Evaluation of Motor Impairment in C. elegans Models of Amyotrophic Lateral Sclerosis
Summary
This protocol describes two sensitive assays for discriminating among mild, moderate, and severe motor impairment in C. elegans models of amyotrophic lateral sclerosis, with general utility for C. elegans strains, with altered motility.
Abstract
The neurodegenerative disease amyotrophic lateral sclerosis (ALS) features progressive loss of motor neurons accompanied by muscle weakness and motor impairment that worsens with time. While considerable advances have been made in determining genetic drivers of ALS for a subset of patients, the majority of cases have an unknown etiology. Further, the mechanisms underlying motor neuron dysfunction and degeneration are not well understood; therefore, there is an ongoing need to develop and characterize representative models to study these processes. Caenorhabditis elegans can adapt their movement to the physical constraints of their surroundings, with two primary movement paradigms studied in a laboratory environment- crawling on a solid surface and swimming in liquid. These represent a complex interplay between sensation, motor neurons, and muscles. C. elegans models of ALS can exhibit impairment in one or both of these movement paradigms. This protocol describes two sensitive assays for evaluating motility in C. elegans: an optimized radial locomotion assay measuring crawling on a solid surface and an automated method for tracking and analyzing swimming in liquid (thrashing). In addition to the characterization of baseline motor impairment of ALS models, these assays can detect suppression or enhancement of the phenotypes from genetic or small molecule interventions. Thus, these methods have utility for studying ALS models and any C. elegans strain that exhibits altered motility.
Introduction
Amyotrophic Lateral Sclerosis (ALS) is a debilitating, aging-related neurodegenerative disease with a particular impact on motor neurons. The disease features loss of motor neurons in the brain and spinal cord and progressive motor impairment. This results in major functional disability and premature death, typically within 3-5 years after diagnosis1. Mutations in at least 38 genes can cause ALS; however, most patients with ALS accumulate ubiquitinated inclusions of the protein TDP-43 as their primary pathology in neurons and glial cells2,3,4. A number of animal models have been developed to study the underlying mechanisms causing or contributing to ALS in vivo (reviewed in5). In C. elegans, these models include genetic loss-of-function mutations in homologs of ALS-causing genes or transgenic expression of human ALS genes. There are numerous advantages to modeling ALS in C. elegans. C. elegans are a tractable simple animal with a differentiated nervous system, well-characterized behavioral paradigms, and considerable genetic homology to humans6,7. Many tools exist for working with C. elegans, including robust genome editing capabilities, in vivo fluorescent reporters of neurodegeneration, RNAi screening paradigms, tractable genetics, and established behavioral and phenotypic assays. C. elegans models of ALS recapitulate aspects of human disease, including accumulation of insoluble protein, neurodegeneration, and early death8,9. Further, motor dysfunction featuring disturbance of both crawling and swimming behaviors is present in many C. elegans ALS models.
This protocol describes two methods to characterize C. elegans motor phenotypes: the radial locomotion assay for evaluating crawling on a solid surface and the assessment of swimming in liquid (thrashing) using the WormLab automated tracking and analysis. These sensitive methods for characterizing motor deficits allow comparisons of severity and offer tools for measuring suppression and enhancements of motor phenotypes. The radial locomotion assay quantifies differences in crawling motility (sinusoidal movement on a solid surface) among populations of worms. This assay takes advantage of C. elegans natural unstimulated exploration behavior by placing worms in a single location on a plate and marking their final location after a given period of time10. Alternatively, swimming in liquid (thrashing) assays count body bends of individual worms over a set period of time. The manual counting of body bends by the human eye is time-intensive and typically exhibits considerable variability between experimenters. The use of computer-assisted automated tracking and analysis can eliminate much of that variability. In addition to the characterization of baseline motor impairment of ALS models, both radial locomotion and swimming assays can detect modulation of distinct locomotor phenotypes from genetic or small molecule interventions. These methods have utility for studying ALS models and any C. elegans strain that exhibits altered motility.
Protocol
1. Radial locomotion assay
- Prepare 100 mm or 150 mm diameter Petri dishes about half full of nematode growth media (NGM) and seeded with a uniform lawn of OP50 bacteria to cover the entire surface of the agar.
- Flip the NGM assay plates upside-down and label the bottom with an identifier for the C. elegans strains to be assayed, 2 plates per strain (Figure 1A).
- Make a small dot with the marker in the center of the upside-down plate (Figure 1A).
- While working with a dissecting microscope, transfer 15-20 staged matched11 worms to the center of the assay plate directly over the center dot (visible through the NGM) using a platinum-wire pick (Figure 1B).
NOTE: Try to transfer all the worms at the same time or as quickly as possible.- Set a timer for 30 min after worms are placed in the center of the plate. Put the lid back on the plate and set it aside (Figure 1C).
- Continue transferring worms until all strains are on the designated assay plates, making a note of approximately how long it takes to transfer between plates (aim for ~ 1 min between plates). Keep all plates in the same order that they were set up.
- After 30 min, begin scoring the first plate.
- Remove the lid and place the plate face down under the dissecting microscope, such that the labeled back of the plate is facing up, and the NGM agar is between the line of sight and the worms. The plate will be upside-down from the normal usage.
- Adjust the microscope focus until worms are visible through the agar.
- Using a different colored felt tip pen from the center point, put a small dot at the location of each worm – follow the worm tracks through the bacterial lawn to find the most distal animals.
- Check the edge of the plate as some worms may end up there. Also, count and record the number of worms in the center point dot (Figure 1D).
- Continue marking all assay plates in order, such that all plates have 30 min of activity time, being careful to account for the time it took to set up the plate. Then, perform manual or digital measurements as described in steps 1.8 or 1.9.
- Perform manual measurements using a ruler.
- Use a ruler to measure in mm, the distance from the center point to the final location markings for each worm and record the distance. To help keep track of progress during scoring, label the first scored point with a small marking line.
