotope-labeled membrane protein samples and analyze them at high-resolution with modern solid-state NMR spectroscopy methods.
Abstract
Membrane proteins are vital for cell function and thus represent important drug targets. Solid-state Nuclear Magnetic Resonance (ssNMR) spectroscopy offers a unique access to probe the structure and dynamics of such proteins in biological membranes of increasing complexity. Here, we present modern solid-state NMR spectroscopy as a tool to study structure and dynamics of proteins in natural lipid membranes and at atomic scale. Such spectroscopic studies profit from the use of high-sensitivity ssNMR methods, i.e., proton-(1H)-detected ssNMR and DNP (Dynamic Nuclear Polarization) supported ssNMR. Using bacterial outer membrane beta-barrel protein BamA and the ion channel KcsA, we present methods to prepare isotope-labeled membrane proteins and to derive structural and motional information by ssNMR.
Introduction
Structural and motional studies of membrane proteins in physiologically relevant environments pose a challenge to traditional structural biology techniques1. Modern solid-state nuclear magnetic resonance spectroscopy (ssNMR) methods offer a unique approach for the characterization of membrane proteins2,3,4,5,6,7 and has long been used to study membrane proteins, including membrane embedded protein pumps8, channels9,10,11, or receptors12,13,14,15. Technical advances such as ultra-high magnetic fields >1,000 MHz, fast magic angle spinning frequencies >100 kHz, and hyperpolarization techniques16 have established ssNMR as a powerful method for the study of membrane proteins in environments of ever-increasing complexity from liposomes to cell membranes and even whole cells. For example, DNP has become a powerful tool for such experiments (see reference17,18,19,20,21,22,23,24,25). More recently, 1H-detected ssNMR offers increasing possibilities to study membrane proteins at high spectral resolution and sensitivity25,26,27,28,29. This work highlights two bacterial membrane proteins that are involved in essential functions, i.e., protein insertion and ion transport. The corresponding proteins, BamA25,30,31,32,33 and KcsA23,27,28,34,35,36,37,38,39 (or chimeric variants thereof10,40) have been examined by ssNMR methods for more than a decade.
A representative protocol for the preparation and ssNMR characterization of bacterially originating membrane proteins is presented here. The different steps of the protocol are shown in Figure 1. First, the expression, isotope-labeling, purification, and membrane-reconstitution of BamA is explained. Then, a general workflow for the characterization of the membrane protein by ssNMR is presented; specifically, the assignment of membrane protein backbones using 1H-detected ssNMR at fast magic angle spinning. Finally, basic setup and acquisition of dynamic nuclear polarization-(DNP)-supported experiments, which significantly boost ssNMR signal sensitivity, are detailed.
Protocol
1. Production of uniformly labeled 2H, 13C, 15N-labeled BamA-P4P5
NOTE: While this protocol requires working with non-pathogenic Gram-negative bacteria, adherence to basic biological safety procedures is a must, namely, wearing safety glasses, lab coats, gloves, and following institutional standard operating procedures for work with microorganisms.
- Use a single colony of E. coli BL 21 Star (DE3) containing the pET11aΔssYaeT plasmid encoding for E. coli BamA-P4P5 to inoculate 50 mL of Lysogeny broth supplemented with 50 µg/L of ampicillin.
- Grow the culture at 37 °C at 200 rpm until an OD600 of 0.6 is reached. Spin at 2,000 x g for 10 min. Resuspend the pellet in 50 mL of M9 (see Table 1) to a maximum OD600 of 0.1.
NOTE: All future growing steps take place at the conditions stated above unless otherwise noted. - Grow the culture until OD600 of 0.5. Spin at 2,000 x g for 10 min at room temperature. Resuspend the pellet in 50 mL of M9 containing 90% D2O (see Table 1 for recipe) to a maximum OD600 of 0.1. Grow the culture overnight at 30 °C at 200 rpm.
- Spin down the culture at 2,000 x g for 10 min at room temperature. Resuspend the pellet in the prewarmed 50 mL 90% D2O M9 to a maximum OD600 of 0.1. Grow until an OD600 of 1.0 is reached.
- Spin down the culture at 2,000 x g for 10 min at room temperature. Prepare 100 mL of 100% D2O M9 medium with isotope-enriched amounts of non-enriched glucose and ammonium chloride. Resuspend in 100% D2O M9 medium to a maximum OD600 of 0.1. Grow until an OD600 of 0.7 is reached.
- Spin down the culture at 2,000 x g for 10 min at room temperature. Discard the supernatant and resuspend the pellet in 500 mL of 100% D2O M9 medium with isotopes (see Table 1) to a maximum OD600 of 0.1.
- Allow the culture to grow until an OD600 of 0.6-0.9 is reached. Induce with 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). Express for 4 h and harvest cells at 4,000 x g for 15 min at room temperature.
2. Purification, refolding, and BamA-P4P5 proteo-liposome formation
NOTE: All the steps of this section should be conducted in a fume hood. Special care must be taken when opening tubes post-centrifugation limits harmful aerosols.
- Thaw the pellet on ice. Resuspend the pellet in 20 mL of cold Buffer 1. See Table 2 for the Buffers.
- Spin down the solution at 4,000 x g for 20 min at 4 °C. Resuspend the pellet in 20 mL of cold distilled H2O. Allow the suspension to incubate on ice for 10 min.
- Spin down the suspension at 4,000 x g for 20 min at 4 °C. Resuspend in 10 mL cold Buffer 2 and allow to incubate on ice for 30 min.
- Supplement the mixture with an additional 10 mL of cold buffer 2 and incubate for a further 30 min on ice. Add 0.5% n-Dodecyl-B-D-Maltoside (DDM) and swirl gently.
- Sonicate the sample on ice at 13 kHz until vicious-use 10 s long on/off pulses. Spin down at 25,000 x g for 20 min at 4 °C. Wash three times with 20 mL of Buffer 3. Centrifuge at 25,000 x g for 20 min at 4 ˚C.
- Resuspend the pellet in 20 mL of Buffer 4. Incubate at 37 °C for 30 min. Sonicate the sample on ice at 13 kHz, until clear-use 10 s long on/off pulses.
- Spin down the sample at 25,000 x g for 20 min at 4 °C. Repeat steps 2.4 to 2.6 omitting addition of protease inhibitor and incubation at 37 °C.
- Wash the pellet with 20 mL of water and twice with Buffer 3. Spin down at 25,000 x g for 20 min at 4 °C. Aliquot the suspension into 1 mL microcentrifuge tubes and spin the microcentrifuge tubes at maximum speed on a bench-top centrifuge for 20 min. Inclusion body purity is assessed by 10% SDS-Page gel, see Figure 2A. Expected yield for purified BamA-P4P5 triply (2H,13C,15N) labeled is 30 mg/L.
