Bacteriophage Effectiveness for Biocontrol of Foodborne Pathogens Evaluated via High-Throughput Settings
Summary
This protocol describes a robust method for using high-throughput settings to screen the antibacterial efficacy of bacteriophage cocktails.
Abstract
Bacterial pathogens continually challenge food safety systems worldwide. With increasing concerns about the emergence of heat- and sanitizer-resistant bacteria, novel antibacterial agents, are urgently needed. A bacteriophage-based biocontrol strategy is the therapeutic use of phages to control bacterial pathogens in agricultural settings. Phage biocontrol is increasingly accepted as a sustainable technology, effective at decontaminating foodborne pathogens. To ensure effective biocontrol outcomes, systematic screening of phage combinations against targeted bacteria under required environmental conditions is crucial. Antibacterial efficacy of phage cocktails may be affected by phage genera and combination, targeted bacterial strains, the multiplicity of infection, temperature, and time. To formulate a phage cocktail with superior efficacy, the proposed method was to systematically evaluate the effectiveness of individual phages and phage cocktails in killing foodborne bacterial pathogens under targeted conditions. Bacterial killing efficacy was monitored by measuring optical density at desired temperatures and durations. Superior phage efficacy was determined by complete inhibition of bacterial growth. The proposed method is a robust, evidence-based approach to facilitate formulating phage cocktails with superior antibacterial efficacy.
Introduction
Bacteriophages (phages) are viruses that naturally invade bacterial cells, disrupting bacterial metabolism and causing lysis of the bacterium. In contrast to conventional antimicrobials (e.g., antibiotics), phage host spectrums are relatively narrow, only capable of infecting a targeted set of bacterial species or strains and thus should minimize collateral effects on microbiota that benefit animal and human health. With the rise of antimicrobial resistance (AMR), phages and their derivatives lead to alternative antimicrobials to control bacterial infectious diseases, including AMR bacterial infections in humans and animals1,2. Phages have confirmed therapeutic potential against >20 bacterial pathogens that cause superficial infections and infections of the upper respiratory system and gastrointestinal tract of humans3.
In agricultural settings, a phage-based biocontrol strategy is the therapeutic use of phages to control bacterial pathogens. Phage biocontrol is well accepted as a green technology, effective at decontaminating foodborne pathogens (e.g., Shiga-toxin producing Escherichia coli (STEC), Salmonella, and Listeria) in various foods4,5. In addition, phages can be used as sanitizers to disinfect food processing surfaces and animal hides, which can be integrated into conventional antimicrobial systems (e.g., chemicals, steam, and hot water pasteurization) to enhance desired outcomes and reduce environmental impacts. The use of phages to reduce zoonotic bacteria in animals is also promising1. However, there is a need to address the technical challenges to improve outcomes from the phage biocontrol approach to be popularly applied in diverse food production systems. The main challenge is the impaired effectiveness of phages due to the development of bacteria-resistant mutants5 and changes in bacterial physiology due to exposure to environmental stressors6.
For minimizing the risk of phage resistance, phage cocktails (i.e., a combination of multiple phages) are proposed and have improved biocontrol potency in agriculture and aquaculture settings7. However, from several studies, it has been proved that phage cocktails did not always offer better effectiveness than the administration of a single phage. For instance, a cocktail of 3 T4-like phages had a narrower host range against E. coli strains8. Furthermore, AKFV33, a member of Tequintavirus, had greater efficacy than a cocktail of four phages in removing E. coli O157 from beef, despite the incubation temperatures applied4. Recently, it has been reported that the effectiveness of individual phages does not predict the efficacy of phage cocktails for control of O1579, as interactions among multiple phages can alter efficacy. Most importantly, numerous factors, such as phage genera and combinations, targeted strains and MOIs, and incubation temperatures and times, may impact interactions between phages. Therefore, carefully screening combinations of phages against specific bacteria to assess phage synergy or facilitation, or at least to ensure minimal phage antagonism under specific environmental conditions, is vitally important for optimal outcomes. Here, a method is described to systematically assess the efficacy of various phage combinations against foodborne pathogens under a range of environmental conditions. The benefit of this approach is to enable screening of all possible biotic and abiotic factors predicted to affect the antibacterial efficacy of phages in natural settings. In the protocol, STEC O157 and their infecting phages are employed as an example.