- Record length data for each dot consecutively by rotating the plate in a clockwise fashion (Figure 1E).
- Perform digital measurement using a scanner and ImageJ.
- Using a flatbed scanner, scan the back of the assay plate(s) next to a ruler (numbers facing the scanning surface) (Figure 2A).
- Open the scanned plate image in ImageJ or a similar program.
NOTE: ImageJ is a free Java-based image processing software hosted by the NIH. - Click on the Straight-Line Tool and then draw a line connecting the 1 cm and 2 cm points on the scanned image of the ruler (Figure 2B).
NOTE: Holding the Shift computer key will force the line to the nearest 45° angle. - Navigate to the Analyze menu and use it to set the scale (Analyze | Set Scale) (Figure 2C).
- Enter the Known Distance (10) and Units of Length (mm) in the appropriate boxes, do not adjust the distance in pixels number. Check Global to apply the same scale to all images being analyzed in each ImageJ session; otherwise, set the scale for each image upon opening (Figure 2D). Click Ok.
NOTE: Steps 1.9.2-1.9.5 are performed to set the measurement scale. Measurements for this image will now automatically relate the length of lines to the defined pixel: length relationship. Images scanned at 300 dpi will have an approximate relationship of 11.8 pixels per mm. - Use the Paintbrush Tool to mark just above the first point to be scored.
NOTE: This is a visual indicator for the first worm scored. - Use the Straight-Line Tool to draw a line from the center point to the first worm data point.
- Click the M key on the keyboard to measure the distance. Alternatively, navigate to the Analyze menu and choose Measure (Analyze | Measure). In both cases, the measurement will appear in a new window; this window will collect all measurements per image.
- Keeping the center point constant, move the end of the line touching the first worm point to the next worm point, moving clockwise, and click the M key to measure.
- Continue measuring each point in a clockwise fashion until all points are scored.
- Copy and Paste length measurement data into a spreadsheet to save it. Then, repeat the process for each scanned plate image.
NOTE: Steps 1.9.6 – 1.9.11 measure the worm displacement values (Figure 2E-F).
- Perform statistical analysis.
- Combine replicates within a single experiment so that a total number scored is between 30-40 worms.
- Perform independent experimental replicates on different days and with independent populations of worms to end up with 3 independent experimental replicates.
- Analyze data using appropriate statistical analyses such as Student's t-test for 2 strains or 1-way ANOVA for three or more strains.
2. Computer-analyzed swimming assay
NOTE: This protocol contains detailed instructions for the commercially available WormLab hardware and software system (see Table of Materials). However, the workflow can be applied to other computer-analyzed swimming assay systems.
- Setting up and recording videos.
- Raise the shielding on the imaging hardware (Figure 3) and verify that the camera height is set to the expected location.
NOTE: The optimal height for capturing worms on a 35 mm plate is such that the third from the bottom threaded screw bore is visible below the adjustable camera mounting bracket, but no additional mounting hole is seen above the screw bore (approximately 30.5 cm above the stage) (Figure 3C). This will result in a capture ratio of 11.43 µm/pixel. Consider adding a "correct height" indicator so that this is easier to confirm prior to beginning the assay. See protocol step 2.18 to account for height adjustments. - Open the associated software (Supplementary Figure 1A).
- Click the Video Capture icon (film with a right-pointing triangle in the middle) to open a new Video Capture window. The camera will be in the Live View, but the display will be black (Supplementary Figure 1B).
- Press down on the dimmer knob and turn it clockwise to adjust the light.
NOTE: The dimmer knob is on the stage of the imaging system near the bright field light source. The Video Capture live view display will turn from black to illuminated. - Back in the Video Capture window, click on the Settings tab, adjust as follows: Video mode: 2456 x 2052_Mono8, Frame rate: 14, Output: monochrome, Exposure: 0.00300 s, Gain: 1 dB , Gamma: 1, Rotate 180 (check on).
- Move back to the Capture tab. Select the recording folder (Supplementary Figure 1B).
NOTE: This is where all recordings and project files will be stored. Create unique folders for individual experiments to keep data organized. - Assign a file name by entering it into the File prefix text box.
NOTE: The software will automatically append a progressive number to the end of the prefix in the format "0001". It is advisable to have a consistent naming scheme for the files such as: "YYYYMMDD_treatmentName_rep#_". - Set other Capture settings as follows: Buffer: 128 frames (default), Duration (check on): 00 h:01min:00 s. Ignore or turn off other settings in this tab.
- Place the assay plate, lid up, on the device stage, and center it in the Video Capture screen; remove the lid.
NOTE: Prepare assay plates ahead of time. Each plate consists of a 35 mm Petri dish approximately half full of unseeded (no bacterial lawn) NGM. - Use a micropipette to wash the stage synchronized11 worms in 1 mL of M9 onto the assay plate – aim for about 50 worms (Figure 3D). Gently swirl the plate to bring the animals to the center or use a micropipette to add a few drops of M9 to separate animals (Figure 3E). Set a timer for 60 s to allow worms to acclimate to swimming.
NOTE: The plate will not be 100% visible on the screen. Animals are not accurately tracked while overlapping. - While the timer counts down, manually adjust the camera focus by turning the focusing ring on the lens body of the camera while watching the display (Figure 3F). It is possible to zoom in on the worms using a scroll wheel mouse allowing for more precise focusing.
- After setting the focus, adjust the light knob such that the display is as bright as possible without overexposure (shows up as red pixels on display).
- After 60 s, press the Record button.
NOTE: The video will go from the Live View a Recording and a timer will be visible. When the capture is complete, the Video Capture display will return to the Live View (Supplementary Figure 1B). - Remove the plate after recording and set up the next plate. Rename the prefix as needed. Wash worms into the assay plate, incubate for 60 s, and record each treatment.
- After completing all recordings for an experiment, close any open video tabs, and turn off the light source by depressing the dimmer knob.