- Solubilize inclusion bodies in 200 µL of Buffer 5 and 300 µL of H2O. Add 6M guanidium chloride (GdnCl). Adjust the volume of microcentrifuge tube to 1 mL with H2O. Vortex the mixture and incubate for 4 h at room temperature. Vortex the microcentrifuge tubes periodically (every 30 min).
- Spin at 100,000 x g for 1 h at 4 °C. Use Beer-Lambert's Law to determine protein concentration by 280 nm. The molar extinction coefficient is 117,120 M-1cm-1. If concentration is >100 µM dilute with 1x Buffer 5 with 6 M GdnCl. Rapidly dilute the protein 10x in Buffer 6. Incubate the mixture overnight at room temperature.
NOTE: For ultracentrifugation, use ultracentrifugation grade 1.5 mL tubes. - Determine refolding efficiency using a semi-native SDS-Page gel. Take two 10 µL aliquots from step 2.10 and add 10 µL of Lameili buffer (buffer 8) to each aliquot. Boil one sample for 10 min at 95 °C; leave the other aliquot on ice. Afterwards, run both samples at 14 mA on a 10% SDS-Page gel. The stacking and running portions of the gels lack reducing agent and SDS. Protein staining is achieved using Coomassie dye. The expected results are shown in Figure 2B.
- Spin down the sample at 4,000 x g for 20 min at 4 °C. Concentrate the sample to a working volume of 8 mL using a 30 kDa centrifugal filter. Spin the sample at 4,000 x g for 20 min at 4 °C. Measure protein absorbance at 280 nm. As in step 2.10, determine the protein concentration.
- Calculate the amount of lipid required for a 10:1 lipid to protein ratio (LPR-mol/mol). From a chloroform stock, add the required amount of lipids, as calculated in step 2.12, into a 100 mL round bottom flask (RBF). Evacuate the chloroform from the RBF under a gentle stream of nitrogen. Place RBF on high vacuum for 3 h. Hydrate the lipid film with 1 mL of Buffer 7. Incubate RBF for 10 min at 37 °C.
- Add the protein mixture (from step 2.12) to the RBF.
- Dilute the protein/lipid mixture (from step 2.14) to the final volume of 50 mL in the RBF. Use Buffer 7 supplemented with 1x protease inhibitor dissolved in DMSO and incubate for 30 min at 37 °C.
- Dialyze against 100 volumes of Buffer 7 for 2 weeks (use 12-14 kDa molecular weight cut-off dialysis tubing). For the first 24 h, perform the dialysis at room temperature with addition of fresh Buffer 7 every 8 hrs. Afterwards, change the buffer once daily, and perform dialysis at 4 °C.
3. Filling of the ssNMR rotor
- After step 2.16, centrifuge for 30-60 min at 10,000 x g at 4 °C so that a pellet is formed. Remove the supernatant with a pipette.
NOTE: The technique discussed below has been optimized for a 3.2 mm rotor but is applicable for almost any rotor diameter. - Pipette the sample into the rotor and gently spin the same down with a table centrifuge.
NOTE: Depending on sample consistency, one may either use a densification tool to compact the sample, or a spatula instead of a pipette to put the sample into the rotor. - Repeat this process 2-3x until the rotor is filled to the correct height and check if there is enough space for the cap left, as indicated by the marker line on the densification tool. Close the rotor with the cap positioning tool on an even surface. Mark half the bottom edge of the rotor.
NOTE: New ssNMR rotor generations come with a durable laser mark at the bottom edge for the optical detection of the rotor spinning frequency. Otherwise, the use of a sharpie pen is suggested; as the MAS detector has been optimized specifically for sharpie ink. - Use a magnifying glass to check whether the cap is well placed.
4. Sample characterization by 2D 13C- 13C ssNMR spectroscopy
- Insert the rotor into the magnet, using the automatic spinning routine of the MAS unit, spin the sample up to the desired MAS frequency between 10 and 20 kHz MAS while cooling the sample to 270 K set temperature. Tune and match the 1H and 13C channels.
NOTE: For the choice of the MAS frequency, avoid accidentally matching the chemical shift distance between the spectral Cα region and the Carbonyl carbon region to the spinning frequency, i.e., avoid the first order rotational resonance condition41. - Optimize a 1H-13C cross-polarization42 (CP) experiment by varying the rf power on the proton channel from 25-80 kHz. Typical parameters are 45 kHz rf power on 13C, 80-100 kHz heteronuclear 1H-decoupling43 during acquisition, 1.8-2.0 s recycle delay, and 64 scans.
- Optimize the CP contact time for maximal signal sensitivity on the aliphatic carbons.
- Determine 1H 90° pulse length in the 13C-CP experiment by multiplying the starting pulse by four, i.e., by optimizing a 360° pulse, and vary the length until the signal has disappeared. Include a rectangular pulse directly after the CP transfer and optimize the 13C 90° pulse length analogously.
- Run a 2D 13C-13C proton-driven spin diffusion (PDSD)-type experiment such as PARIS with 30 ms magnetization transfer time to detect intra-residual correlations and gauge the sample quality. Transfer the power-levels, the contact time, and 90° pulse lengths from the previously optimized 13C-CP experiment. Use an acquisition time of 4-6 ms in the indirect dimension. Process the spectrum with squared sin bell functions (QSIN) or, for insensitive samples, exponential line broadening in both dimensions.
- Extract slices of isolated cross-peaks and determine the linewidth at half heights. A linewidth between ~0.3-1.0 13C ppm is indicative of a well-ordered sample, ideal for ssNMR characterization, whereas a linewidth above 1.0 13C ppm is indicative of conformational heterogeneity that may compromise ssNMR characterization. The 2D CC PARIS spectrum of BamA shown in Figure 3A is an example of a well-ordered protein that gives high-quality spectra.
5. Backbone assignment by 1H-detected 3D ssNMR spectroscopy
- Acquisition of 2D NH spectral fingerprints
- Fill the sample in a 1.3 mm rotor, as described above. Insert the rotor into the magnet, spin the sample to 10-20 kHz MAS and cool the sample to 240-250 K set temperature or lower, depending on the specifications of the ssNMR probehead. Use the MAS unit interface to increase the MAS frequency gradually in steps of 5 kHz to 60 kHz MAS.
- Optimize the CP amplitudes, contact times, and 90° pulse lengths in 13C- and 15N CP experiments. Use amplitudes around 30-50 or 70-100 kHz on the heteronuclear channels and vary the amplitude on the 1H-channel from 80-180 kHz. Use a recycling delay of 0.7-1.2 s, along with 15 kHz low-power heteronuclear proton decoupling44. Also, see reference45 for further practical details on how to set up CP ssNMR experiments.