Protocol
1. Preparation of buffer and reagents
- Make 500 mL of tryptic soy broth (TSB) (15 g of TSB powder and 500 mL of ultrapure water) and autoclave.
NOTE: This can be stored at room temperature for up to 3 months or at 4 °C for up to 6 months. - Make 500 mL of tryptic soy agar (TSA) (20 g of TSA powder and 500 mL of ultrapure water) and autoclave.
NOTE: This can be stored at 4 °C for up to 3 months. - Make 500 mL of Phosphate Buffered Saline (PBS; 4 g of NaCl, 0.1 g of KCl, 0.77 g of Na2HPO4, and 0.12 g of KH2PO4, 500 mL of ultrapure water), measure and adjust the pH to 7.2 ± 0.2 at 25 °C and autoclave.
NOTE: This can be store at 4 °C for up to 6 months. - Make 200 mL of 1 M MgSO4 (49.294 g and 200 mL of ultrapure water) and sterilize via 0.22 µm polyethersulfone filter.
- Make 500 mL of TSB with 10 mM MgSO4 (mTSB; 15 g of TSB powder, 500 mL of ultrapure water, and 5 mL of 1 M MgSO4) and autoclave; let the autoclaved media cool to room temperature. Apply the aseptic technique. Using a serological pipette, carefully transfer 5.0 mL of 1 M MgSO4; swirl gently to mix.
2. Preparation of bacterial culture
- To prepare Bacterial Research Laboratory Culture (BRLC), remove glycerol stock vial(s) from the freezer inventory and transfer to the laboratory.
- Use a disposable inoculation loop or equivalent, dip into the vial, remove a scraping of inoculum (frozen slush), and streak onto a TSA plate or equivalent within a level II biological safety cabinet. Incubate the plate at 37 ± 2 °C for 15−18 h.
- Bag the prepared BRLC plates and store at 4 °C.
NOTE: General expiry period for BRLC: 14 days at 4 °C after generation. - Inoculate a single colony of each E. coli O157 strain from the BRLC plates to be tested in 10 mL of TSB and statically incubate at 37 °C for 18 h to reach 9 log10 CFU/mL.
- Prepare a serial dilution of the overnight cultures (the following morning), each E. coli O157 strain using mTSB containing 10 mM of MgSO4 to achieve 4-5 log10 CFU/ml (or other desired inoculum level).
- Mix an equal volume of an overnight culture of each strain to achieve 4-5 log10 CFU/mL in total (or as desired) to prepare the bacterial mixture.
- Immediately place the diluted bacterial culture at 4 °C for pending use.
- Dilute the inoculum culture 10 or 100-fold and plate 0.1 mL aliquots of these dilutions on the TSA plates to obtain the isolated colonies.
3. Preparation of phage working solutions
- Propagate high-titer working stocks for each phage to be screened (≥108 PFU/mL) by following the standard methods4.
NOTE: The general expiry period for the phage stocks is 3 months in a plastic bottle at 4 °C after generation. - To achieve the desired titer of, e.g., ~108 PFU/mL for lysis kinetics, dilute individual phage preparations with mTSB containing 10 mM of MgSO4. Prepare phage cocktails by mixing equal volumes of each working stock with the same titer in all the possible combinations.
4. Preparation of in-vitro lysis kinetics for individual phage and phage cocktails
- Prepare serial 10-fold dilutions of each phages in Columns 1-8 in sterile 96-well microplates to set up the microplate assay.
NOTE: Four phages against one bacterial strain can be tested in duplicate, in adjacent columns, on each plate. The remaining four columns are for controls, which are without phage, as well as mTSB blank (Figure 1). - Place 180 µL of mTSB to wells from Columns 1 to 12 of the 96-well microplate.
- Add 20 µL of diluted individual phages or phage cocktails (~108 PFU/mL) to wells 1-8 of the top row of the microplate (Row A).
