Culture and Imaging of Human Nasal Epithelial Organoids
Summary
A detailed protocol is presented here to describe an in vitro organoid model from human nasal epithelial cells. The protocol has options for measurements requiring standard laboratory equipment, with additional possibilities for specialized equipment and software.
Abstract
Individualized therapy for cystic fibrosis (CF) patients can be achieved with an in vitro disease model to understand baseline Cystic Fibrosis Transmembrane conductance Regulator (CFTR) activity and restoration from small molecule compounds. Our group recently focused on establishing a well-differentiated organoid model directly derived from primary human nasal epithelial cells (HNE). Histology of sectioned organoids, whole-mount immunofluorescent staining, and imaging (using confocal microscopy, immunofluorescent microscopy, and bright field) are essential to characterize organoids and confirm epithelial differentiation in preparation for functional assays. Furthermore, HNE organoids produce lumens of varying sizes that correlate with CFTR activity, distinguishing between CF and non-CF organoids. In this manuscript, the methodology for culturing HNE organoids are described in detail, focusing on the assessment of differentiation using the imaging modalities, including the measurement of baseline lumen area (a method of CFTR activity measurement in organoids that any laboratory with a microscope can employ) as well as the developed automated approach to a functional assay (which requires more specialized equipment).
Introduction
Introduction to the technique
Ex vivo culture-based assays are an increasingly utilized tool for precision medicine and the study of disease pathophysiology. Primary human nasal epithelial (HNE) cell culture has been used in numerous studies of cystic fibrosis1,2,3,4,5,6,7,8,9,10,11,12,13, an autosomal recessive disease that affects epithelial cell function in multiple organs. HNE culture provides a renewable source of airway epithelia that may be obtained prospectively and recapitulates electrophysiological and biochemical qualities to test Cystic Fibrosis Transmembrane conductance Regulator (CFTR) activity. HNE cells can be sampled with minimal side effects14, similar to common viral respiratory swabs. Research work describing a model for cystic fibrosis study derived from HNE brush biopsies has been recently published11,13. While similar to other models using primary HNE2,3 and intestinal tissue15,16,17,18,19, detailed characterization of the differentiation and imaging of this model are described here for use in CF research and for aiding in the studies of other airways diseases13. The organoid model is not unlimited like immortalized cell lines but can be expanded by conditional reprogramming (using irradiated and inactivated feeder fibroblasts and Rho-kinase inhibitors) to a more stem cell-like state20,21,22,23. The processing of HNE brush biopsies using this method yields large numbers of epithelial cells for use in multiple applications at higher throughput while still retaining the ability to differentiate fully. While this protocol was developed using feeder cells, other methodologies may be used by investigators wishing to avoid feeder cell technology14,24.
Importance of the technique to pulmonary biology
A significant study has been devoted to understanding how the absence of regular, functioning CFTR in the cell membrane of epithelial cells results in dysfunction in the lungs, pancreas, liver, intestine, or other tissues. Dysfunctional epithelial ion transport, particularly that of chloride and bicarbonate, results in a decreased volume of the epithelial lining fluids and changes in mucous secretions, leading to mucous stasis and obstruction. In other airway diseases, such as primary ciliary dyskinesia, altered ciliary motion impairs mucociliary clearance and leads to mucous stasis and obstruction25. Therefore, the current HNE organoid model has been developed for various applications, depending on the investigator's experimental design and resources. This includes live-cell imaging using live-cell stains; fixation and sectioning to characterize the morphology; immunofluorescence staining with antibodies and whole-mount confocal imaging to avoid disrupting intraluminal structures; and bright-field imaging and micro-optical coherence tomography for quantitative measurements of ciliary beat frequency and mucociliary transport13. To facilitate expansion to other investigators, commercially available reagents and supplies were used for culturing. A functional assay was developed that used common microscope techniques and more specialized equipment. Overall, while the present model was designed to assess CFTR activity at baseline or in response to therapeutics, the techniques described in this protocol can be applied to other diseases involving epithelial cell function, especially epithelial cell fluid transport.
