Improved Enzyme Protection Assay to Study Staphylococcus aureus Internalization and Intracellular Efficacy of Antimicrobial Compounds
Summary
This protocol aims to describe how to study the extent of Staphylococcus aureus internalization and its ability to survive inside the human host cell, as well as the intracellular efficacy of antimicrobial compounds.
Abstract
Staphylococcus aureus expresses virulence factors to trigger its internalization into eukaryote cells and to survive inside different subcellular compartments. This paper describes an enzyme protection assay to study the extent of S. aureus internalization and its intracellular survival in adherent non-professional phagocytic cells (NPPCs) as well as the intracellular efficacy of antimicrobial compounds. NPPCs are grown in a multi-well plate until they reach 100% confluence. S. aureus cultures are grown overnight in cell culture medium. The bacterial suspension is diluted according to the number of cells per well to inoculate the cells at a controlled multiplicity of infection. Inoculated cells are incubated for 2 h to allow the bacteria to be internalized by the NPPCs, following which lysostaphin is added to the culture medium to selectively kill extracellular bacteria. Lysostaphin is present in the culture medium for the rest of the experiment.
At this point, the infected cells could be incubated with antimicrobial compounds to assess their intracellular activities against S. aureus. Next, the cells are washed three times to remove the drugs, and intracellular S. aureus load is then quantified by culturing on agar plates. Alternatively, for studying staphylococcal virulence factors involved in intracellular survival and cell toxicity, lysostaphin could be inactivated with proteinase K to eliminate the need for washing steps. This tip improves the reliability of the intracellular bacterial load quantification, especially if cells tend to detach from the culture plate when they become heavily infected because of the multiplication of intracellular S. aureus. These protocols can be used with virtually all types of adherent NPPCs and with 3D cell culture models such as organoids.
Introduction
Staphylococcus aureus is both a life-threatening pathogen and a commensal bacterium of the skin and the mucosa that colonizes two billion individuals around the world1. In humans, nasal carriers of S. aureus have an increased risk of infection with their own strain of carriage; however, the multifactorial determinants of S. aureus mucosal carriage are still unclear1,2. In addition to acute infections, patients can also develop chronic S. aureus infections that are often challenging to cure3. A better understanding of host-pathogen interactions during colonization and infection is crucial for developing novel therapeutic strategies and improving patient management.
In vitro, S. aureus can trigger its internalization into host cells expressing the α5β1 integrin4. The tripartite interaction between the staphylococcal fibronectin-binding proteins anchored to the cell wall of S. aureus, the fibronectin, and the β1 integrin expressed at the host cell surface is well known as the main pathway of S. aureus internalization in NPPCs such as keratinocytes, osteoblasts, fibroblasts, and epithelial and endothelial cells4. Recent studies show that S. aureus can be found inside human cells during nasal colonization5,6 and infection7. However, the role of the intracellular reservoir in the pathogenesis of S. aureus infection remains unclear. The host cells could act as a shelter for S. aureus, which is protected from both the immune system8 and most antimicrobial compounds6,9.
The lysostaphin protection assay, described by Proctor10 earlier in the 1980s, enables the study of bacterial and host factors involved in the internalization of S. aureus isolates. Lysostaphin is a bacteriocin produced by Staphylococcus simulans, which exhibits potent activity against almost all S. aureus isolates, including antibiotic-resistant strains11. Lysostaphin has been used to destroy only extracellular S. aureus to enable the counting of only viable intracellular bacteria12. This technique has been widely used and has contributed to the discovery of several virulence factors of S. aureus. Gentamycin, alone and combined with lysostaphin, is also widely used to study intracellular bacteria.