- Close the Video Capture window by clicking the red x button in the window's upper right corner; alternatively, close the entire software package and reopen.
- Prepare videos for tracking.
- From the Workflow sidebar menu, select the first button on this menu, Import Image Sequence, and find and double click on a video. The video will open in the middle window (Supplementary Figure 2A).
NOTE: The default display for the software is the Workflow sidebar menu; this menu can also be accessed through the view top dropdown menu under View [View | Workflow]. The Workflow menu has several view options; the Track view is appropriate at this point in the protocol. If the Set Sequence Info button is not visible in the menu, then this is not the correct view. First, look at the bottom of the Menu and check that the Workflow tab is active, next check that the Track view is active by clicking on the Track icon. Where menus appear in the window is dependent on monitor size and the location of other opened menus. Menus can be moved by drag and drop, resized, and closed. Most of the menus can also be opened through the View dropdown menu. - From the Workflow menu, select Set Sequence Info (Supplementary Figure 2B). In the new menu window, check the naming scheme, add notes, and validate metadata.
- Check that the scaling is set correctly; Scale: 11.43 µm/pixel and Measure will auto fill as 1000 µm is 87 pixels. If the camera height has been adjusted differently than prescribed here, follow the instructions in this menu to update the scale. Click the Save button.
- In the Workflow menu, select Adjust Image, and a new popup menu called Image Adjustments will open (Supplementary Figure 2C). Select Dark worms on light background (bright field).
- Adjust additional settings. Recommended Image Processing Settings are as follows: Background smoothing: 10, Gaussian smoothing: 5, Fill holes: 2, Small object filter: 0, and skip Integral Derivative Segmentation.
- Adjust the Threshold level such that the animals are completely filled with green but still distinct from the background. Click Apply.
NOTE: It is permissible to pick up some objects that are not worms. - From the Workflow menu, select Detect and Track (Supplementary Figure 2D).
NOTE: A menu will appear to the side of the video with 3 tabs: Detecting, Tracking, and Repair. The cursor will look like a pointing finger when over the video display. Alternatively, click the Select icon (cursor arrow) in the icon menu in the upper left of the software.- Use the finger selector tool to select 3-7 worms and click the Detect worms button in the Detection tab.
NOTE: This selection helps the software accurately identify worms for given conditions and must be done for each video. After a few seconds, most of the worms will be highlighted in green and have a number in yellow. Worms that are touching or curled up will likely not be highlighted; however, the program will most likely find them during tracking. There is an option to adjust the detection parameters; however, the defaults work well under most conditions. If adjustments to the detection parameters are made, make sure to be consistent between samples and replicates.
- Use the finger selector tool to select 3-7 worms and click the Detect worms button in the Detection tab.
- Still in the Detect and Track window, move to the Tracking tab (Supplementary Figure 2E). In the Tracking Parameters check Use backtracking, uncheck Track worms at the edge of the image.
- Set the Max tracked hypothesis to 5. Set the Tracking mode a Swimming.
NOTE: The Max tracked hypothesis setting adjusts how many potential worm-tracks a single tracked worm is allowed to have and gives the program some leeway in how accurately to track a single worm without losing track of it. 5 is a good setting for swimming. - Move to the Advanced Settings section and set the following: Frames worms can touch boundary: 50, Frames worms can overlap: 500, Position tolerance: 0.50, Shape tolerance: 0.50. Save these settings and deploy them for all videos in an experiment (and beyond) by saving them as a configuration.
NOTE: Track filtering is not recommended and can easily be done post-processing, leave off. - Navigate to the Configuration Manager (gear icon) in the upper-left icon menu. When clicked, the Configuration Manager sidebar menu will open. Next, click the Save (gear +) icon, give this configuration a name and description, then click the OK button.
- To access this configuration in the future, open the Configuration menu, select the desired configuration, and click the Load (gear up arrow) icon. Then add sequence information, possibly fine-tune the threshold level, and select worms for detection. All other settings will be the same.
- In the Workflow menu, click Save Project, verify the save location and add a project file name (matching the project file name to the video name helps with continuity). Close the video.
NOTE: These projects are saved with the .wpr filetype. - Repeat for all videos: Import Image Sequence (2.18), Load saved configuration (2.28.1), adjust the threshold (2.23), set sequence info (2.19), detect worms (2.24.1), save the project (2.29), close video.
- Track videos
- To track the project files, go to the Workflow menu and click the Batch (folder containing worm pages) icon (Supplementary Figure 2F).
NOTE: The Batch Processing panel will be displayed (Supplementary Figure 2G). Sometimes it is hidden but can be accessed at the very bottom of the Batch Workflow menu; the window may need to be expanded to see it. - Click the Add button under the File Selection section of this Batch Processing menu. Navigate to and select all the project files (.wpr) to be processed, click Open.
NOTE: The Batch Processing menu will fill with the selected file path, a status indicator and a gray progress bar for each file. - Click the Start (play button icon). Note the green progress indicator on the first file. Allow all the files to be processed. When all the files read as finished (green), the software can be closed (Supplementary Figure 2G).
- If a project file gets stuck, consider running the hung-up file by itself, this usually fixes the problem. If this does not fix the problem, open the project file, delete the selected worms (Detect and Track menu), and reselect from a new training set. Then reprocess the project file.
- To track the project files, go to the Workflow menu and click the Batch (folder containing worm pages) icon (Supplementary Figure 2F).
- Repairing tracks (recommended)
NOTE: Repairing tracks can increase the accuracy of the tracks. Common fixes are combining tracks that clearly belong to the same worm as viewed by the human eye as, deleting tracks that are not accurate or do not capture real worm data, and splitting tracks that are misrepresented because worms are touching (remove the bad section and combine tracks from before and after the poor section if practical).- Open the software and open a project file [File | Open Project] of interest. Click Open and click yes to use this file. Wait for the temporal filter to load. The processed video will be on the screen; worms will be highlighted in green and outlined in yellow. Use the video player to scrub through or play the video.