- For the phage-free and Blank control, add 20 µL of mTSB to the top well of Columns 9, 10, and 11.
- With a 12-channel pipette, dilute down the plate. Transfer 20 µL from row to row. Mix the well contents by gentle, repeated aspiration, ejection (at least five times), and changing tips between dilutions. Remove 20 µL from the last row (Row H).
NOTE: Using tips with filters is recommended to prevent cross-contamination. - Set up a reservoir for each strain, use 1 mL pipette to transfer 2-3 mL of the diluted culture into the reservoir. For columns 1-10, use a multichannel pipette to add 20 µL of the diluted bacterial culture to each well. Change tips between each addition.
- Cover and incubate the microplate at desired conditions (e.g., 37 °C for 10 h or 22 °C for 22 h).
- At 2 h or other desired intervals, remove the microplates from the incubator.
5. Determination of optical density
- Examine for optical density at 600 nm (OD600nm) using a Microplate Reader.
NOTE: It is advised to set up and save the assay protocol in the program before test plate preparation. - Turn on the microplate reader and open program.
- Select the Simple mode in the Startup Options window and select Create a New Protocol di Task Manager (Supplementary Figure 1a,b).
- From the Select Plate Type window, choose 96 Well Plate from the drop-down list (Supplementary Figure 2).
- Select Absorbance for detection method, the Endpoint/Kinetic option for read type, and Monochromators for optics type. Click on OK (Supplementary Figure 3).
- For Read Step, enter 600 nm for wavelengths and select Normal for read speed. Click on OK (Supplementary Figure 4).
- To set the temperature for the assay, click on the Incubate checkbox and select Incubator On. Enter the desired temperature in the Temperature box. Click on OK. To prevent condensation on the plate lid during incubation, set a temperature gradient by entering a value (1-3) in the Gradient box. Click on OK (Supplementary Figure 5a).
NOTE: Click on the Preheat Before Continuing With the Next Step checkbox for the 37 °C incubation temperature. This step is not required for the test condition at 22 °C (RT). - Next, click on the Kinetic checkbox to open the Kinetic Setup. Set the run time for 10 h for 37 °C incubation or 22 h for RT. Enter 2 h for reading interval. Click on OK (Supplementary Figure 5b).
- Click on the Shake checkbox in the Procedura window to set up the shake condition, if needed, for each kinetic read (Supplementary Figure 6a).
- Select Linear for Shake Modes and change Duration to 30 s. Set Linear Frequency value to 731 cpm (2 mm), and then click on OK (Supplementary Figure 6b).
- In the Protocol Summary Dialogue window, click on Plate Layout and select Blanks, Assay Controls, and Samples. Click on Next (Supplementary Figure 7).
NOTE: Enter 1 di Number of Different Control Types for negative control. - After defining the settings for each well type, click on Finish.
- Select a Well ID in the left-hand side interface, and then assign it to the matrix shown in the plate layout widow. Click on OK (Supplementary Figure 8).
- Click on the Read Plate button and save the protocol as a .prt file and click on Save (Supplementary Figure 9).
- Insert the plate and click on OK.
- Once the experiment is completed, save the experiment as a .xpt file and click on Save (Supplementary Figure 10).
- Export data to Excel by clicking on the Yes button in the message window box for further analysis (Supplementary Figure 11).
- Click on the Save Yes/No selection and the Don't Ask Again checkbox to save preference.
6. Data analyses
- Repeat at least two independent experiments as described above. Compile results from all the independent trials. Calculate the average and standard deviation of the OD600 from each phage-treated and -free culture.
- Perform square root of the OD values at 600 nm and analyze them using an appropriate statistical model for each bacterial strain and temperature.
NOTE: For SAS software, the MIXED model and the least-squares to differentiate means (P < 0.05) are selected. For each strain, assign panels A−G to each phage treatment of which overall anti-O157 efficacy across time and MOIs differed (P < 0.05). - Define superior phage efficacy based on OD600nm value ≤ 0.01 corresponding to no detectable bacterial growth (limit of detection:300 CFU/mL).