- Acquire a 2D NH experiment to gauge the 1H-resolution. Transfer the 15N-CP parameters and use approximately 15 and 25 ms acquisition time in the direct and indirect dimensions. For the first 1H to 15N CP transfer, use the previously optimized contact time. For the 15N to 1H backtransfer, use 0.7-1.0 ms contact time to minimize interresidual magnetization transfer.
- Use 50-250 ms water MISSISSIPPI suppression46, dependent on sample water content. Process the spectrum with squared sin bell functions (QSIN) in both dimensions. Extract slices of isolated cross-peaks and determine the linewidth at half heights. An example for a high-quality 2D NH spectrum25 is shown for membrane-embedded BamA-P4P5 in Figure 3B.
- Backbone assignments
- Optimize a 1D 15N to 13Cα specific CP experiment47. Place the 13C carrier in the middle of the Cα region around 55 ppm. Use a 13C rf amplitude of 20 kHz and vary the 15N amplitude from 35 to 45 kHz for maximal transfer to the Cα signals and minimal transfer to the carbonyl signals. Optimize the contact time from 3 to 10 ms in increments of 1 ms.
- Run a 3D CαNH experiment. Transfer all CP amplitudes, contact times, and pulse lengths from the previously optimized steps. Use approximately 10 ms and 30 ms in the indirect and direct acquisition dimensions. Process the spectrum with squared sin bell functions (QSIN) in all three dimensions.
- Optimize a 1D Cα to CO DREAM magnetization transfer48. Start with a 15N to 13Cα specific CP transfer. Then, transfer the magnetization from Cα to CO using a spin lock on the 13C channel. Vary the rf amplitude from 20 to 35 kHz for optimal transfer. Optimize the transfer time from 4 to 10 ms.
- Run a 3D Cα(CO)NH experiment. Transfer all rf amplitudes, contact times, and pulse lengths from the previously optimized steps. Use approximately 10 ms and 30 ms in the indirect and direct acquisition dimensions. Process the spectrum with squared sin bell functions (QSIN) in all three dimensions.
- Repeat and adapt steps 1-4 for the analogous 3D CONH and CO(Cα)NH experiments. Use this quartet of 3D experiments for sequential Cα and CO walks to assign the protein backbone, as shown on the example of KcsA in Figure 428.
6. Protein dynamics by 1H-detected ssNMR spectroscopy
- For 15N T1 studies, use the previously optimized 2D NH experiment at 60 kHz MAS and include a delay after the first 1H to 15N cross-polarization step. Acquire a series of 2D NH experiments and vary the delay from 0 to 16 s using steps of, e.g., 0, 2, 4, 8, and 16 s.
- Plot the normalized intensities for resolved signals as a function of the increasing delay time. Fit the T1 decay to single exponentials according to y = exp(-x/T1), with y being the signal intensity at decay time x.
- Analogously, for 15N T1rho studies, include a spin-lock pulse in the pulse program on the 15N channel with an rf amplitude between 15-20 kHz after the first 1H to 15N cross-polarization step26,49. Acquire a series of 2D NH experiments and vary the delay from 0 to 150 ms and fit the T1rho decay of resolved signals to single exponentials. Illustrative 15N T1rho data is shown for membrane-embedded K+ channels in Figure 527,28.
7. Dynamic nuclear polarization
NOTE: The following preparative steps relate to the use of a commercial DNP setups using 3.2 mm sapphire MAS rotors (Figure 6)20. Use of the zirconia rotors or other DNP equipment may lead to lower DNP signal enhancements.
- Resuspend membrane protein pellet (from step 3.1) in 50 µL of deuterated DNP buffer (60% (v/v) deuterated d8- and glycerol, and 15 mM AMUPol50. For experiments at 800 MHz, we recommend using 10 mM NATriPOL-351. Also see reference52 for further practical aspects to DNP ssNMR experiments.
- Centrifuge for 10-20 min at 100,000 x g at 4 °C so that a pellet is formed. Remove the supernatant with a pipette.
- Precool the centrifugal adaptor and the 3.2 mm sapphire MAS rotor components to <4 °C. The following steps (7.4-7.7) need to be done as quickly as possible to prevent the DNP agents from getting reduced.
- Place the 3.2 mm sapphire rotor in the centrifuge adapter and fill it with the suspension of proteo-liposomes from step 7.2.
- Pipette the resuspended membrane protein sample against the inner wall of the rotor carefully using a 200 µL pipette tip. Allow the solution to slide down to the bottom of the rotor. Ensure that air bubbles are not formed.
- Spin down the sample in the rotor for 5-10 mins at 4,500 x g at 4 °C. Remove the supernatant containing excess DNP juice by pipetting it out carefully without disrupting the cell pellet. Repeat steps 6.4-6.6 until the rotor is full.
- Carefully position the polytetrafluoroethylene (PTFE) spacer using the spacer screw on top of the rotor.
- Place the rotor, cap side down in a rotor plunger53 and plunge the rotor into liquid nitrogen. Allow at least 30-60 s for the rotor and the sample to freeze.
- Start up the heat exchanger unit and cool down the DNP probe to ~90K.
- Set the MAS unit to Manual mode and set the variable temperature (VT) to 135 l/h and bearing and drive gas flows to ~6 l/h.
- Engage Eject on the MAS console.
- Transfer the frozen sample from Step 7.8 from the liquid N2 into the sample catcher and place it in the probe (also see reference53).
- Manually increase the probe bearing pressure (Bp) to ~500 mbar before engaging the "Insert". This is advisable for stable and safe spinning of the rotor.
- Increase MAS rate to the desired value using the procedure described in reference53.
- Check for DNP enhancements with and without microwaves using a standard 1D CP experiments (also see, e.g., reference51,54) (Figure 7) before proceeding with multidimensional experiments, e.g., 2D CC experiments described in section 3 of this article.
Representative Results
Figure 2 shows representative gels for inclusion body purity (Panel A) and refolding of inclusion bodies (Panel B3). Figure 2 confirms the successful purification of 13C,15N-labeled BamA-P4P5.
Figure 3A shows a typical 2D 13C-13C spectrum of a well-ordered membrane protein, and Figure 3B shows a typical, high-quality 2D 15N1H spectrum of a perdeuterated membrane protein25. Note that this membrane protein expressed as inclusion bodies, so that the transmembrane part is also accessible by perdeuteration. For membrane proteins that do not express in inclusion bodies, one usually needs to employ H2O-based growth media27,55,56,57.
Figure 4 exemplifies how to assign the backbone of membrane proteins using 1H-detected 3D experiments28. Figure 5 demonstrates the powerof ssNMR relaxation measurements that can provide detailed, site-resolved information on the dynamics of membrane proteins28,49.
Eventually, Figure 6 and Figure 7 illustrate how DNP can boost the sensitivity of ssNMR experiments of membrane-embedded proteins25. Figure 6 shows high-resolution DNP-supported ssNMR information on the structure of the outer-membrane protein BamA.