However, a recent study showed that gentamycin enters eukaryotic cells and reaches internalized bacteria in a time- and concentration-dependent manner13. This study also demonstrated that lysostaphin does not enter eukaryotic cells, confirming that a lysostaphin-based enzyme protection assay (EPA) is the most accurate assay for quantifying intracellular S. aureus load by culture13. Regardless of which compound is used to destroy extracellular bacteria (e.g., lysostaphin or gentamycin), it should be removed by washing the cells before plating intracellular S. aureus on agar plates. Successive washes may result in the detachment of cells, especially poorly adherent cells (e.g., heavily infected cells), which would lead to an underestimation of the intracellular S. aureus load. This paper describes in detail how EPA can be used to quantify the intracellular S. aureus load and to measure the intracellular efficacy of antimicrobials compounds using an in vitro model. Of note, a simple method has been proposed to improve the reliability of intracellular load quantification by avoiding intensive washes.
Protocol
1. Culture of human epithelial cells
- Prepare complete culture medium with Dulbecco's modified Eagle medium (DMEM) high glucose with phenol red, supplemented with 10% fetal bovine serum (FBS) without antibiotics.
- Grow A549 epithelial cells in complete culture medium at 36 ± 1 °C in 5% CO2. Ensure the use of an appropriately sized culture vessel to have enough cells for subsequent steps (see step 1.10).
NOTE: One 75 cm2 (T-75) flask is sufficient to seed two 24-well plates and subculture the cells. - Two days before infection, prepare a single 24-well plate.
- Remove and discard the spent culture medium from the T-75 flask and wash the cells once with 10 mL of Dulbecco′s phosphate-buffered saline (DPBS).
- Add 5 mL of trypsin-EDTA and incubate the cells for 5 min at 36 ± 1 °C in 5% CO2.
- Add 5 mL of complete culture medium and transfer the cells into a tube.
- Centrifuge the cells for 5 min at 300 × g.
- Discard the supernatant and resuspend the cells in 10 mL of fresh complete culture medium.
- Count the cells with an automatic cell counter (or a counting chamber).
- Dilute the cells in complete culture medium to prepare 30 mL of cell suspension at a concentration of 2.0 × 105 cells/mL.
- Add 1 mL of the cell suspension to each well of a 24-well plate, which corresponds to a cell density of approximately 1.0 × 105 cell/cm² for a well area of 2 cm².
- Incubate the cells for 48 h at 36 ± 1 °C in 5% CO2 until they reach 100% confluence.
NOTE: In addition to the conditions to be tested, three wells should be reserved for cell counting on the day of infection (see step 3.1.4). According to the number of conditions to be tested, Up to two 24-well plates can be prepared simultaneously. Volumes indicated in the protocol should be increased accordingly.
2. Culture of S. aureus strains
- Two days before infection, prepare complete infection medium with DMEM high glucose without phenol red, supplemented with 10% FBS without antibiotics.
- Thaw S. aureus strains to be tested on agar plates.
- Incubate the agar plates for 18-24 h at 36 ± 1 °C.
- The day before inoculation, inoculate one colony of the S. aureus strain to be tested in 10 mL of complete infection medium.
- Incubate the bacteria for 18-24 h at 36 ± 1 °C with shaking at 160 rpm. Use 50 mL tubes held at 45° to avoid the bacteria settling.
NOTE: Before starting with a new strain, it is recommended to verify its lysostaphin susceptibility in the same conditions of culture that will be used for further experiments (media, bacterial loads, and lysostaphin concentration and incubation time). It is also important to determine the bacterial load corresponding to an OD600nm of 0.5 because it could vary slightly from one strain to another. Culture conditions of bacterial strains could be adapted according to the experimental aim.
3. Infection assay with S. aureus
- Determination of cell density and viability
- Remove and discard the spent culture medium from the three wells dedicated for counting A549 cells.
- Add 1 mL of complete infection medium containing 5 µg/mL of Hoechst 33342 and 1 µg/mL of propidium iodide.
NOTE: Hoechst 33342 is a known mutagen and should be handled with care. Propidium iodide, a potential mutagen, must be handled with care and disposed of safely according to applicable regulations. - Incubate the cells for 30 min at 36 ± 1 °C in 5% CO2.