- From the Workflow menu, select Detect and Track (Supplementary Figure 2D). Switch to the Repair tab. Activate the Selector cursor by clicking on the Selector tool icon (cursor arrow). Scrub through the video, finding and selecting a worm track, and then use the repair menu to create the desired change.
NOTE: To strategically fix most problems quickly, zoom in such that about 1/6th of the plate is visible, start with the area surrounding track #1. Quickly scrub through the video checking for worms that mysteriously appear out of nowhere, tracks where the numbers change, or worms bumping into each other, editing as problems arise. Follow worms in numerical order from the start position. Scrub back and forth to follow each worm as necessary until the whole plate is repaired. Pay careful attention to the edges of the plate; often, fake worms appear in the shadows of plate edges.
- Data analysis
- From the Workflow menu, select Analyze Data (Supplementary Figure 3A).
NOTE: A new window named Data Analysis and Plotting will appear (Supplementary Figure 3B). This window has many ways to assess and view the movement data of a specific project file's worm population. Most of these analyses allow for the selection of specific, multiple, or all worms from a given video allowing for track information to be plotted in different ways. - To analyze turn count, navigate to the Track Summary [Position & Speed | Track Summary] (Supplementary Figure 3C). Use the Export button in the bottom right to export data in a spreadsheet readable format (.csv), choose a path to the desired location of the spreadsheet, including renaming it to something memorable. The default settings are a good choice (Supplementary Figure 3D).
- Paste this data, as well as any other relevant project datasets into a consolidated spreadsheet workbook (Supplementary Figure 3E).
- Use the Turn Count and Track Duration to calculate turns per min for each track in using spreadsheeting functions (=turns/track duration * 60) or use a different metric as desired (Supplementary Figure 3F).
NOTE: Turns per minute captures very similar information as a manual thrashing assay which is a widely used metric of activity in liquid, but there are many options, and it is quite possible that a different metric may identify other interesting data for a given treatment or comparison. - If desired, screen for worm size, track duration, and/or some other chosen metric(s) using conditional formatting tools to highlight tracks for removal if they are potentially confounding. Maintain consistent data processing between treatments and replicates.
- Analyze data using appropriate statistical analyses such as Student's t-test for 2 strains, or 1-way ANOVA for 3 or more strains. Perform independent experimental replicates on different days and with independent populations of worms for at least 3 independent experimental replicates.
- From the Workflow menu, select Analyze Data (Supplementary Figure 3A).
Representative Results
Both radial locomotion and swimming assays offer sensitive detection of motility impairment (Figure 4 and Figure 5). To investigate the mechanisms underlying pathological TDP-43 in ALS, C. elegans models have been developed that express wild-type or ALS-mutant human TDP-43 pan-neuronally. These animals display molecular and cellular characteristics reminiscent of ALS, including motor dysfunction9. Importantly, they exhibit moderate motility impairment with the expression of wild-type human TDP-43 and more severe motility impairment in animals expressing ALS-mutant TDP-43 using both radial locomotion and swimming assays. Some mutant or transgenic animals will have greater impairment in crawling than swimming, or vice versa. By using two different motility assays, a clearer picture of phenotypic differences between strains is obtained.
Radial locomotion
When placed on a seeded agar plate, C. elegans explore their surroundings, including seeking out the boundaries of their food source. Radial locomotion assays are one way to take advantage of that behavior as a metric for physical fitness. By analyzing the locomotor behavior (crawling on a solid surface) in a controlled and quantifiable way, the radial locomotor assay offers a simple and effective tool for assessing the severity of motor deficits and other motor-related phenotypes. Radial locomotion assays capture differences in motility in moderately or severely impaired ALS model worms and offer a baseline to compare modulation of motility phenotypes or changes in motility with age (Figure 6). This strategy can be applied to quantify the crawling behavior of any strain that has altered movement from wild type (N2) or control worms. However, this method may not be a good choice to assess animals unable to crawl normally, such as roller mutants or animals that are paralyzed. Typically, wild-type worms will present an average displacement between 200-300 µm/min when raised and tested at 20 °C. The example data presented in Figure 4 show expected results comparing N2, two different transgenic strains expressing wild-type human TDP-43 with a mild phenotype [TDP-43(WT-mild), CK402 (bkIs402[Psnb-1::TDP-43; Pmyo-3::GFP])] or stronger phenotype [TDP-43(WT-moderate), CK410 (bkIs402[Psnb-1::TDP-43; Pmyo-2::GFP])], a transgenic strain expressing mutant human TDP-43 with a severe phenotype [TDP-43(M337V), CK423 (bkIs423[Psnb-1::TDP-43; Pmyo-2::GFP]), and a transgenic strain expressing another neurodegenerative-disease associated protein, wild-type human tau [tau(WT), CK144 (bkIs144[Paex-3::tau(4R1N); Pmyo-2::GFP])]. The two strains expressing wild-type TDP-43 have different degrees of impairment as detected by radial locomotion. TDP-43(WT-low) is not significantly different from N2, while TDP-43(WT-high) does exhibit clear differences in motility. TDP-43(M337V) and tau (WT) strains have more severe impairments in motility.