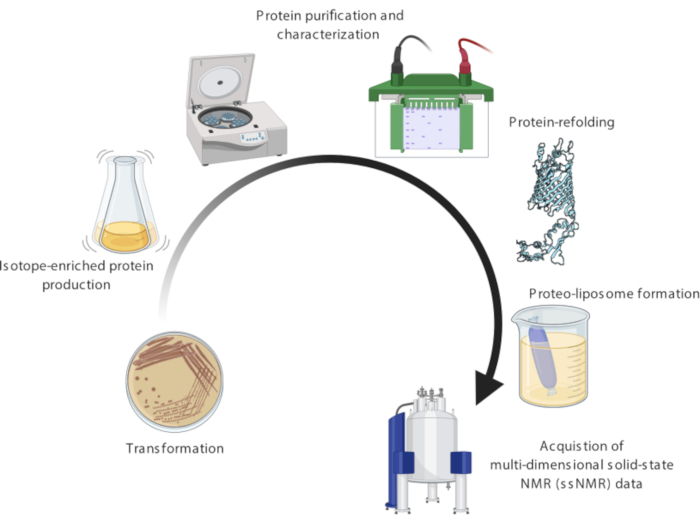
Figure 1: Experimental methodology for atomic-level multidimensional solid-state nuclear magnetic resonance (ssNMR) studies of membrane proteins embedded in native-membrane environments. Specifically, the workflow details robust procedures for membrane protein production, purification, refolding, proteo-liposomes formation, and most crucially setup/acquisition/analysis of cutting-edge multidimensional ssNMR spectra. Please click here to view a larger version of this figure.
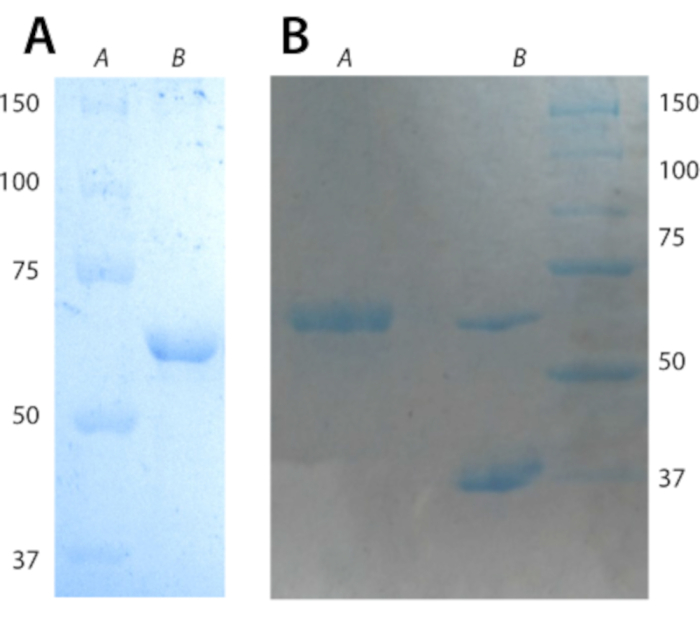
Figure 2: Representative gels for inclusion body purity and refolding. (A) 10% SDS-Page gel of BamA-P4P5 inclusion bodies. Lane A has the molecular weight ladder; Lane B has BamA-P4P5 inclusion bodies. The expected molecular weight of BamA-P4P5 is 61.1 kDa. (B) 10% SDS-page semi native gel for assessing BamA-P4P5 refolding efficiency. Lane A contains the boiled sample; as expected, it migrates to the molecular weight of 61.1 kDa. Lane B contains the un-boiled sample; the top band is protein that has remained unfolded and the bottom band near 37 kDa is the folded population. The folding efficiency can be determined using image analysis software-in this specific case, folding efficiency is >70%. Please click here to view a larger version of this figure.
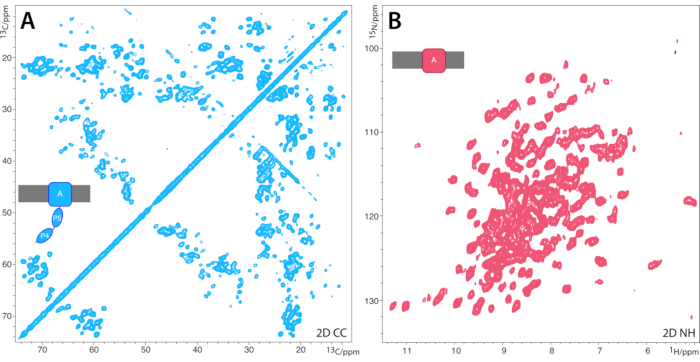
Figure 3: Representative 2D 13C13C PARIS and 1H-detected NH spectra for BamA-P4P5 proteo-liposomes. (A) 2D 13C13C PARIS spectrum of 13C,15N-labeled BamAP4P5 in liposomes. The spectrum was acquired at 700 MHz (1H-frequency) magnetic field using 13 kHz MAS and a 13C13C magnetization transfer time of 30 ms. (B) 1H-detected 2D NH spectrum of the 2H,13C,15N-labeled transmembrane part of BamA in liposomes. The spectrum was acquired at 800 MHz (1H-frequency) and 60 kHz MAS. This figure has been modified from reference25. Please click here to view a larger version of this figure.
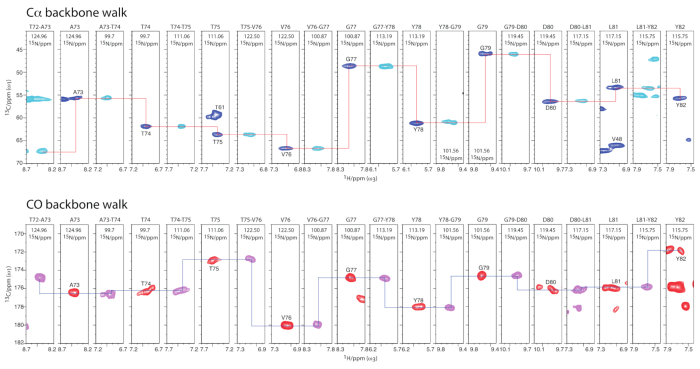
Figure 4: Sequential ssNMR assignments of membrane-embedded ion channels with 1H-detected 3D experiments. Upper panel: Cα-Cα+1 backbone walk showing full connectivity for residues T72-Y82 in KcsA mutant E71A. Dark blue signals show CαH planes from a 3D CαNH experiment, cyan CαH planes were taken from a 3D CαcoNH experiment. Lower panel: CO-1-CO backbone walk showing full connectivity for residues T72-Y82 in E71A. Magenta signals show COH planes from a 3D CONH experiment, red COH planes were taken from a 3D COcαNH experiment. This figure has been modified from reference28. Please click here to view a larger version of this figure.