- Count the cell number and calculate the cell viability using a wield-field fluorescence microscope.
NOTE: If a fluorescence microscope is not available, the cell density and viability can be calculated with trypan blue staining by using a cell counting chamber.
- Preparation of the bacterial suspension
- Dispense 25 mL of complete infection medium in a tube and pre-warm at 36 ± 1 °C.
- Adjust the S. aureus suspension to anOD600nm of 0.5 in complete infection medium using a cell density meter.
- Prepare 20 mL of bacterial suspension for cell inoculation by diluting the 0.5 OD600nm in complete infection medium to achieve a multiplicity of infection (MOI) of 1 according to the number of cells per well.
NOTE: The MOI corresponds to the number of bacteria added per cell in each well. For example, to achieve an MOI of 1 with 1.0 × 106 cells per well, prepare a bacterial suspension at 2.0 × 106 CFU/mL so that 106 CFU can be added in a volume of 500 µL (see step 3.3.3). The MOI can be adjusted according to the cell types and bacterial strains to be tested. - Use an automatic spiral plater to determine the S. aureus load of the diluted bacterial suspension to be used for the cell inoculation step.
- Incubate the agar plates for 18-24 h at 36 ± 1 °C.
- The next day, count the number of colonies with a colony counter to calculate the accurate MOI for each strain tested.
NOTE: If no automatic spiral plater is available, the bacterial load could be determined by serial dilution on an agar plate. See the bacteriological analytical manual for details14.
- Cell inoculation
- Observe every well of the 24-well plate by low magnification microscopy to ensure that the cells are healthy and growing as expected.
- Remove and discard the spent cell culture medium from the 24-well plate.
- Add 500 µL of the bacterial suspension for inoculation to each well with 100% confluent cells.
- Incubate the cells for 2 h at 36 ± 1 °C and 5% CO2.
NOTE: it is recommended to use three wells of the plate for each condition to be tested (triplicate) and to perform at least three independent experiments. The delay of incubation can be adapted according to the experimental aim.
- Quantification of intracellular bacteria with improved enzyme protection assay (iEPA)
- Prepare 7 mL of 4x lysis buffer with 3.5 mL of 2% Triton X-100 in sterile water and 3.5 mL of trypsin-EDTA.
- Prepare a lysostaphin stock solution at 10 mg/mL in acetate buffer and aliquot 25 µL into cryovials. Store at -80 °C for up to 6 months.
- Prepare 250 µL of a fresh lysostaphin working solution at 1 mg/mL by mixing 25 µL of the lysostaphin stock solution (10 mg/mL) and 225 µL of 0.1 M Tris-HCl. Store at 4 °C for up to 48 h.
- Prepare 6.25 mL of complete infection medium supplemented with lysostaphin by adding 6 mL of complete infection medium to 250 µL of the lysostaphin working solution.
- Add 250 µL of complete infection medium supplemented with lysostaphin into each well and gently agitate the plate by swiveling the plate by hand.
- Incubate the cells for 1 h at 36 ± 1 °C in 5% CO2 to let the lysostaphin kill the extracellular bacteria.
- At the end of the incubation time, add 10 µL of proteinase K at 20 mg/mL into each well to inactivate the lysostaphin.
- Incubate the cells for 2 min at room temperature.
- Add 250 µL of 4x lysis buffer to lyse the cells by osmotic shock.
- Incubate the cells for 10 min at 36 ± 1 °C.
- Mix thoroughly by pipetting up and down ten times all over the bottom of the well to ensure that the cells are fully lysed and homogenized.
- Use an automatic spiral plater to determine the S. aureus load of each well.
- Incubate the agar plates for 18-24 h at 36 ± 1 °C.
- The next day, count the number of colonies with a colony counter to calculate the intracellular S. aureus load of each well.