Comparison to other methodologies
Recently the utility of this organoid model was developed by correlating in vitro CFTR modulator responses of patients' organoids with their clinical response11. Notably, it is also demonstrated that the present model paralleled short-circuit current responses, the current gold standard for assessing CFTR function, in the same patients. Short-circuit current differs from the swelling assay because the former measures CFTR function via ion transport26. In contrast, this assay measures a more downstream effect with fluid transport, providing additional information about the overall function of CFTR27,28,29,30,31,32. Short-circuit current measurements have continued to be a common and reliable method for determining CFTR chloride channel activity1,33. These electrophysiological assays require specialized, expensive equipment, require many times more cells for each experimental replicate than the organoid assay, cannot be easily automated, and are not amenable to scaling up for higher throughput applications. Another organoid model derived from intestinal epithelia has additional advantages15,16,17,18, such as more excellent replicative capability, but is neither derived from an airway tissue nor is universally available. HNE brushings are obtained with inexpensive cytology brushes without the need for sedation and at minimal risk. Getting the brushing does not require a clinician and can be performed by trained research coordinators and other research staff14. The HNE organoid model can be cultured by any laboratory with primary cell culture capabilities, and some of the applications can be performed with standard microscopy techniques. Altogether, these advantages provide additional access to technology for assessing airway epithelial function that might otherwise be unavailable to some laboratories. Furthermore, HNE organoids can be utilized to study other disease states that affect the airway, such as primary ciliary dyskinesia25 or viral infection, which intestinal organoids cannot.
Protocol
HNE samples were collected at the Children's of Alabama hospital. All procedures and methods described here have been approved by the IRB University of Alabama at Birmingham (UAB IRB #151030001). To facilitate the expansion and improve the function of human nasal epithelial cells (HNEs), the present culturing methods are adapted from the well-known air-liquid interface (ALI) culture method28,34. HNEs were initially collected by brush biopsy as previously described12,14, with the only difference being the use of a cytology brush. All sample processing steps and cell culture were performed in the biosafety cabinet.
1. Cell culture and expansion of nasal epithelial cells
- After brushing, store the nasal biopsy sample in 8 mL of RPMI media in a 15 mL conical tube on ice and transfer it to the lab within 4 h (not more than 24 h).
- Dissociate the nasal brush biopsy into 8 mL of RPMI media in a 15 mL conical tube by passing the cytology brush through a 1 mL large bore pipette tip (cut off the tip) several times until the brush is clear of tissue.
- Centrifuge the cells at 500 x g for 5 min at 4 °C, remove the supernatant, and then re-suspend the cell pellet in 3 mL of cell detachment solution (see Table of Materials) and incubate at room temperature for 16 min to digest.
- Use 5 mL of expansion media (Table 1) to wash the cells twice. Then, add cells to a T75 flask pre-seeded with irradiated and inactivated 3T3 fibroblasts22 (50%-60% confluence) to co-culture the cells and expand in the expansion media for 7-14 days (see Figure 1 for the appropriate colony). Before use, test the irradiated 3T3 fibroblasts to ensure they cannot proliferate, which will negatively influence the epithelial cell expansion.
NOTE: Discard the cells if the nasal epithelial cell confluence does not reach 70% within 14 days. - For the cells derived from patients with CF, introduce four antibiotics (100 µg/mL of tobramycin, 2.5 µg/mL of amphotericin B, 100 µg/mL of ceftazidime, 100 µg/mL of vancomycin, see Table of Materials) into the expansion media for disinfection of the cells for the first 3 days of culture, and then replace the media with antibiotic-free expansion media changing the media every 2 days.
NOTE: This limited use of antibiotics is intended to reduce culture loss due to bacterial colonization while encouraging proliferation after the additional antibiotics are removed. These antibiotics can be tailored to the patient's specific microbiology results, with sensitivities, if needed. - Harvest HNEs from co-culture using the double trypsinization method22 once the cells reach approximately 80% confluence. This method ensures that the irradiated and inactivated 3T3 fibroblasts are removed from the flask, and they do not contaminate subsequent organoid seeding.