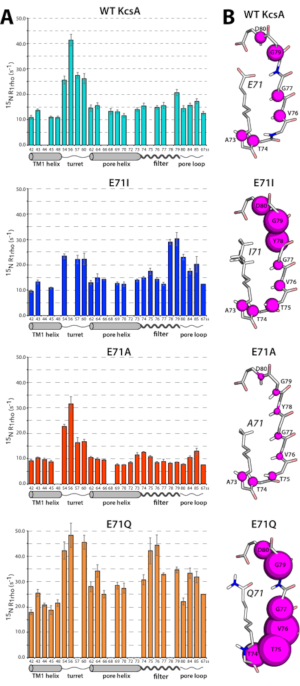
Figure 5: SSNMR 15N T1rho data reveal differential dynamics in membrane-embedded point-mutants of the bacterial potassium channel KcsA. (A) 15N rotating frame ssNMR relaxation rates (R1rho) that report on slow molecular motions in WT KcsA (cyan), E71A (red), E71I (blue), and E71Q (orange) measured at 700 MHz and 58 kHz MAS. The error bars show the standard error of the fit. (B) Illustration of the site-resolved selectivity filter dynamics. The size of the magenta spheres represents the R1rho relaxation rates. This figure has been modified from reference28. Please click here to view a larger version of this figure.
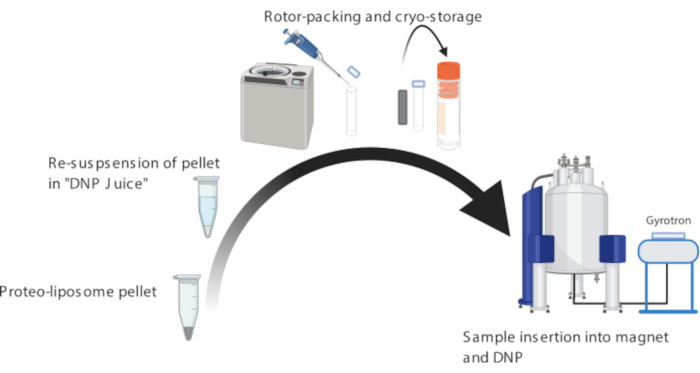
Figure 6: Preparation of proteo-liposomes for dynamic nuclear polarization (DNP) NMR experiments. Important steps are highlighted, including resuspension of proteo-liposomes in DNP juice, packing the sample into a sapphire rotor, and ultimately execution of DNP measurements. Please click here to view a larger version of this figure.
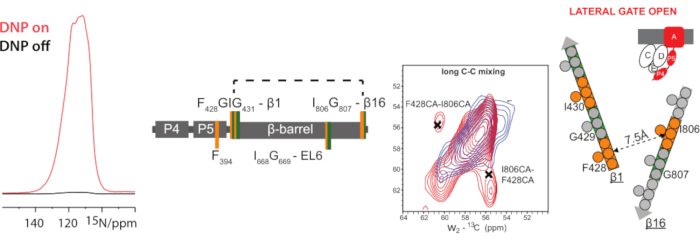
Figure 7: DNP spectra of 13C and 15N-labeled BamA-P4P5. Left: A DNP signal enhancement of about 110 could be obtained for membrane-embedded 13C,15N-IFG-labeled BamAP4P5 using a 400 MHz/263 GHz DNP system. Right: Close-up of the Cα-Cβ region of the DNP enhanced 15N-edited carbon-carbon correlation experiment58 using a long (1 s) mixing time, measured on 13C,15N-IFG-labeled BamAP4P5, in the presence (red spectrum) and absence (blue spectrum) of unlabeled BamCDE. Crosses are the tentative assignments for the residues targeted by these experiments. Schematic representation of the correlations observed between β-strand 1 and 16 in 15N-edited CxCx DNP spectra for the case of a lateral gate open. The distance between Cα residues of the closest residue in β1 to the I806 of β16 is indicated. This figure has been modified from reference25. Please click here to view a larger version of this figure.
| Chemical | Amount/Volume for 1L of Medium |
| Disodium phosphate | 6.0 g |
| Monopotassium phosphate | 3.0 g |
| Sodium Chloride | 0.5 g |
| Ammonium Chloride – both nitrogen-15 enriched and not enriched | 0.5 g |
| Magnesium sulphate (1 M) | 2 mL |
| Calcium dichloride (1 M) | 10 µL |
| Iron sulphate (0.01 M) | 1 mL |
| Isotope enriched glucose* | 2 g |
| Glucose | 5 g |
| Thiamine (0.5 mg/mL) | 10 mL |
| Micronutrients | 1 mL |
| Vitamin Supplements | 1 mL |
| Ampicillin (50 mg/mL) | 1 mL |
Table 1: M9 minimal medium recipe. The medium can be made up in H2O or varying amounts of D2O as directed by the protocol. *In case of perdeuterated protein production D-Glucose-13C6,1,2,3,4,5,6,6-d7 is used.
| Chemical | Final Concentration |
| Buffer 1 | |
| Tris-HCl pH 8.0 | 50 mM |
| EDTA | 40 mM |
| Buffer 2 | |
| Tris-HCl pH 8.0 | 50 mM |
| EDTA | 40 mM |
| Lysozyme | .2 mg/mL |
| Sucrose | 25% |
| Buffer 3 | |
| Tris-HCl pH 8.0 | 10 mM |
| Buffer 4 | |
| Tris-HCl pH 8.0 | 10 mM |
| MgCl2 | 2 mM |
| Benzonase (>90%) | 40 units |
| Sucrose | 25% |
| DDM | 0.10% |
| Protease inhibitor cocktail in DMSO | 1x |
| Buffer 5 | |
| Tris-HCl pH 8.0 | 100 mM |
| Glycine | 500 mM |
| Buffer 6 | |
| Sodium Phosphate pH 7 | 50 mM |
| lauryl-dimethylamine oxide (LDAO) | 1% |
| Protease inhibitor cocktail in DMSO | 1x |
| Buffer 7 | |
| Sodium Phosphate pH 7 | 20 mM |
| MgCl2 | 5 mM |
| Buffer 8 | |
| Tris-HCl pH 6.8 | 62.5 mM |
| Sodium dodecyl sulfate | 0.10% |
| Glycerol | 10% |
| Bromophenol blue | 0.00% |
Table 2: Buffers used for the sample preparation of membrane embedded BamA.
| Chemical | Concentration (mol/L) |
| Ammonium molibdate | 3 x 10-6 |
| Boric acid | 4 x 10-4 |
| Cobalt chloride | 3 x 10-5 |
| Copper sulphate | 1 x 10-5 |
| Manganese chloride | 8 x 10-5 |
| Zinc chloride | 1 x 10-5 |
| Note: The stock can be made up in water or varying amounts of D2O as directed by the protocol. | |
Table 3: Micronutrients stock (1,000x). The stock can be made up in water or varying amounts of D2O as directed by the protocol.
| Chemical | Amount (mg) for 100 mL of Supplement |
| D-Biotin | 100 |
| Choline chloride | 50 |
| Folic acid | 50 |
| Myoinositol | 100 |
| Nicatinamide | 50 |
| Panthotenic acid | 50 |
| Pyridoxal-HCl | 50 |
| Riboflavin | 5 |
| Thiamine-HCl | 50 |
| Note: The supplement can be made up in H2O or varying amounts of D2O as directed by the protocol. | |
Table 4: Vitamin Supplement (1,000x) for 100 mL. The supplement can be made up in H2O or varying amounts of D2O as directed by the protocol.