- Measurement of intracellular efficacy of antimicrobial compounds with enzyme protection assay (EPA)
- Prepare 25 mL of 1x lysis buffer with 3.125 mL of 2% Triton X-100 in sterile water, 6.25 mL of trypsin-EDTA, and 15.625 mL of sterile water.
- Prepare 250 µL of a fresh lysostaphin working solution at 1 mg/mL by mixing 25 µL of a lysostaphin stock solution (10 mg/mL) and 225 µL of 0.1 M Tris-HCl.
- Prepare 25 mL of complete infection medium supplemented with lysostaphin by adding 24.75 mL of complete infection medium to 250 µL of the lysostaphin working solution.
- For each antimicrobial compound to be tested, prepare 3.1 mL of complete infection medium supplemented with lysostaphin and the antimicrobial compound at the concentration to be studied.
- Remove and discard the spent cell culture medium from the 24-well plate.
- Add 1 mL of complete infection medium supplemented with lysostaphin.
- Incubate the cells for 1 h at 36 ± 1 °C in 5% CO2 to let the lysostaphin kill the extracellular bacteria.
- Remove and discard the medium supplemented with lysostaphin from the 24-well plate.
- Fill three wells with 1 mL of medium supplemented with lysostaphin plus the antimicrobial compound to be tested.
- Repeat step 3.5.9 for each antimicrobial compound to be tested.
- For the control condition, fill three wells with 1 mL of medium supplemented with lysostaphin without any antimicrobial compound.
- Incubate the cells for 24 h at 36 ± 1 °C in 5% CO2.
- At the end of the incubation period, remove and discard the spent medium and gently wash each well three times with sterile DPBS with CaCl2 and MgCl2.
- Add 1 mL of 1x lysis buffer to each well to detach and lyse the cells by osmotic shock.
- Incubate the cells for 10 min at 36 ± 1 °C.
- Mix thoroughly by pipetting up and down ten times all over the well to ensure that the cells are fully lysed and homogenized.
- Use an automatic spiral plater to determine the S. aureus load of each well.
- Incubate the agar plates for 18-24 h at 36 ± 1 °C.
- The next day, count the number of colonies with a colony counter to calculate the intracellular S. aureus load of each well.
NOTE: The intracellular activity of each antimicrobial compound should be calculated according to the bacterial load of the control condition. It is also important to check the cytotoxicity of all antimicrobial compounds to prove that the differences observed between the control and the compounds are not due to cell death.
Representative Results
The results of S. aureus internalization by A549 epithelial cells are depicted in Figure 1A. A549 cells were inoculated with S. aureus SF8300 WT and SF8300 ΔfnbA/B, which lacks fibronectin-binding proteins A and B, at an MOI of 1 for 2 h. To destroy extracellular S. aureus, lysostaphin was added to the culture medium, and the cells were incubated for 1 h. Next, lysostaphin was either removed by washing for EPA or inactivated with proteinase K for iEPA. Then, the cells were disrupted in lysis buffer, and the bacterial load was quantified by culture. By using EPA, the mean intracellular loads were 4.46 and 0.49 Log CFU/mL for SF8300 WT and SF8300 ΔfnbA/B, respectively (Figure 1A, green bars). Using iEPA, the mean intracellular loads were 4.53 and 0.56 Log CFU/mL for SF8300 WT and SF8300 ΔfnbA/B, respectively (Figure 1A, red bars). It is interesting to note that both EPA and iEPA showed similar results, which can be explained by the ease of performing the washes when the cells are in good condition and because the S. aureus-induced cytotoxicity is very low in these experimental settings (data not shown).