- Wash the cells with 1x DPBS, and then add 1.5 mL of 0.05% trypsin-EDTA into the T75 flask for 4 min at 37 °C to remove the irradiated and inactivated 3T3 fibroblast from culture.
- Rinse the T75 flask with 1x DPBS twice to thoroughly wash away any remaining 3T3 fibroblasts; add 1.5 mL of 0.05% trypsin-EDTA into the T75 flask for 10 min at 37 °C to detach HNEs.
- Neutralize the trypsin with soybean trypsin inhibitor (see Table of Materials) at a 1:1 ratio. Centrifuge the cells at 500 x g for 5 min, and then remove the supernatant. After washing the cells with 5 mL of expansion media once, the cells are ready for seeding to grow organoids.
NOTE: HNEs with a passage number of three are recommended to be used for further experiments.
2. Growth and differentiation of organoids in slides and culture inserts
- Thaw the organoid culture extracellular matrix (ECM) overnight at 4 °C. Cool pipette tips to 4 °C and 15-well angiogenesis slides (see Table of Materials) overnight at 4 °C.
NOTE: ECM should be put at 4 °C the night before the cell harvesting needs to be done. - Coat the slides with 5 µL of cold 100% ECM on ice (pipette 5 µL of cold 100% ECM with a cold pipette tip into each well of the 15-well slide), and then place them into a cell culture incubator at 37 °C for at least 30 min for consolidation.
- Count the HNEs harvested from co-culture using a hemocytometer and dilute the cells to 500 cells/µL in total number with 20% ECM diluted by differentiation media (Table 2) on ice. Then, seed 5 µL of the cold cell/ECM mixture into each well of the slides coated with ECM.
- Immediately transfer the slides into a culture incubator at 37 °C for 1 h to consolidate the cell/ECM mixture.
- Feed the cells in each well of the 15-well angiogenesis slides with 50 µL of differentiation media. Change the media every other day until the organoids are ready for further experiments.
NOTE: Organoids can usually be visualized after 1-2 days. There are 20-90 organoids commonly formed in each well of the slides. The organoids can typically survive for 40-60 days in ECM with feeding every other day when kept in a humidified incubator at 37 °C. - Culture the organoids in the culture inserts (see Table of Materials) for greater quantity for specific applications (such as sectioning for histology or immunofluorescence) as per the steps mentioned below.
- To grow organoids in the culture inserts, prepare the organoid culture ECM, pipette tips, and inserts as mentioned in steps 2.1-2.2. Coat the inserts with 100 µL of cold 100% ECM.
- Seed 60 µL of cell/ECM mixture (500 cells/µL with 20% ECM in differentiation media) on top of the ECM coating in the insert.
NOTE: All the other steps for making organoids in the inserts are the same as those conducted in the slides, steps 2.4-2.5. - Add 600 µL of the differentiation media into the bottom part of the insert. Change the media every alternate day until the organoids are used for experiments, typically 2-3 weeks.
3. Preparation and isolation of organoids for whole-mount immunofluorescence
- Isolation and fixation of organoids
- Pre-treat 8-well glass-bottom chamber slides with cell adhesive (see Table of Materials) for 30 min following Reference35. After discarding the solution, air-dry the wells for 30 min.
- To harvest the organoids, remove the media from the top of the ECM, and then add 50 µL of cold 1x PBS into each well of the 15-well slides on ice (1:1 with ECM volume).
- Pipette up and down 3-5 times using 200 µL of the large-bore pipette tip, and then dispense the solution onto the center of a well of the 8-well chamber slides.
- Immediately remove excess liquid from the wells by a fine-tip pipette. Then, place the chamber slide into a 37 °C incubator for 40 min to enhance the organoid adhering to the glass bottom.