Discussion
Membrane proteins are key players in the regulation of vital cellular functions both in prokaryotic and eukaryotic organisms; thus, understanding their action mechanisms at atomic levels of resolution is of vital importance. The existing structural biology techniques have pushed scientific understanding of membrane proteins quite far but have heavily relied on experimental data gathered from in vitro systems devoid of membranes. In this article, an experimental approach is presented that allows to obtain atomistic insight into the structure and function of two bacterial membrane-proteins embedded in native-like membrane by utilizing cutting-edge solid-state NMR techniques, namely, fast-MAS and DNP. This method is broadly applicable to other bacterial membrane proteins.
The protocol begins by describing a robust methodology to produce and purify triply isotope labeled BamA in high yield and purity (Figure 2A) -other stable-isotope labeling schemes can be employed. The purification protocol makes use of synergy between surfactant activity and sonication to remove cellular debris from BamA inclusion bodies. The subsequent refolding sees formation of BamA-LDAO micelles (Figure 2B). Further addition of lipids and dialysis aids in the removal of LDAO and residual surfactant molecules thereby facilitating formation of proteo-liposomes suitable for solid-state NMR studies. The formation of proteo-liposomes is a crucial step and must be done with adherence to the buffer exchange schedule detailed above. This is mandatory for a successful preparation.
The acquisition of 2D spin diffusion-based CC spectra (Figure 3A) of membrane proteins is routine and often among the first experiments conducted to gauge the sample's quality and secondary structure content. By using long CC mixing times, these experiments can also be conveniently used to derive structural information. Note that spin diffusion transfer efficacy inversely scales with the MAS frequency. Hence, longer mixing times are required with increasing MAS frequency.
If the membrane-protein is grown in D2O-based buffers as in the representative example of Figure 3B, it is necessary to back-exchange the amino-protons in H2O-based buffers afterwards59. This may prevent the detection of the water-inaccessible transmembrane-part. This limitation can be overcome by the use of dedicated H2O-based growth media27 or fully-protonated membrane proteins in combination with MAS frequencies above 100 kHz55,56,57,60.
Spectral assignments of small membrane proteins can be conveniently performed with the quartet of 1H-detected 3D experiments that is shown in Figure 4 for the selective filter of the K+ channel E71A KcsA28. This quartet can be supplemented by a 3D CαCαNH experiment or 2D CC experiments to obtain sidechain information that simplifies the assignments. For larger membrane proteins, the use of 4D or higher order experiments is necessary to disambiguate the assignments. These spectra are usually acquired with dedicated non-uniform sampling strategies in order to shorten experimental time.
Detailed relaxation data is shown in Figure 5 for the K+ channel KcsA and three KcsA point mutants with different gating behaviors, hence correlation membrane protein dynamics to function28. For example, compared to the WT KcsA channel, the constitutively active mutant E71A shows a striking rigidification, and the so-called "flicker" mutant E71Q shows a marked increase in global protein dynamics. For relaxation measurements, a high sensitivity is indispensable because signals toward the end of the relaxation series also need to be acquired with robust signal-to-noise ratios.
The spectacular advantages of DNP are shown in Figure 6 and Figure 7. DNP can strongly improve sensitivity in ssNMR experiments that enable otherwise infeasible experiments27,61,62,63. In Figure 7B, we utilized DNP to probe residue-specific, through-space contacts in a membrane-embedded BamA (547 residues)-Bam CDE complex. A combination of amino-acid specific labeling and DNP-supported 15N-edited 13C-13C correlation spectroscopy58 allows to probe through-space contacts between α-strands 1 and 16, suggesting the so-called lateral gate that is critical for function (see, e.g., reference64) is open.
In the future, we expect modern solid-state NMR methods to continue to play a major role for structural and dynamical studies of membrane proteins, especially in complex media such as cellular preparations and whole cells.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work is part of the research programs ECHO, TOP, TOP-PUNT, VICI, and VIDI with project numbers 723.014.003, 711.018.001, 700.26.121, 700.10.443, and 718.015.00, which are financed by the Dutch Research Council (NWO). This article was supported by iNEXT-Discovery (project number 871037).