The results of intracellular activity of vancomycin, rifampicin, and levofloxacin against S. aureus are depicted in Figure 1B. To measure the intracellular activity of these antibiotics, HaCaT cells were inoculated with S. aureus ATCC 29213 at an MOI of 1 for 2 h. The cells were incubated with lysostaphin, with or without the antimicrobial compounds to be tested, for 24 h. Next, lysostaphin and the antimicrobial compounds were removed by washing. The cells were disrupted in lysis buffer, and the bacterial load was quantified by culture. The mean intracellular loads were 4.57, 4.51, 3.03, and 2.91 log CFU/mL for control, vancomycin (50 µg/mL), rifampicin (7 µg/mL), and levofloxacin (10 µg/mL), respectively (Figure 1B).
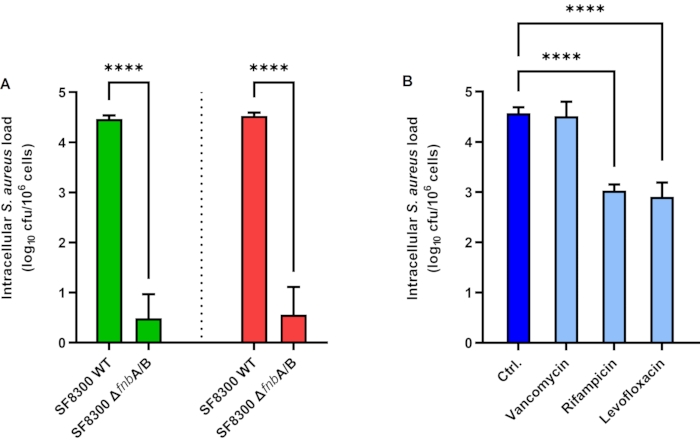
Figure 1: Intracellular Staphylococcus aureus load in epithelial cells. (A) Enzyme protection assay (green bars) and improved enzyme protection assay (red bars) in A549 cells infected with S. aureus SF8300 WT and ΔfnbA/B. (B) Intracellular activity of antimicrobial compounds in HaCaT cells infected with S. aureus ATCC 29213. Bars represent the mean values of three independent experiments performed in triplicate. Error bars represent the standard deviations. **** p < 0.0001. Abbreviations: Ctrl = control; cfu = colony-forming units. Please click here to view a larger version of this figure.
Discussion
The assays described here are valuable for studying the extent of internalization and the intracellular survival of S. aureus in NPPCs, as well as the intracellular efficacy of antimicrobial compounds6,15,16. Some steps in both assay protocols can be critical. The health condition and the density of the cells must be perfectly controlled and consistent between independent experiments. The bacterial inoculum must be carefully standardized to obtain a real MOI close to the targeted theoretical MOI. In general, care must be taken not to detach any of the cells while pipetting. The washes to remove lysostaphin and antibiotics are critical steps in the EPA. The use of proteinase K has been found to improve this step when no antibiotic is used (see below). Last but not least, the cells should be fully detached in each well and thoroughly homogenized after the incubation with the lysis buffer to reliably quantify the S. aureus intracellular load.
In some instances, issues may be encountered, and several points must be checked first. In case of a lack of reproducibility, it must be kept in mind that S. aureus can form clumps, making quantification by absorbance inaccurate. The clumping of bacteria can be increased by centrifugation and washing steps if the culture medium is to be replaced (e.g., for eliminating a secreted protein). The bacterial suspension should be used rapidly because bacteria continue to grow at room temperature. The lysostaphin efficacy could decrease because of incorrect storage conditions, suboptimal pH for enzyme activity in the culture media, variability in the enzymatic activity between batches and providers, and lack of lysostaphin sensitivity of some strains in specific growth conditions. Phenol red could have a slight bacteriostatic effect, especially when the culture medium is relatively poor in nutrients compared to the typical broths used for growing bacteria. Thus, it is advisable to use a cell culture medium without phenol red, which also improves fluorescence microscopic observations by reducing the background noise.