- After gently washing with 1x PBS twice, fix the organoids with 300 µL of 4% paraformaldehyde in each well for 30 min at room temperature (RT).
- Wash twice with 1x PBS and store the organoids in 1x PBS at 4 °C for immunostaining for up to 2 weeks.
- Immunofluorescence staining
- To reduce auto-fluorescence, add 250 µL of 50 mM NH4Cl in 1x PBS into each well of the slides at RT for 30 min while gently shaking on a shaker at 20 rpm.
- After washing with 1x PBS twice, permeabilize the cells with 0.1% Triton X-100 for 30 min at RT while gently shaking at 20 rpm.
- After washing with 1x PBS twice, add 300 µL of blocking solution, including 5% BSA and 0.1% Triton X-100 in 1x PBS, into each well for 1 h at RT.
- Following washing, add primary antibody into the appropriate wells followed by secondary antibodies (see Table of Materials). Prepare primary and secondary antibody solutions in 2% BSA and 0.3% Triton X-100 in 1x PBS. Incubate all the primary antibodies at 4 °C for 2 days and all the secondary antibodies at 4 °C for 1 day.
NOTE: The final concentrations are dependent on the protein desired for the experiment. Please check the stock concentration on the manufacturer's datasheet. Then, calculate the final concentrations based on the dilutions used in the Table of Materials. - After incubation, wash the wells thoroughly with 1x PBS and add DAPI in 2% BSA and 0.3% Triton X-100 into each well for nuclear staining.
- Image the organoids using a confocal laser scanning microscope, with a 20-60x oil immersion objective (see Table of Materials). Use Z-stack mode to set the upper and lower bounds of the image and use the recommended optimal Z-step size determined by the confocal software.
NOTE: The following four confocal laser excitation wavelengths were used: 408.7 nm, 489.1 nm, 561.7 nm, and 637.9 nm.
4. Preparation and isolation of organoids for histological sectioning
- To harvest the organoids for histological studies, remove the media from the culture and add 50 µL of cold 1x PBS into each well of the slides on ice.
- Pipette up and down three to five times using a 200 µL large-bore pipette tip, combine all the solutions from the 15-well slide or culture inserts into a 15 mL conical tube on ice. Adjust the total solution volume in the tube to 10 mL by adding additional cold 1x PBS.
- Centrifuge the tube at 4 °C, 300 x g for 5 min; aspirate out the supernatant and add 60 µL of warm histogel (see Table of Materials) to mix with the organoid pellet using a 200 µL of the large-bore pipette tip.
- Immediately transfer the suspension into a histology mold. After the consolidation of the histogel at room temperature, put the mold block into 4% paraformaldehyde for fixation overnight at 4 °C.
- After embedding in paraffin, cut the histogel block into 5 µm cross-sections (for example, with a microtome), fix the sections onto glass slides, and stain using hematoxylin and eosin (H&E) or immunofluorescence-labeled antibodies. Take images using a bright-field microscope or inverted epi-fluorescence microscope (see Table of Materials).
5. Imaging of live organoids
NOTE: The following steps are carried out using an automated imaging system (see Table of Materials). Different imaging systems need to adapt these steps following their specific manufacturer's instructions. Regardless of the equipment utilized, imaging live organoids require a temperature-controlled and humidified environmental chamber with an accompanied CO2 gas controller.
- To monitor the differentiation of organoids before performing a functional swelling assay36, capture the whole slide images manually with any bright-field microscope or with an automated imaging system, as detailed below.
- Turn on the power to the automated imaging system and the CO2/O2 gas controller and allow the system to complete the automated calibration (~30 min).
- After completion, set the imaging system temperature to 37 °C; add 15 mL of sterile water to the humidification reservoir, open the CO2 valves, close the lids, and let the imaging system pre-incubate for a minimum of 30 min before imaging.
- Open the automated imaging software to set up a protocol for imaging and choose the location to save the raw experimental data.