Materials
| Ammonium molibdate | Merck | 277908 | |
| Ammonium-15N Chloride | Cortecnet | CN80P50 | |
| Ampicillin | Sigma Aldrich | A9518 | |
| AMUpol | Cortecnet | C010P005 | |
| Benzonase | EMD Millipore Corp | 70746-3 | |
| Boric acid | Merck | B6768 | |
| bromophenol blue | Sigma | B0126 | |
| calcium dichloride | Merck | 499609 | |
| Choline chloride | Sigma | C-1879 | |
| Cobalt chloride | Merck | 449776 | |
| Copper sulphate | Merck | C1297 | |
| D-Biotin | Merck | 8512090025 | |
| Deuterium Oxide | Cortecnet | CD5251P1000 | |
| Dimethyl sulfoxide | Merck | D9170 | |
| Ethylenediaminetetraacetic acid | Sigma Aldrich | L6876 | |
| Folic acid | Sigma | F-7876 | |
| Glucose 13C + 2H | Cortecnet | CCD860P50 | |
| Glycerol | Honeywell | G7757 | |
| Glycerol (12C3, 99.95% D8, 98%) | Eurisotope | CDLM-8660-PK | |
| glycerol (non-enriched) | Honeywell | G7757-1L | |
| Glycine | Sigma Aldrich | 50046 | |
| Guanidine hydrochloride | Roth Carl | NR.0037.1 | |
| Iron sulphate | Merck | 307718 | |
| isopropyl β-D-1-thiogalactopyranoside | Thermofisher | R0392 | |
| Lysogeny Broth | Merck | L3022 | |
| Lysozyme | Sigma Aldrich | L6876 | |
| Magnesium chloride – hexahydrate | Fluka | 63064 | |
| magnesium sulphate | Merck | M5921 | |
| monopotassium phosphate | Merck | 1051080050 | |
| Myoinositol | Sigma | I-5125 | |
| n-Dodecyl-B-D-maltoside | Acros Organics | 3293702509 | |
| N,N-Dimethyldodecylamine N-oxide | Merck | 40236 | |
| Nicatinamide | Sigma | N-3376 | |
| Panthotenic acid | Sigma | 21210-25G-F | |
| protease inhibitor | Sigma | P8849 | |
| Pyridoxal-HCl | Sigma Aldrich | P9130 | |
| Riboflavin | Aldrich | R170-6 | |
| Sodium Chloride | Merck | K51107104914 | |
| Sodium dihydrogen phospahte – monohydrate | Sigma Aldrich | 1,06,34,61,000 | |
| Sodium dodecyl sulfate | Thermo-scientific | 28365 | |
| Sodium hydroxide | Merck | 1,06,49,81,000 | |
| Sucrose | Sigma Life Science | S9378 | |
| Thiamine-HCl | Merck | 5871 | |
| Tris-HCl | Sigma Aldrich | 10,70,89,76,001 | |
| Zinc chloride | Merck | 208086 | |
| E.coli BL21 DE3* | New England Biolabs | C2527 | |
| 1.5 mL Ultra-tubes | Beckman Coulter | 357448 | |
| 30 kDa centrifugal filter | Amicon | UFC903024 | |
| 3.2 mm sapphire DNP rotor with caps | Cortecnet | H13861 | |
| 3.2 mm teflon insert | Cortecnet | B6628 | |
| 3.2 mm sample packer/unpacker | Cortecnet | B6988 | |
| 3.2 mm Regular Wall MAS Rotor | Cortecnet | HZ16913 | |
| 3.2 mm Regular Wall MAS rotor | Cortecnet | HZ09244 | |
| Tool Kit for 3.2 mm Thin Wall rotor | Cortecnet | B136904 | |
| 1.3 mm MAS rotor + caps | Cortecnet | HZ14752 | |
| 1.3 mm filling tool | Cortecnet | HZ14714 | |
| 1.3 mm sample packer | Cortecnet | HZ14716 | |
| 1.3 mm cap remover | Cortecnet | HZ14706 | |
| 1.3 mm cap set tool | Cortecnet | HZ14744 | |
| Dialysis tubing 12-14 kDa | Spectra/Por | 132703 | |
| Sharpie – Black | Merck | HS15094 |
Riferimenti
- Weingarth, M., Baldus, M. Solid-state NMR-based approaches for supramolecular structure elucidation. Accounts of Chemical Research. 46 (9), 2037-2046 (2013).
- Kaplan, M., Pinto, C., Houben, K., Baldus, M. Nuclear magnetic resonance (NMR) applied to membrane-protein complexes. Quarterly Reviews of Biophysics. 49, 15 (2016).
- Brown, L. S., Ladizhansky, V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Science. 24 (9), 1333-1346 (2015).
- Ladizhansky, V. Applications of solid-state NMR to membrane proteins. Biochimica Et Biophysica Acta. Proteins and Proteomics. 1865 (11), 1577-1586 (2017).
- Hong, M., Zhang, Y., Hu, F. H. Membrane protein structure and dynamics from NMR spectroscopy. Annual Review of Physical Chemistry. 63, 1-24 (2012).
- Shahid, S. A., et al. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nature Methods. 9 (12), 1212-1217 (2012).
- Wang, S. L., et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nature Methods. 10 (10), 1007-1012 (2013).
- Herzfeld, J., Lansing, J. C. Magnetic resonance studies of the bacteriorhodopsin pump cycle. Annual Review of Biophysics and Biomolecular Structure. 31 (1), 73-95 (2002).
- Ketchem, R. R., Hu, W., Cross, T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 261 (5127), 1457-1460 (1993).
- Lange, A., et al. Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature. 440 (7086), 959-962 (2006).
- Cady, S. D., et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 463 (7281), 689-692 (2010).
- Park, S. H., et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 491 (7426), 779-783 (2012).
- Luca, S., et al. The conformation of neurotensin bound to its G protein-coupled receptor. Proceedings of the National Academy of Sciences of the United States of America. 100 (19), 10706-10711 (2003).
- Joedicke, L., et al. The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors. Nature Chemical Biology. 14 (3), 284-290 (2018).
- Krug, U., et al. The conformational equilibrium of the neuropeptide Y2 receptor in bilayer membranes. Angewandte Chemie (International Edition in English). 59 (52), 23854-23861 (2020).
- Maly, T., et al. Dynamic nuclear polarization at high magnetic fields. Journal of Chemical Physics. 128 (5), (2008).
- Bajaj, V. S., Mak-Jurkauskas, M. L., Belenky, M., Herzfeld, J., Griffin, R. G. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 106 (23), 9244-9249 (2009).
- Linden, A. H., et al. Neurotoxin II bound to acetylcholine receptors in native membranes studied by dynamic nuclear polarization NMR. Journal of the American Chemical Society. 133 (48), 19266-19269 (2011).
- Ong, Y. S., Lakatos, A., Becker-Baldus, J., Pos, K. M., Glaubitz, C. Detecting substrates bound to the secondary multidrug efflux pump EmrE by DNP-enhanced solid-state NMR. Journal of the American Chemical Society. 135 (42), 15754-15762 (2013).
- Koers, E. J., et al. NMR-based structural biology enhanced by dynamic nuclear polarization at high magnetic field. Journal of Biomolecular NMR. 60 (2-3), 157-168 (2014).
- Becker-Baldus, J., et al. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 112 (32), 9896-9901 (2015).
- Kaplan, M., et al. EGFR dynamics change during activation in native membranes as revealed by NMR. Cell. 167 (5), 1241-1251 (2016).
- Visscher, K. M., et al. Supramolecular organization and functional implications of K+ channel clusters in membranes. Angewandte Chemie (International Edition in English). 56 (43), 13222-13227 (2017).
- Joedicke, L., et al. The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors. Nature Chemical Biology. 14, 284 (2018).
- Pinto, C., et al. Formation of the beta-barrel assembly machinery complex in lipid bilayers as seen by solid-state NMR. Nature Communications. 9, (2018).
- Good, D. B., et al. Conformational dynamics of a seven transmembrane helical protein Anabaena Sensory Rhodopsin probed by solid-state NMR. Journal of the American Chemical Society. 136 (7), 2833-2842 (2014).
- Medeiros-Silva, J., et al. (1)H-detected solid-state NMR studies of water-inaccessible proteins in vitro and in situ. Angewandte Chemie (International Edition in English). 55 (43), 13606-13610 (2016).
- Jekhmane, S., et al. Shifts in the selectivity filter dynamics cause modal gating in K(+) channels. Nature Communications. 10 (1), 123 (2019).
- Schubeis, T., Le Marchand, T., Andreas, L. B., Pintacuda, G. 1H magic-angle spinning NMR evolves as a powerful new tool for membrane proteins. Journal of Magnetic Resonance. 287, 140-152 (2018).