Although this method is a valuable tool to study the intracellular fate of different strains, some limits of the method should be considered. The use of a very high MOI can overload the capability of internalization by NPPCs and level out the differences between the different strains tested. The extent of internalization of the most cytotoxic strains may be underestimated because lysostaphin (or antibiotics) rapidly destroys S. aureus that is released by damaged cells. Thus, experiments with extended durations (i.e., to study intracellular survival or intracellular activity of antibiotics) are easier to set up with strains with low cytotoxicity. Therefore, the incubation time and the MOI should be accurately adjusted according to the strain virulence, the cell type, and the experimental aim.
The method described here with the use of lysostaphin is more reliable than those based on gentamicin because, unlike lysostaphin, gentamicin tends to be internalized by host cells13. The other advantage is the possibility to inactivate the lysostaphin. Inhibition of lysostaphin activity was reported by Kim et al.13 with the use of EDTA to chelate zinc ions or 1,10-phenanthroline; however, intensive washes are still required to remove the enzyme before plating of the bacteria. Here, proteinase K enables rapid inactivation of lysostaphin. We observed that cells tend to detach from the culture plate when they become heavily infected because of the multiplication of intracellular S. aureus. By skipping the final washing step, the iEPA method greatly simplified technical handling and enabled the recovery of the internalized bacteria in loosely adherent or already detached cells.
The more concentrated reagents and buffers used in iEPA also helped reduce pipetting effort and minimize the loss of cells. In addition, iEPA can be used with cells in suspension, as well as with organoids that are difficult to wash. In conclusion, enzyme protection assays enable the study of the extent of internalization and the intracellular fate of S. aureus, as well as the intracellular activity of antimicrobials drugs with different in vitro models. Improvements should be made to better characterize the relationship between internalization and cytotoxicity to better appreciate the importance of developing drugs capable of reaching S. aureus inside the cell.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
S. aureus strains SF8300 WT and SF8300 ΔfnbA/B were generously gifted by Prof. Binh Diep (University of California, San Francisco, USA). This work was supported by a grant of the FINOVI association (#AO13 FINOVI) under the aegis of the Foundation for the University of Lyon.
Materials
| 24-well plate | CORNING-FALCON | 353047 | |
| A549 cell line | ATCC | CCL-185 | |
| Acetate buffer solution pH 4.6 | Fluka | 31048 | Used to prepare lysostpahine stock solution at 10 mg/mL in 20 mM sodium acetate. |
| AMBICIN (Recombinant lysostaphin) | AMBI | LSPN-50 | Lyophilized recominant lysostaphin. Freeze at -80 °C for long-term storage. |
| COS – Colombia agar + 5% sheep blood | Biomerieux | 43049 | Any agar plate suitable for growing staphylococci can be used instead. |
| Densitometer WPA CO8000 | Biochrom Ltd. | 80-3000-45 | Cell density meter |
| Dulbecco’s Modified Eagle’s Medium, high glucose with phenol red | Sigma-Aldrich | D6429 | |
| Dulbecco’s Modified Eagle’s Medium, high glucose without phenol red | Sigma-Aldrich | D1145 | |
| Dulbecco’s Phosphate Buffered Saline | Sigma-Aldrich | D8537 | |
| Dulbecco′s Phosphate-buffered Saline with MgCl2 and CaCl2, sterile-filtered | Sigma-Aldrich | D8662 | |
| Dulbecco′s Phosphate-buffered Saline, sterile-filtered | Sigma-Aldrich | D8537 | |
| Easyspiral dilute | Interscience | 414000 | Automatic diluter and spiral plater |
| Fetal bovine serum | Gibco | 10270-106 | |
| HaCaT cell line | Cell lines service (CLS) | 300493 | |
| Hoechst 33342, Trihydrochloride, Trihydrate, 10 mg/mL Solution in Water | Fisher scientific | 11534886 | |
| Propidium iodide, 1.0 mg/mL solution in water | Invitrogen | P3566 | |
| Proteinase K, recombinant 20 mg/mL | Eurobio | GEXPRK01-B5 | > 30 U/mg, lot 901727 |
| Scan 4000 | Interscience | 438000 | Automatic colony counter |
| Sterile water | OTEC | 600500 | |
| T-75 culture flask | CORNING-FALCON | 353136 | |
| TC20 Automated cell counter | Biorad | 1450102 | Automatic cell counter |
| Tris 1 M pH 8.0 | Invitrogen | AM9855G | Used to prepare lysostaphine working solution at 1 mg/mL in 0.1 M Tris-HCl. |
| Triton X-100 | Sigma-Aldrich | T8787 | |
| Trypsin – EDTA solution | Sigma-Aldrich | T3924 | 0.05% porcine trypsin and 0.02% EDTA in Hanks′ Balanced Salt Solution with phenol red |
| Wide-field fluorescence microscope | Nikon | Ti2 |
Riferimenti
- Verhoeven, P. O., et al. Detection and clinical relevance of Staphylococcus aureus nasal carriage: an update. Expert Review of Anti-Infective Therapy. 12 (1), 75-89 (2014).