NOTE: An example protocol file (Supplementary File 1), specific to the imaging system, has been provided as a template for the automated imaging of organoids to monitor organoid differentiation. - Once the environmental settings are met, immediately transfer up to two 15-well slides with lids into the environmental chamber in the automated image system's slide holder insert (see Table of Materials).
- Select the wells to be imaged and start imaging to complete imaging of the desired wells.
NOTE: The basic recommended settings are: 4x and 10x air objective, bright-field channel, 2 x 2 montage for 4x objective to cover the whole well area, 4 x 4 montage for a 10x objective. Z-stack settings: three to six Z-stack slices, Z-step size = 50-100 µm, one to two slices below autofocus point, and three to five slices above.
- To image live organoids for use in a forskolin-induced swelling (FIS) assay, use a microscope with an automated stage equipped with an environmentally controlled imaging chamber that allows temperature and CO2 control.
- Begin with steps 5.1.1-5.1.4, substitute the protocol file in step 5.1.3 with the one provided in the example protocol file (Supplementary File 2) containing settings specific for performing a FIS assay.
- Before each experiment, adjust the exposure settings by evaluating at least three wells for each slide (far left, middle, and far right). Apply the X and Y coordinate offsets to ensure that the objective will be in the center of the well for all the wells.
NOTE: The basic recommended settings are: 4x air objective, Channel 1 = Bright Field, Channel 2 = DAPI, 2 x 2 montage (four image tiles). Z-stack settings: three to four Z-stack slices, Z-step size = 50-100 µm, one slice below autofocus point and two to three slices above. Image acquisition time: 8 h with 20 min intervals (Total reads = 25; Read 1 is T = 0). - Once all the settings are appropriately selected, choose the wells to image for both slides, or select all, and begin the run as per the equipment's instructions.
- After running, save the experiment, close the imaging software, and shut down the imaging system37.
6. Baseline lumen measurements
NOTE: This is done using manual imaging analysis software (see Table of Materials). A similar methodology can be followed using an open-source software38 or any software that can measure the area of a region on an image.
- In the software, open the Automated Measurement Panel by right-clicking the bottom of the screen, select Measurement, and then Automated Measurement Results. The area of each region of interest (ROI) measured will appear there.
- Open the organoid image in the software and select 5-10 organoids with visible lumens.
NOTE: Lumens will be a circular area in the middle of the organoid that is visibly different in color than the rest of the organoid (Figure 2A). - Using the polygon ROI measurement feature, hold right click on the image to open the menu and select Polygonal ROI to outline the full organoid to obtain the organoid's total surface area (TSA). Then using the same feature, outline the lumen area (LA) (Figure 2B).
- Repeat for the remaining organoids in the well and all the wells in the assay.
- Export the data to excel. Divide the LA by the TSA and average all the organoids from the sample to get the Baseline Lumen Ratio (BLR)11,13.
NOTE: Typically, ~87% of non-CF organoids will have a BLR over 0.6, and 97% will be over 0.5, while only 14% of CF organoids will have a BLR over 0.6, and 31% will be over 0.5.
7. Pre-treatment and automated imaging of HNE organoids
NOTE: All pre-treatment steps are carried out in a clean biosafety cabinet. Pre-setup the automated imaging system and the software for recording the assay before step 7.1. The incubation with DAPI is optional but is recommended as a fail-safe if the quality of bright field images is compromised. In this case, the DAPI channel (377 nm) can be analyzed instead.
- Pre-incubate the organoids in a well of 15-well slides with 50 µL of differentiation media containing DAPI with or without 100 µM of CFTRinh-172 (see Table of Materials) in a 37 °C incubator for 1 h. While the organoids incubate, perform step 5.2.1 using a swelling assay custom protocol following Supplementary File 2.
- Remove the pre-incubation medium using a glass Pasteur pipette with aspiration. Add 10 µM of forskolin and 100 µM of IBMX (stimulation cocktails) (see Table of Materials) for a total volume of 50 µL differentiation media into each well.