- Analyze effects of time, incubation temperature, E. coli O157 strains, MOIs, and phage types on phage efficacy using an appropriate statistical model.
NOTE: For SAS software, use GLIMMIX with random measures. - Calculate Odds Ratios to compare superior efficacy for different environmental and biological factors of interest.
- Analyze effects of time, incubation temperature, E. coli O157 strains, MOIs, and phage types on phage efficacy using an appropriate statistical model.
Representative Results
Following the protocol, a comparison of the anti-O157 phage efficacy with various phage combinations, temperatures, times, and MOIs was performed. Impacts of incubation temperature and times, MOIs, phages, and bacterial strains used on enhancing anti-E. coli O157 efficacy is presented in Table 1 (odds ratio table)9. The percentage (%) of optical turbidity measurement ≤ 0.01 yielded was analyzed by each phage preparation under each experimental condition. Based on this analysis, anti-O157 phage efficacy was maximized after 14, 16, or 18 h of incubation at 22 °C (P < 0.001) and MOI = 1000 (P < 0.001), with 75% and 89% of the growth of phage-treated culture being completely inhibited, respectively. In general, among 11 phage preparations, a cocktail of T1, T4, and rV5 was most effective (P < 0.05) against O157. In addition, across incubation temperatures, times, MOIs, and phages tested, sensitivity to phages varied, with O157 strains tested with strain CO281-31N being most sensitive (P < 0.001) and 3081 (P < 0.001) being least sensitive to phages.
For understanding the phage-killing kinetics of each bacterial strain tested, OD600 value was plotted against each sampling time, MOIs, and phage treatment at each incubation temperature. Representative results from the efficacy of phages against E. coli O157 3081 at 37 °C are shown in Figure 2. It is ensured that the growth curve of phage-free control culture is normal before assessing inhibition of growth curve of phage-treated culture. According to Figure 2, the inclusion of T4 completely inhibited bacterial growth at 37 °C, at each sampling time at an MOI as low as 1. To overview how phage inhibited bacterial growth over time at various temperatures, irrespective of MOIs, the mean OD600nm value averaged from all the MOIs (MOI > 0) was plotted against each sampling time and phage treatment (Figure 3). Compared to 37 °C, particular phage killing efficacy, e.g., phages T4 and T1 + T4, was enhanced over 22 °C. Another approach to summarize and compare overall anti-O157 effectiveness among phages is shown in Table 2 and Table 39. OD600 value was averaged from each sampling time and MOI and ranked phage efficacy from highest (A) to lowest (F: 37 °C and D: 22 °C). This table facilitates the identification of the best phage treatment. For example, for strain 3081, phages T4 and T5 + T4 (Panel A) were the most effective treatment at 37 °C, whereas in addition to phage T4, phages T1 + T4, T1 + T4 + rV5, and 4 phage cocktails (Panel A) were the most effective treatment at 22 °C.
This protocol also enabled us to compare phage efficacy against various bacterial strains targeted and customize phage preparation. For example, when selecting phages, at 37 °C, excluding strain 3081, phage T1 + T4 + rV5 had the most significant effectiveness against all other strains, regardless of their phage types. At 22 °C, the 4-phage cocktail was the most effective against all strains tested. The best MOIs resulted in complete lysis (OD600 ≤ 0.01), which warranted potential superior efficacy under experimental conditions (Table 2 and Table 3).