Swimming assay
C. elegans engage in stereotypical swimming (thrashing) movement when immersed in liquid. Immediately upon submersion, worms begin to characteristically bend head and tail towards each other with a bend angle of approximately 45°, with the angle vertex being the midpoint of the worm. Worms alternate bending in the ventral and dorsal directions. One thrash, as measured by the software, represents the movement of going from straight to a body bend angle of 20° or greater, irrespective of directionality (angle thresholding can be adjusted post-processing within the Analysis and Plotting window [Workflow | Analyze Data | Body Shape | Bending Angle | Mid-Point | Amplitude Threshold: # degrees]. The swimming assay described here uses automated computer-based tracking and analysis to provide unbiased scoring of swimming activity. It is expected that wild-type (N2) worms will average between 150-200 thrashes per min when raised and recorded at 20 °C. The example data presented in Figure 5, shows expected results comparing N2, two different transgenic strains expressing wild-type human TDP-43 with a mild phenotype [TDP-43(WT-mild), CK402 (bkIs402[Psnb-1::TDP-43; Pmyo-3::GFP])] or stronger phenotype [TDP-43(WT-moderate), CK410 (bkIs402[Psnb-1::TDP-43; Pmyo-2::GFP])], and a transgenic strain expressing another neurodegenerative-disease associated protein, wild-type human tau [tau(WT), CK144 (bkIs144[Paex-3::tau(4R1N); Pmyo-2::GFP])]. The transgenic strain expressing ALS-mutant TDP-43 [TDP-43(M337V)] does not thrash in liquid, and so it is denoted on the graph as ND (no data). This assay can discriminate different phenotypes than the radial locomotion assay shown in Figure 4. For example, in the radial locomotion assay (Figure 4), TDP-43(WT-low) was not significantly different from N2. However, in the swimming assay (Figure 5), both TDP-43(WT-low) and TDP-43(WT-high) are significantly different from N2, as well as significantly different from one another. Also, despite both TDP-43(M337V) and tau(WT) having severe crawling impairments by radial locomotion (Figure 4), only tau(WT) is able to thrash enough to be tracked by the software (Figure 5). TDP-43(M337V) animals are unable to thrash, and software-based analysis does not detect or track these worms accurately. Thus, data on these worms were not collected (ND, no data).
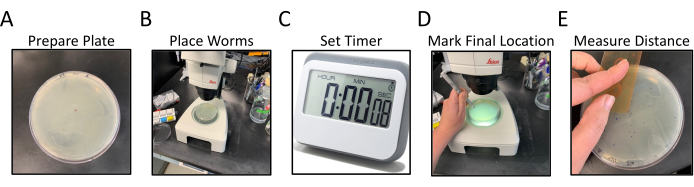
Figure 1: Radial locomotion assay workflow. Panels A-E show the generalized steps of the radial locomotion assay. The steps are as follows: (A) NGM Plates, with OP50 seeded to the edges, are prepared by labeling and marking central dot, (B) worms are placed in the center of the agar as marked by the central dot, (C) worms are allowed to move freely for a set amount of time, (D) plate is flipped bottom up and the final location of each worm is marked in a different color than the central dot, (E) The distance from the center dot to each final worm location in measured either by hand or digitally. Please click here to view a larger version of this figure.
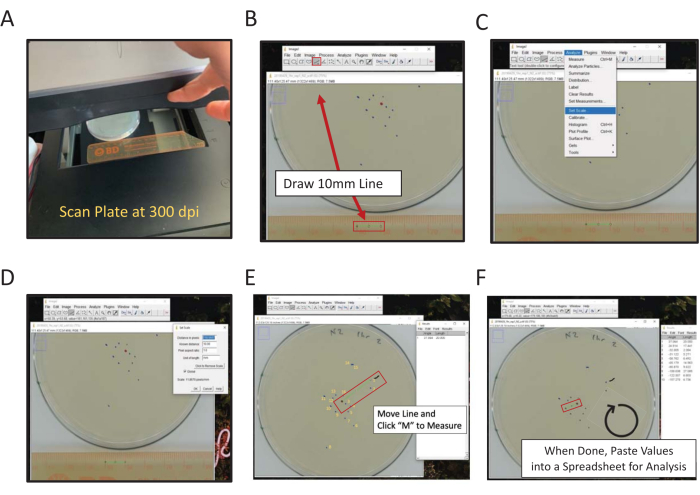
Figure 2: Digital measurement of final location using ImageJ. Distance from center measurements can be measured by hand or digitally, to measure digitally using ImageJ (A) scan backside of the plate with a ruler in the frame. (B) Draw a known length using the line tool. (C) Use the known length drawn in (B) to set scale [Analyze | Set Scale…]. (D) Use the line tool to draw a line from the center point to a final location mark, repeat for each mark. (E) A paintbrush line marking the first mark measured aids in scoring (squiggly black line near the 1 mark). Final location marks are numbered in yellow to illustrate the directionality of scoring. (F) Measurements are recorded in the Results window. Save results elsewhere for statistical analysis. Please click here to view a larger version of this figure.
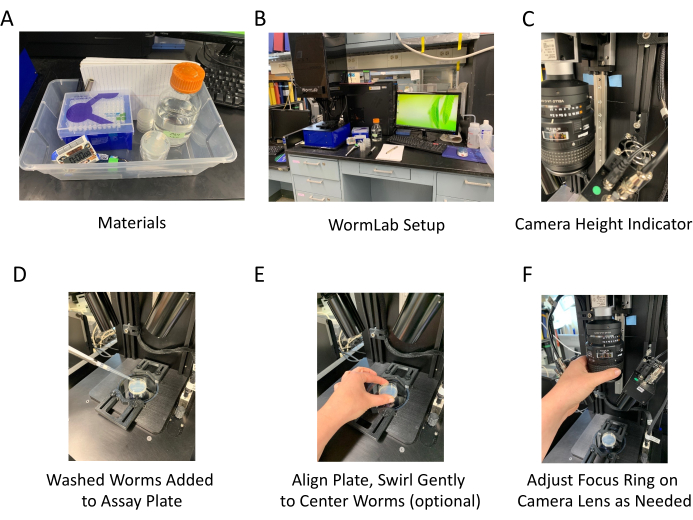
Figure 3: Materials and hardware adjustments for using the software. (A) Materials used for the software-assisted swimming assay. (B) General set up of the software and needed materials, note that the housing is in the raised position. (C) The lens mounting bracket height can be adjusted. This image shows the adjustable track and a tape indicator marking the preferred height for performing swimming assays. It is necessary to adjust the brightfield light and camera focus before recording. (D) A 35 mm assay plate is placed on the stage, animals in M9 are added to the plate, and a 1 min timer is started. (E) It is sometimes advantageous to gently swirl the plate to move worms closer to the center – observe location on the live capture screen; alternatively, a few drops of M9 from a pipettor can be used to separate worms. (F) Adjust the focus ring on the camera lens, observe worms on the screen to determine optimal focus. Please click here to view a larger version of this figure.