NOTE: Ensure no bubbles are introduced into the wells. Bubbles on the images will affect the automated analysis. - Without delay, begin the FIS imaging protocol following step 5.2.2 to 5.2.4. Acquire images every 20 min in each well, with a total runtime of 8 h.
8. Automated analysis of forskolin-induced swelling assay on HNE organoids
- Open the automated imaging analysis software, find the experiment previously saved from step 7.3, and bring up the images of the experiment.
- Choose the vessel window to be evaluated and select the option containing the processed images that have been stitched and z-projected. This step should provide a picture of the entire well with all organoids within the imaging frame for masking assessment and measurements.
- Choose the well image to assess. Select Analyze. Repeat this process for other images to ensure appropriate masking for all images included in the automated measurements. Save the setting parameters.
- After completing the analyzing settings, apply the changes. The software will change the measurements based on the settings.
NOTE: After the initial image pre-processing is complete, quality control (QC) measures should be performed to ensure consistent masking. These include manually reviewing all wells to ensure organoids are within the imaging frame, bubbles or debris are not inappropriately masked, and checking the masking in bright field and DAPI channels. - Export the data for the summary analysis (including graphing and statistical analysis).
Representative Results
HNEs expansion is essential for a thriving organoid culture. HNEs from a successful sample collection should expand to over 70% confluence around 10 days. An example of successful and unsuccessful samples is shown in Figure 1A and Figure 1B, respectively. The cells must be discarded if they cannot reach 70% confluence by 14 days after co-culture with irradiated 3T3 cells. Any contaminated cells are to be immediately discarded if unable to rescue with additional antimicrobial agents quickly.
The growth of the organoids was compared in 15-well slides and culture inserts. The culture inserts are thicker and further from the objective than the optically optimized slides, impacting the image and resolution. Despite this, no significant difference in the morphology was observed in these two culture methods, as shown in Figure 2. Morphological differences can be seen between non-CF and CF organoids, as shown in Figure 3A. Non-CF organoids tend to have a larger lumen containing more fluid within it. In contrast, CF organoids usually have a smaller lumen with less fluid and sometimes are filled with mucus and debris. Lumen size was measured manually (Figure 3B), and the baseline lumen ratio was calculated and shown in Figure 3C. Cross-sectioned organoids were characterized using H&E and immunofluorescence staining. The representative images are shown in Figure 4A,B. Airway epithelial markers such as cilia, mucus, and tight junction are demonstrated in organoids by whole-mount immunofluorescent staining shown in Figure 5A–D. Depending on the application, sectioned or whole-mount immunofluorescence can be employed. The whole-mount method maintains the three-dimensional nature of the organoid, keeping the organoid's interior intact, as shown in the previously published work13.
CFTR function was assessed by forskolin-induced swelling (FIS) assay using an automated imaging system. Only 15-well slides are used for functional assays due to the better image resolution. A representative forskolin dose-response experiment of non-CF volunteers (n = 5 subjects) is shown in Figure 6A to illustrate the rationale of the optimized imaging time and analysis. Data comparing non-CF and CF organoid responses are detailed in previous publications11,13. A dose-response shows the incremental change in CFTR activity to demonstrate the best approach to measurements. Assay duration of 1 h and 8 h were evaluated (Figure 6B,C) as well as analysis using average fractional change (AFC) versus area under the curve (AUC) is seen in Figures 6C,D. Based on our previous experience, swelling for most subjects and conditions plateau after 8 h, and in some cases, results in bursting of the organoids over that time. Therefore, assays were limited to 8 h only. At this extended assay length, swelling becomes non-linear. The use of the AUC also considers both the changes in size and the rate of change. Therefore, the AUC over 8 h was used for all the FIS assays in the final methodology.