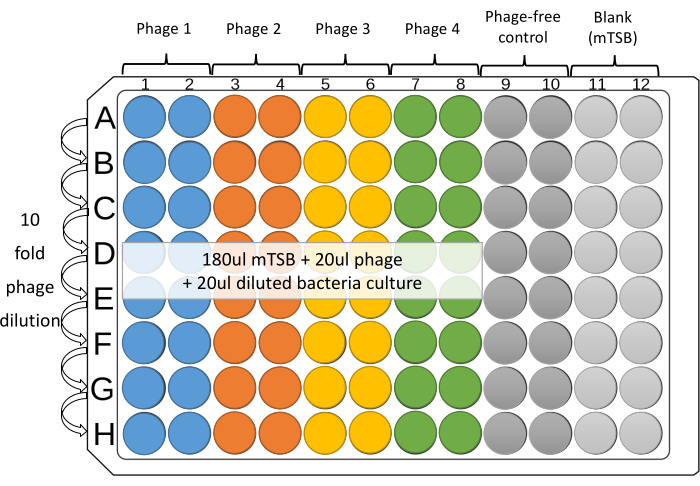
Figure 1: Suggested layout of 96-well microplate assay. Serial 10-fold dilutions of each phage are prepared in Columns 1-8 of sterile 96-well microplates. Four phages against one bacterial strain can be tested in duplicate, in adjacent columns, on each plate. The remaining columns are for controls. Please click here to view a larger version of this figure.
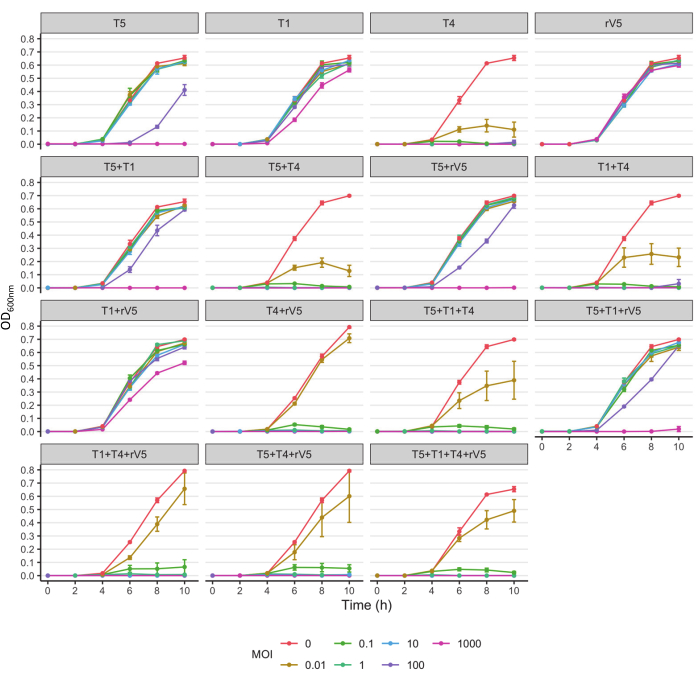
Figure 2: Growth curves of E. coli O157 strain 3081 at 37 °C treated and not treated with phages at each MOIs. The data were compiled from two independent trials. Bars present standard deviation. Please click here to view a larger version of this figure.
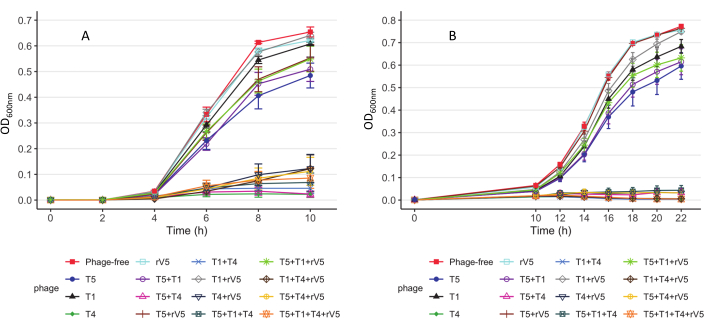
Figure 3: Growth curves of E. coli O157 strain 3081 strains. (A) Growth curves at 37 °C. (B) Growth curves at 22 °C. Each curve represents individual and mixed cultures, treated and not treated with phages across MOIs. The data were compiled from two independent trials. Bars present the standard error of the mean (adapted from Reference9 with permission). Please click here to view a larger version of this figure.
Table 1: Odds ratios comparing the likelihood of superior phage efficacy against E. coli O157 at incubation temperatures, incubation times, MOIs, phages, and strains of E. coli O157 (adapted from Reference9 with permission). Please click here to download this Table.
Table 2: Overall phage efficacy against E. coli O157 at 37 °C (adapted from Reference9 with permission). Please click here to download this Table.