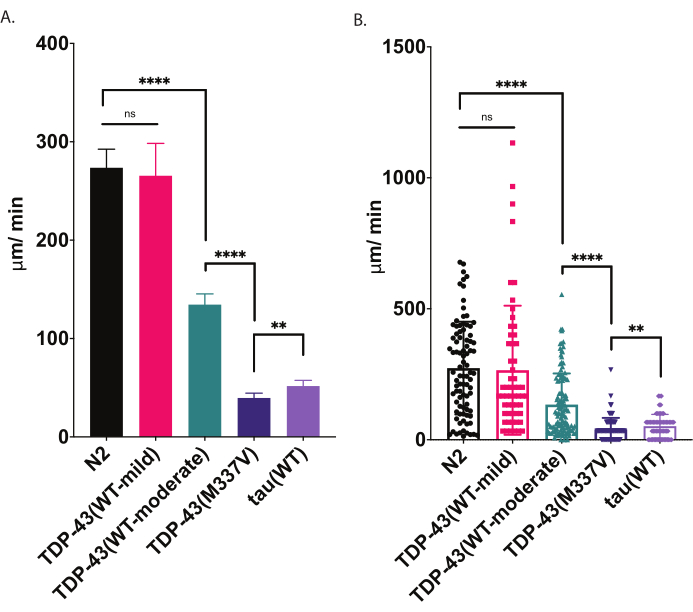
Figure 4: Radial locomotion assays detect differences in crawling speed. Unstimulated dispersion of developmentally staged L4 larvae was measured using the radial locomotion assay described above and graphed as µm/min traveled (A-B). The same data plotted in (A) and (B) demonstrate two different possible graphical presentations. In (A), data are displayed as a bar graph, making relative differences between strains clearer. In (B), the final displacement of each worm isplotted within the graph, allowing the variation within the population to be better visualized. To evaluate significance among strains tested, one-way analysis of variance (ANOVA) with Tukey's multiple comparison test was used. **p=0.0022, ****p<0.0001, ns=not significant. TDP-43(M337V) and tau(WT) are also significantly different from N2, p<0.0001. Error bars in (A) are standard error of the mean (SEM) and in (B) are standard deviation. Please click here to view a larger version of this figure.
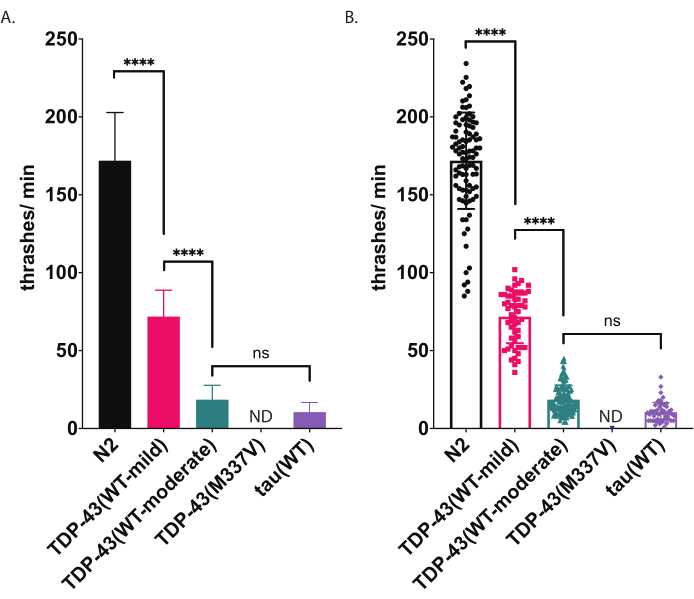
Figure 5: Swimming assays detect differences of thrashing in liquid. Rates of swimming (thrashing or undulation frequency in liquid) were measured using unbiased computer-assisted scoring and analysis described above and graphed as thrashes/min (A-B). Same data plotted in (A) and (B). In (A), data are graphed as a bar graph, which makes relative differences between strains easier to see. In (B), data from each individual worm scored are plotted within the graph, allowing the variation within the population to be better visualized. To evaluate significance among strains tested, a one-way analysis of variance (ANOVA) with Tukey's multiple comparison test was used. ****p<0.0001, ns=not significant. TDP-43(WT-moderate) and tau(WT) are also significantly different from N2, p<0.0001.Error bars in (A) are standard error of the mean (SEM) and in (B) are standard deviation. Please click here to view a larger version of this figure.
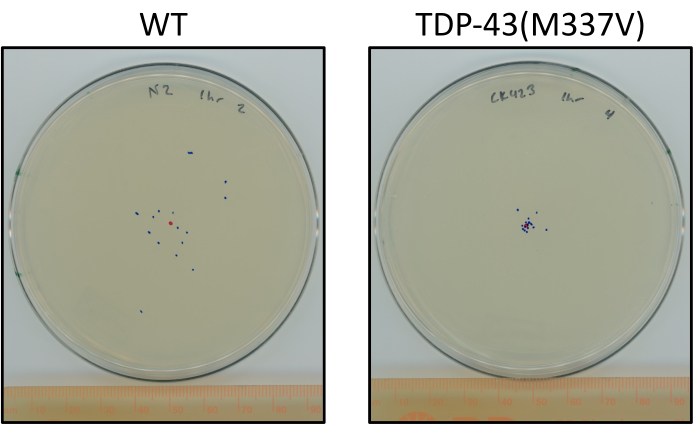
Figure 6: Mutant TDP-43 worms travel less than wild-type worms. Plates showing a representative difference in the final location of staged L4 N2 (wild-type) and TDP-43(M337V) (CK423 (bkIs423[Psnb-1::TDP-43; Pmyo-2::GFP]), a strain expressing mutant human TDP-43 with severely impaired motor function after 1 hour of unstimulated crawling at room temperature. Plates are marked with a red dot for the center point and blue dots for the final location of the animals. Please click here to view a larger version of this figure.