- Sinnige, T., et al. Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. Journal of Molecular Biology. 426 (9), 2009-2021 (2014).
- Sinnige, T., et al. Insight into the conformational stability of membrane-embedded BamA using a combined solution and solid-state NMR approach. Journal of Biomolecular NMR. 61 (3-4), 321-332 (2015).
- Sinnige, T., et al. Conformational plasticity of the POTRA 5 domain in the outer membrane protein assembly factor BamA. Structure. 23 (7), 1317-1324 (2015).
- Renault, M., Bos, M. P., Tommassen, J., Baldus, M. Solid-state NMR on a large multidomain integral membrane protein: the outer membrane protein assembly factor BamA. Journal of the American Chemical Society. 133 (12), 4175-4177 (2011).
- Doyle, D. A., et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280 (5360), 69-77 (1998).
- Wylie, B. J., Bhate, M. P., McDermott, A. E. Transmembrane allosteric coupling of the gates in a potassium channel. Proceedings of the National Academy of Sciences of the United States of America. 111 (1), 185-190 (2014).
- Xu, Y., Zhang, D., Rogawski, R., Nimigean, C. M., McDermott, A. E. Identifying coupled clusters of allostery participants through chemical shift perturbations. Proceedings of the National Academy of Sciences of the United States of America. 116 (6), 2078-2085 (2019).
- vander Cruijsen, E. A. W., Prokofyev, A. V., Pongs, O., Baldus, M. Probing conformational changes during the gating cycle of a potassium channel in lipid bilayers. Biophysical Journal. 112 (1), 99-108 (2017).
- Varga, K., Tian, L., McDermott, A. E. Solid-state NMR study and assignments of the KcsA potassium ion channel of S. lividans. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics. 1774 (12), 1604-1613 (2007).
- Zhang, D., Howarth, G. S., Parkin, L. A., McDermott, A. E. NMR studies of lipid regulation of the K+ channel KcsA. Biochimica et Biophysica Acta (BBA) – Biomembranes. , 183491 (2020).
- Schneider, R., et al. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. Journal of the American Chemical Society. 130 (23), 7427-7435 (2008).
- Raleigh, D. P., Levitt, M. H., Griffin, R. G. Rotational resonance in solid-state Nmr. Chemical Physics Letters. 146 (1-2), 71-76 (1988).
- Schaefer, J., Stejskal, E. O. C-13 nuclear magnetic-resonance of polymers spinning at magic angle. Journal of the American Chemical Society. 98 (4), 1031-1032 (1976).
- Fung, B. M., Khitrin, A. K., Ermolaev, K. An improved broadband decoupling sequence for liquid crystals and solids. Journal of Magnetic Resonance. 142 (1), 97-101 (2000).
- Weingarth, M., Bodenhausen, G., Tekely, P. Low-power decoupling at high spinning frequencies in high static fields. Journal of Magnetic Resonance. 199 (2), 238-241 (2009).
- Loquet, A., Tolchard, J., Berbon, M., Martinez, D., Habenstein, B. Atomic scale structural studies of macromolecular assemblies by solid-state nuclear magnetic resonance spectroscopy. Journal of Visualized Experiments: JoVE. (127), (2017).
- Zhou, D. H., Rienstra, C. M. High-performance solvent suppression for proton detected solid-state NMR. Journal of Magnetic Resonance. 192 (1), 167-172 (2008).
- Baldus, M., Petkova, A. T., Herzfeld, J., Griffin, R. G. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Molecular Physics. 95 (6), 1197-1207 (1998).
- Verel, R., Baldus, M., Ernst, M., Meier, B. H. A homonuclear spin-pair filter for solid-state NMR based on adiabatic-passage techniques. Chemical Physics Letters. 287 (3-4), 421-428 (1998).
- Lewandowski, J. R., Sass, H. J., Grzesiek, S., Blackledge, M., Emsley, L. Site-specific measurement of slow motions in proteins. Journal of the American Chemical Society. 133 (42), 16762-16765 (2011).
- Sauvee, C., et al. Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency. Angewandte Chemie (International Edition in English). 52 (41), 10858-10861 (2013).
- Zhai, W., et al. Postmodification via thiol-click chemistry yields hydrophilic trityl-nitroxide biradicals for biomolecular high-field dynamic nuclear polarization. The Journal of Physical Chemistry. B. 124 (41), 9047-9060 (2020).
- Ghosh, R., Kragelj, J., Xiao, Y., Frederick, K. K. Cryogenic sample loading into a magic angle spinning nuclear magnetic resonance spectrometer that preserves cellular viability. Journal of Visualized Experiments: JoVE. (163), (2020).
- Narasimhan, S., et al. Characterizing proteins in a native bacterial environment using solid-state NMR spectroscopy. Nature Protocols. , (2020).
- Ni, Q. Z., et al. High frequency dynamic nuclear polarization. Accounts of Chemical Research. 46 (9), 1933-1941 (2013).
- Weingarth, M., et al. Quantitative analysis of the water occupancy around the selectivity filter of a K+ channel in different gating modes. Journal of the American Chemical Society. 136 (5), 2000-2007 (2014).
- Lalli, D., et al. Proton-based structural analysis of a heptahelical transmembrane protein in lipid bilayers. Journal of the American Chemical Society. 139 (37), 13006-13012 (2017).
- Schubeis, T., et al. A beta-barrel for oil transport through lipid membranes: Dynamic NMR structures of AlkL. Proceedings of the National Academy of Sciences of the United States of America. 117 (35), 21014-21021 (2020).
- Baker, L. A., Daniels, M., vander Cruijsen, E. A. W., Folkers, G. E., Baldus, M. Efficient cellular solid-state NMR of membrane proteins by targeted protein labeling. Journal of Biomolecular NMR. 62 (2), 199-208 (2015).
- Linser, R., et al. Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angewandte Chemie (International Edition in English). 50 (19), 4508-4512 (2011).
- Shukla, R., et al. Mode of action of teixobactins in cellular membranes. Nature Communications. 11 (1), 2848 (2020).
- Kaplan, M., et al. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nature Methods. 12 (7), 649-652 (2015).
- Visscher, K. M., et al. Supramolecular organization and functional implications of K(+) channel clusters in membranes. Angewandte Chemie (International Edition in English). 56 (43), 13222-13227 (2017).
- Narasimhan, S., et al. DNP-supported solid-state NMR spectroscopy of proteins inside mammalian cells. Angewandte Chemie (International Edition in English). 58 (37), 12969-12973 (2019).
- Wu, R., Stephenson, R., Gichaba, A., Noinaj, N. The big BAM theory: An open and closed case. Biochimica et Biophysica Acta (BBA) – Biomembranes. 1862 (1), 183062 (2020).

.