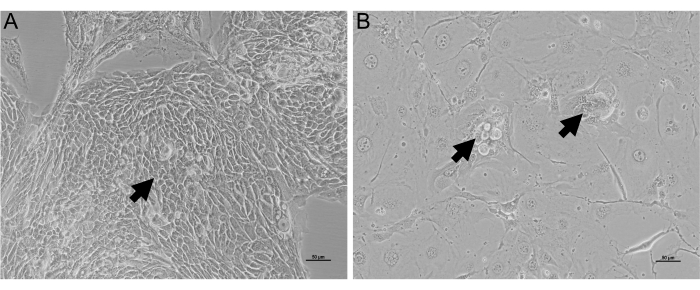
Figure 1: Bright-field images of HNEs in co-culture. HNEs expand in expansion media with irradiated and inactivated 3T3 fibroblasts for 10 days. An inverted bright-field microscope is used for imaging the cells. (A) HNEs grow well in a large cluster (black arrow). In contrast, in (B), the HNEs grow poorly in two small clusters (black arrows) surrounding irradiated 3T3 cells. Scale bar = 50 µm. Please click here to view a larger version of this figure.
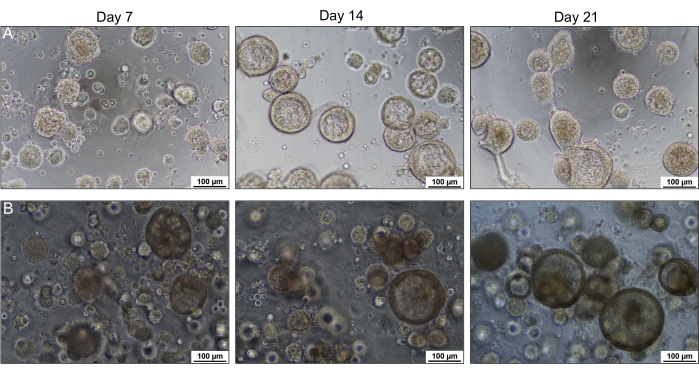
Figure 2: HNE organoid formation in a 15-well slide and culture insert. Bright-field images of organoids were captured using an inverted bright-field microscope over 21 days. Organoids in the 15-well slide (A) have more precise and sharper images than those in the culture insert (B). No morphological difference was observed between the organoids cultured in the slide and insert. Please click here to view a larger version of this figure.
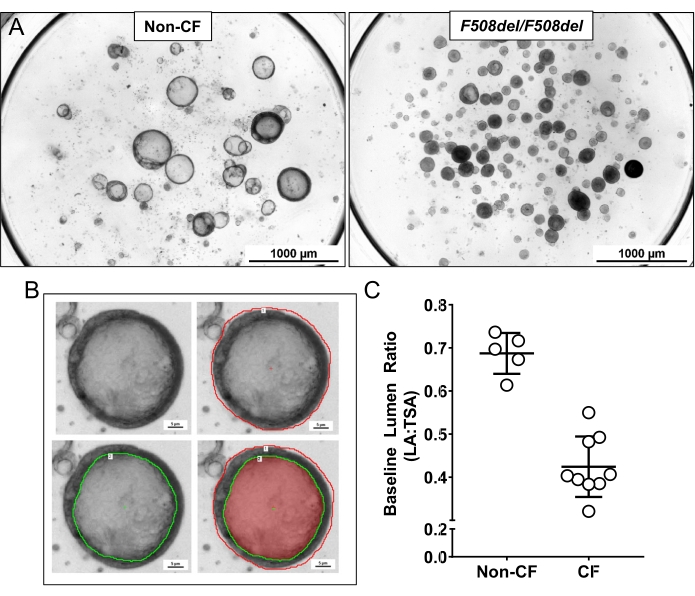
Figure 3: Organoid lumen size (panel A) and lumen measurements (Panel B and C). (A) Non-CF organoids typically have a larger lumen and more fluid than CF (F508del/F508del) organoids. (B) A method to manually measure the total surface area (TSA) indicated by the red outline and lumen area (LA) indicated by the green outline in a single organoid. (C) An example for using the total surface area and the lumen area to calculate the baseline Lumen Ratio (LA: TSA) in organoids from a non-CF vs. a CF subject. Error bars represent standard deviation. Please click here to view a larger version of this figure.