Supplementary Figure 1: Recording swimming behavior using the imaging and tracking software. (A) The tracking workflow and the location of the video capture icon. (B) The Video Capture window is shown in Live View, and the most important aspects are highlighted. The green "Live view" words will change to a red "Recording" when the record button is activated. Please click here to download this File.
Supplementary Figure 2: Preparing and tracking swimming behavior. This figure shows a series of screenshots to aid in setting up a video image sequence for tracking. The general protocol for preparing a video sequence is to open each workflow menu in this order: Import Image Sequence | Set Sequence Info | Adjust Image | Detect and Track | Save Project. The Analyze Data window can only be used after a project file is tracked. (A) shows an opened .avi (video) file after importing the sequence into the software from within the Track workflow. (B) The Set Sequence Info window provides a place for notes, set/change the scale, and to check the metadata for a video sequence. Instructions for changing the scale are found in this window and should be followed when the camera is lowered or raised (B). Image (C) shows the Adjust Image window settings. (D) Screenshot showing the detection tab of the Detect and Track window. Detecting worms is used to train the software and must be set for each video sequence. Additional detection parameters can be set here if desired. (E) Screenshot of Tracking tab of the Detect and Track window with the recommended settings for tracking swimming behavior. This is the last step in setting up a video sequence. Save the sequence as a project using the Save Project window. Repeat these steps for each video sequence. Settings can be saved as a configuration to reduce workload and ensure that all sequences are treated the same (not shown). To track project files for analysis, navigate to the Batch Workflow. (F) shows the icon used to navigate to the Batch workflow, and (G) shows the batch processing window, highlights the add and start buttons, and shows the expected appearance of a project file that has been tracked. Please click here to download this File.
Supplementary Figure 3: Swimming behavior analysis. After a project file has been tracked, it can be opened using [File | Open Project]. Worms will appear in green as shown in (A). Scrubbing through the video offers a quick check that the video processed as expect, green highlighting will disappear while scrubbing. The Data Analysis and Plotting window is opened from the Analyze Data workflow item. (B) shows the Data Analysis and Plotting window with Position view opened (default), all tracks are highlighted. Below the datapoints is a plot showing the recorded track for each highlighted worm. The track summary analysis is used to calculated turns per minute. (C-D) show the track summary report and export details. (E-F) show the track summary in a spreadsheet and how to calculate turns per minute, the output measured in this assay. Please click here to download this File.
Discussion
Radial locomotion:
The resolution of this assay is easily controlled by changing the time variable. Increasing the length of time makes it easier to observe differences between animals with severe phenotypes, thereby identifying subtle differences. However, because this assay measures displacement, if the assay time is extended too long, animals with normal motility, such as N2, will travel to the edges of the plate, and foraging behavior will lead to backtracking. This will artificially decrease the measurement of distance traveled. Time periods that are too long may result in the disappearance of differences between strains, particularly between animals with less severe motor phenotypes, as animals become evenly dispersed across the plate. Shortening the time variable will prevent more active worms from finding the edges of the plate. This method does not track the total distance traveled for each worm but compresses the distance traveled for each worm into a linear distance from the center of the plate. As such, it is inherently less robust than a method that records the total track length of individual worms. However, the radial locomotion assay requires very little researcher training, uses relatively affordable reagents that are commonly available in most worm labs, and is sensitive enough to produce significant and reproducible results. For labs that prefer automated video tracking, several methods have been previously established to track and analyze crawling movements12, or the software parameters used for the swimming assay in this paper could be altered to permit crawling detection and analysis.
This experiment is usually done in triplicate independent replicates, with a set of 30-40 worms per replicate. Each replicate is split onto two different 100 mm or 150 mm plates, with 15-20 worms per plate. Using more worms than recommended per plate can make it hard to score efficiently. A total number scored of 90+ is sufficiently powered to establish significance for mild, moderate, or severe motility impairment. Being consistent with timing between strains scored is essential for accuracy and reproducibility. 30 minutes is generally long enough to establish differences between moderate to severe phenotypes such as transgenic strains expressing human mutant TDP-43 compared to wild-type worms (Figure 4). If the time variable will be extended, it is advisable to also increase the size of the plate from 100 mm to 150 mm. Environmental factors such as temperature and humidity can affect this assay, which is typically conducted at ambient room temperature, so it is important to always use a wild-type (N2) control when comparing across replicates. In addition, this assay can measure the motility of some strains that do not exhibit normal swimming behavior in liquid (thrashing), making it a useful complement to the swimming assay.