Table 3: Overall phage efficacy against E. coli O157 at 22 °C (adapted from Reference9 with permission). Please click here to download this Table.
Supplementary Figures 1–11: Snapshots of the microplate reader operation and procedures. Please click here to download this File.
Discussion
This protocol described a robust approach for systemically evaluating phage efficacy against foodborne pathogens, including STEC9 and Salmonella10. One critical step is when diluting overnight culture of bacteria, using pre-chilled medium and manipulating the dilution with an ice bucket are recommend to minimize potential bacterial growth. In addition, phage dilution was prepared before diluting bacterial culture. The enumeration step 2.8 provided actual numbers of bacterial inoculum for calculation of the final MOI applied. For phage preparation, crude phage lysates prepared by filter phage infected in 4-6 h of bacterial culture is generally used. The critical step associated with phage infectivity is always to use phage work stocks prepared within 3 months. Extremely accurate pipetting (particularly when using a multichannel pipette) and uniformity of approach are also essential to obtain comparable and interpretable results. Modified TSB supplemented with 10 mM of Mg2+ was used to dilute phages, bacterial culture, and base medium to optimize adsorption and infection of phages.
As bacteria proliferate during the log phase, even below incubator temperature, it is recommended to use diluted overnight culture instead of log-phase culture, to minimize potential bacterial growth.
The proposed protocol has limitations. First, because microplate can only hold 200 µL, prolonged incubation may cause substantial evaporation and is not recommended. In this case, the assay may not be suitable for slow-growing bacteria. Second, the proposed protocol was not able to monitor the amplification of phages. Third, this protocol could not monitor the development of phage resistance over time, a critical factor that determines the outcome of phage treatment11,12. Follow-up experiments are required to assess the further performance of the most influential cocktail in the screening in preventing the emergence of anti-phage mutants in an extensive broth culture system and other biological matrices.
In contrast to conventional antimicrobials, the biological nature of phages affects the complexity of biocontrol and therapeutic use in practical settings. Conventionally, rational selection of phage cocktails is primarily based on lytic activity and the host range of phages. Phage candidates with the strongest lytic activity and broadest host range are often recommended13,14. However, based on the current study, phages such as rV5 and T1, although alone not as virulent as T4 and T5, greatly facilitated overall biocontrol outcome when combined with T4 and/or T5. Consequently, to achieve superior efficacy of phage cocktails, systemic screening of antibacterial activity of potential phage combinations against targeted host strains under desired environmental conditions is recommended. In addition, the determination of receptors for phage candidates and the inclusion of phages with various receptors may prevent competition for host attachment, thwart rapid development of anti-phage mutants, and improve biocontrol outcomes13.
This method enabled accurate quantification of phage lysis kinetics in a high-throughput format. Furthermore, it allowed systematic evaluation of various biological and environmental factors on the antibacterial efficacy of an assortment of phages, thereby facilitating the formulation of phage cocktails with optimized outcomes. The method's future applications and development are assumed to involve in situ monitoring the efficacy of each phages within phage cocktails by fluorescence labeling of phages. In addition to the proposed protocol, understanding genetic determinants that promote synergistic and facilitated effects between phages when co-infecting one host would facilitate the formulation of appropriate phage cocktails with superior efficacy.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant, RGPIN-2019-04384), the Canada Foundation for Innovation (Project # 38710), and Major Innovation Fund, Alberta. We thank Dr. John Kastelic for editing the manuscript.