- Gagnaire, J., et al. Epidemiology and clinical relevance of Staphylococcus aureus intestinal carriage: a systematic review and meta-analysis. Expert Review of Anti-Infective Therapy. 15 (8), 767-785 (2017).
- Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., Fowler, V. G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clinical Microbiology Reviews. 28 (3), 603-661 (2015).
- Josse, J., Laurent, F., Diot, A. Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Frontiers in Microbiology. 8, 2433 (2017).
- Hanssen, A. -. M., et al. Localization of Staphylococcus aureus in tissue from the nasal vestibule in healthy carriers. BMC Microbiology. 17 (1), 89 (2017).
- Rigaill, J., et al. Evaluation of the intracellular efficacy of antimicrobial agents used for Staphylococcus aureus decolonization in a cell model mimicking nasal colonization. Journal of Antimicrobial Chemotherapy. 73 (11), 3044-3048 (2018).
- Yang, D., et al. Novel insights into Staphylococcus aureus deep bone infections: the Involvement of osteocytes. mBio. 9 (2), 00415-00418 (2018).
- Tuchscherr, L., et al. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Molecular Medicine. 3 (3), 129-141 (2011).
- Valour, F., et al. Antimicrobial activity against intraosteoblastic Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 59 (4), 2029-2036 (2015).
- Proctor, R. A., Prendergast, E., Mosher, D. F. Fibronectin mediates attachment of Staphylococcus aureus to human neutrophils. Blood. 59 (4), 681-687 (1982).
- Climo, M. W., Ehlert, K., Archer, G. L. Mechanism and suppression of lysostaphin resistance in oxacillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy. 45 (5), 1431-1437 (2001).
- Bur, S., Preissner, K. T., Herrmann, M., Bischoff, M. The Staphylococcus aureus extracellular adherence protein promotes bacterial internalisation by keratinocytes independent of fibronectin-binding proteins. Journal of Investigative Dermatology. 133 (8), 2004-2012 (2013).
- Kim, J. -. H., Chaurasia, A. K., Batool, N., Ko, K. S., Kim, K. K. Alternative enzyme protection assay to overcome the drawbacks of the gentamicin protection assay for measuring entry and intracellular survival of Staphylococci. Infection and Immunity. 87 (5), 00119 (2019).
- Aerobic plate count. Bacteriological Analytical Manual., Edition 8, Revision A Available from: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-3-aerobic-plate-count (2021)
- Kolenda, C., et al. Evaluation of the activity of a combination of three bacteriophages alone or in association with antibiotics on Staphylococcus aureus embedded in biofilm or internalized in Osteoblasts. Antimicrobial Agents and Chemotherapy. 64 (3), 02231 (2020).
- Abad, L., et al. Antibiofilm and intraosteoblastic activities of rifamycins against Staphylococcus aureus: promising in vitro profile of rifabutin. Journal of Antimicrobial Chemotherapy. 75 (6), 1466-1473 (2020).