Swimming assay:
The use of the imaging and acquisition system to automate tracking and analysis of worm swimming allows for rigorous and unbiased data. However, there are several factors during initial setup of the experiment that needs to be controlled between samples. These include the time to acclimate to liquid before beginning recording, ambient conditions (i.e., temperature, humidity), and consistent light and recording settings. On the recording stage, there are several features that help reduce variability between plates. These include an integrated track-mounted camera and bright-field stage that makes recording video consistent between plates, shielding around the stage that prevents reflections, glare, and air movements while recording, and a robust software package that reliably detects worms and allows for manual correction of tracks in video post-processing. In this protocol, videos of a 35 mm plate with worms are recorded for 1 minute and then processed using the software package. After processing, manual correction of tracks ensures that worm behaviors are recorded accurately without confounding tracking errors. Turn count and track duration data are used to determine turns per minute as a final readout. To ensure reproducibility, data is collected over a minimum of 3 independent replicate experiments, each with 40-50 animals scored, to achieve a combined final number of animals 120-150. This number is sufficient to discriminate small differences in swimming behavior from control worms. Some worms have motor deficits too severe to be captured by swimming assays. For example, if the animals placed in a liquid medium curl instead of performing the expected thrashing response, this assay will not record those movements accurately and another movement assay, such as radial locomotion, may better capture those motility defects. The provided protocol uses a commercially available imaging system (see Table of Materials for more details), but other worm tracking systems may provide a similar utility, with some being open source12. Previously published methods describe manual scoring of worm thrashing13. While the automated analysis produces a number of metrics for each individual worm, detection of body bends which is measured in thrashes per minute, provides consistent results between experiments and tracks well with conventional scoring of worm thrashes by eye.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank the reviewers for helpful comments and suggestions. We thank Aleen Saxton, Brandon Henderson, and Jade Stair for outstanding technical assistance. We thank Brian Kraemer and Rebecca Kow for assistance developing these assays. This material is the result of work supported with resources and the use of facilities at the VA Puget Sound Health Care System. This work was supported by a grant from the United States (U.S.) Department of Veterans Affairs (VA) Biomedical Laboratory Research and Development Service [Merit Review Grant #I01BX004044 to N.F.L.]
Materials
| C. elegans | Caenorhabditis Genetics Center (CGC) | – | Aquire your strains as desired, N2 is a useful control strain |
| Disposable pasteur pipets, borosilicate glass | VWR | 14673-010 | Glass pipet used to create worm pick – hold glass pipette in one hand and ~1" of platinum wire (held by pliers) in the other over a flame to join. |
| Disposable petri dishes, 35x10mm | VWR | 10799-192 | Assay plates for WormLab Imaging System |
| Disposable petri dishes, 60x15mm | VWR | 25384-090 | Stock plates for worms |
| Disposable petri dishes, 100x15mm | VWR | 25384-302 | Standard radial locomotion assay plate |
| Disposable petri dishes, 150x15mm | VWR | 25384-326 | Longer time frame radial locomotion assay plate |
| Dissecting microscope | Leica | M80 | Scope for maintaining worms and setting up radial locomotion assays |
| Fine-tipped markers | VWR | 52877-810 | Need at least 2 colors for radial locomotion assays. Fine tips required for accuracy. |
| Flatbed Scanner | Amazon | Epson Perfection V850 | Optional for radial locomotion assay. Protocol assumes a resolution of 300dpi, most scanners would work fine |
| ImageJ | NIH | – | Optional free software provided by the NIH – https://imagej.nih.gov/ij/ |
| M9 buffer | VWR | IC113037012 | Medium used for swimming assay. Can be made from scratch, see WormBook: Maintenance of C. elegans |
| NGM (Nematode Growth Medium) | VWR | 76347-412 | Medium used to cultivate C. elegans. Can be made from scratch, see WormBook: Maintenance of C. elegans |
| OP50 bacteria | Caenorhabditis Genetics Center (CGC) | OP50 | Primary food source for C. elegans |
| p1000 pipettor | VWR | 76207-552 | Pipettor, used in swimming assay |
| p1000 tips | VWR | 83007-384 | Tips for pipettor, used in swimming assay |
| Platinum wire, 0.2032mm diameter | VWR | BT136585-5M | Fine gauge platinum wire used to create worm pick – hold glass pipette in one hand and ~1" of platinum wire (held by pliers) in the other over a flame to join. |
| Ruler | VWR | 56510-001 | Need to score radial locomotion assays |
| WormLab Imaging System | MBF Bioscience | WormLab | The Imaging System includes WormLab hardware (bright field stage, camera, and housing) and WormLab software. https://www.mbfbioscience.com/wormlab-imaging-system |
Riferimenti
- Wijesekera, L. C., Leigh, P. N. Amyotrophic lateral sclerosis. Orphanet Journal of Rare Diseases. 4, 3 (2009).
- Arai, T., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochemical and Biophysical Research Communication. 351 (3), 602-611 (2006).
- Neumann, M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 314 (5796), 130-133 (2006).
- Mejzini, R., et al. ALS genetics, mechanisms, and therapeutics: Where are we now. Frontiers in Neuroscience. 13, 1310 (2019).
- Tan, R. H., Ke, Y. D., Ittner, L. M., Halliday, G. M. ALS/FTLD: Experimental models and reality. Acta Neuropathologica. 133 (2), 177-196 (2017).
- Kim, W., Underwood, R. S., Greenwald, I., Shaye, D. D. OrthoList 2: A new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetica. 210 (2), 445-461 (2018).
- White, J. G., Southgate, E., Thomson, J. N., Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transaction of the Royal Society of London B: Biological Sciences. 314 (1165), 1 (1986).
- Caldwell, K. A., Willicott, C. W., Caldwell, G. A. Modeling neurodegeneration in Caenorhabditis elegans. Disease Models and Mechanisms. 13 (10), (2020).
- Liachko, N. F., Guthrie, C. R., Kraemer, B. C. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. Journal of Neuroscience. 30 (48), 16208-16219 (2010).
- Robatzek, M., Thomas, J. H. Calcium/calmodulin-dependent protein kinase II regulates Caenorhabditis elegans locomotion in concert with a G(o)/G(q) signaling network. Genetica. 156 (3), 1069-1082 (2000).
- Porta-de-la-Riva, M., Fontrodona, L., Villanueva, A., Ceron, J. Basic Caenorhabditis elegans methods: synchronization and observation. Journal of Visualized Experiments. (64), e4019 (2012).
- Husson, S. J., Costa, W. S., Schmitt, C., Gottschalk, A. Keeping track of worm trackers. WormBook. , 1-17 (2013).
- Ringstad, N., Horvitz, B., Koelle, M., Hart, A. C. . WormBook. , 1551 (2006).