Materials
| Essential supplies, reagents, and equipment | |||
| Inoculating loops | VWR | 12000-806 | |
| Magnesium sulfate heptahydrate | Sigma | 1374361 | MgSO4.7H2O |
| Petri Dishes with Clear Lid | Fisher | FB0875713 | Diameter: 100 mm, sterile |
| Phosphate-buffered saline (PBS) | Fisher | 10010023 | |
| Pipet-Lite LTS Pipette L-1000XLS+ | METTLER TOLEDO | 17014382 | |
| Pipet-Lite LTS Pipette L-300XLS+ | METTLER TOLEDO | 17014405 | |
| Pipet-Lite Multi Pipette L12-20XLS+ | METTLER TOLEDO | 17013808 | |
| Pipet-Lite Pipette, Unv. SL-20XLS+ | METTLER TOLEDO | 17014412 | |
| Pipette Tips RT LTS 1000µL FL 768A/8-low retention | METTLER TOLEDO | 30389213 | |
| Pipette Tips SR LTS 20µL F 960A/5 | METTLER TOLEDO | 17005860 | |
| Pipette Tips SR LTS 300µL 768A/4 | METTLER TOLEDO | 17005867 | no filter |
| Reservoir | METTLER TOLEDO | 89094-662 | |
| Sterile, clear, 96-well flat-bottom polystyrene microplates with lids | Fisher | 168055 | |
| Tryptic soy agar (TSA) | Sigma | 105458-0500 | |
| Tryptic soy broth (TSB) | Sigma | 105459-0500 | |
| T-Shaped Cell Spreaders | VWR | 76299-566 | |
| Instruments | |||
| Analog Vortex Mixer | Fisher | 02-215-414 | |
| Compact Microbiological Incubators | Fisher | 50125590H | |
| Magnetic Stirrer Hotplates | FIsher | 13-889-335 | |
| Polygon Stir Bars | FIsher | 14-512-125 | length: 20 mm |
| Synergy Neo2 Hybrid Multi-Mode Reader | Fisher | BTNEO2M | |
| Software | |||
| SAS | SAS Institute | 9.4 |
Riferimenti
- Carvalho, C., Costa, A. R., Silva, F., Oliveira, A. Bacteriophages and their derivatives for the treatment and control of food-producing animal infections. Critical Reviews in Microbiology. 43 (5), 583-601 (2017).
- Czaplewski, L., et al. Alternatives to antibiotics-a pipeline portfolio review. The Lancet. Infectious Diseases. 16 (2), 239-251 (2016).
- Cisek, A. A., Dabrowska, I., Gregorczyk, K. P., Wyzewski, Z. Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages. Current Microbiology. 74 (2), 277-283 (2017).
- Liu, H., et al. Control of Escherichia coli O157 on beef at 37, 22 and 4°C by T5-, T1-, T4-and O1-like bacteriophages. Food Microbiology. 51, 69-73 (2015).
- Moye, Z. D., Woolston, J., Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses. 10 (4), (2018).
- Ly-Chatain, M. H. The factors affecting effectiveness of treatment in phages therapy. Frontiers in Microbiololgy. 5, 51 (2014).
- Dy, R. L., Rigano, L. A., Fineran, P. C. Phage-based biocontrol strategies and their application in agriculture and aquaculture. Biochemical Society Transactions. 46 (6), 1605-1613 (2018).
- Bourdin, G., et al. Coverage of diarrhoea-associated Escherichia coli isolates from different origins with two types of phage cocktails. Microbial Biotechnology. 7 (2), 165-176 (2014).
- Niu, Y. D., Liu, H., Johnson, R. P., McAllister, T. A., Stanford, K. Efficacy of individual bacteriophages does not predict efficacy of bacteriophage cocktails for control of Escherichia coli O157. Frontiers in Microbiololgy. 12, 616712 (2021).
- Niu, Y. D., Liu, H., Johnson, R. P., McAllister, T. A., Stanford, K. Effect of a bacteriophage T5virus on growth of Shiga toxigenic Escherichia coli and Salmonella strains in individual and mixed cultures. Virology Journal. 17, 3 (2020).
- Brüssow, H. What is needed for phage therapy to become a reality in Western medicine. Virology. 434 (2), 138-142 (2012).
- Chan, B. K., Abedon, S. T., Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiology. 8 (6), 769-783 (2013).
- Nikolich, M. P., Filippov, A. A. Bacteriophage therapy: developments and directions. Antibiotics. 9 (3), 135 (2020).
- Niu, Y. D., et al. Host range and lytic capability of four bacteriophages against bovine and clinical human isolates of Shiga toxin-producing Escherichia coli O157:H7. Journal of Applied Microbiology. 107 (2), 646-656 (2009).