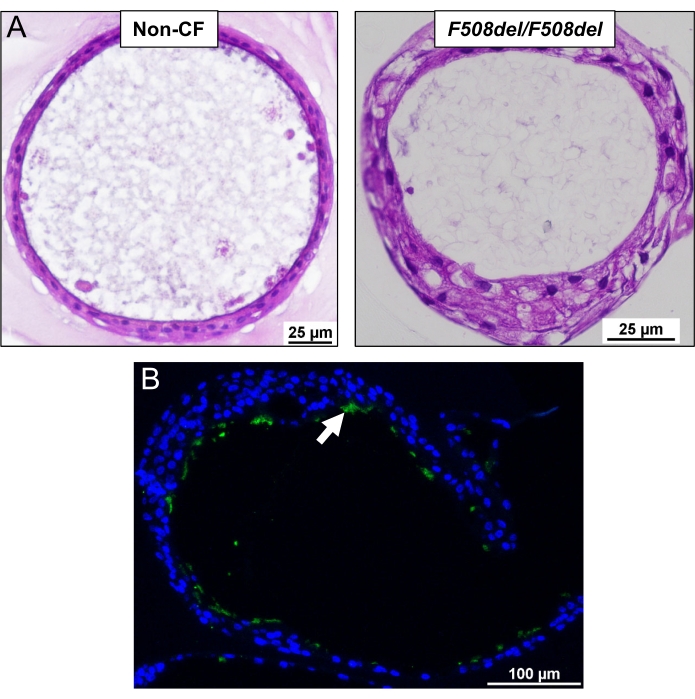
Figure 4: Cross-section of the organoids embedded in paraffin. (A) An example of H&E staining in organoids from a non-CF and CF (F508del/F508del) subject. (B) Immunofluorescent staining of cilia in an organoid. Green is the cilia (white arrow) stained with acetylated-tubulin and FITC labeled secondary antibody, and blue is the nuclei labeled with DAPI. Please click here to view a larger version of this figure.
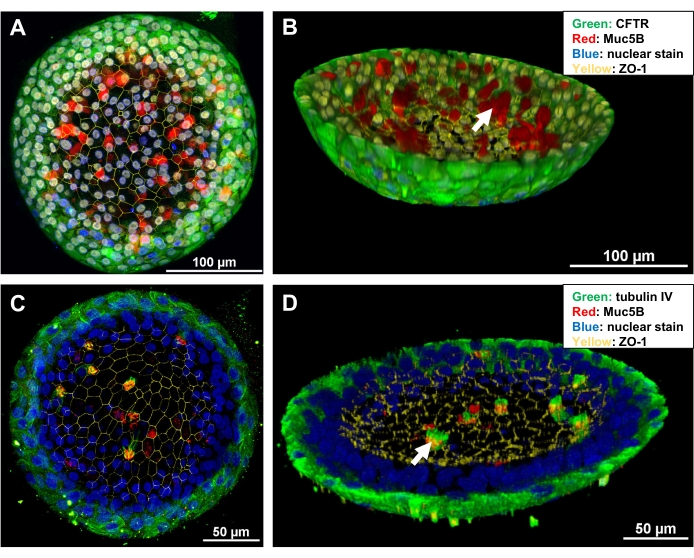
Figure 5: Confocal images of whole-mount immunofluorescence in organoids. (A,C) Maximum projection images of the two representative organoids. (B,D) Three-dimensional reconstruction images of (A) and (C), respectively. An 8-well glass-bottom slide was fitted on the platform of a confocal microscope, and the 40x lens was used to create the photomicrographs. Imaging analysis software was applied for imaging and reconstruction of the images. White arrows indicate mucus (in B) and cilia (in C) within the lumen of the organoids. Please click here to view a larger version of this figure.
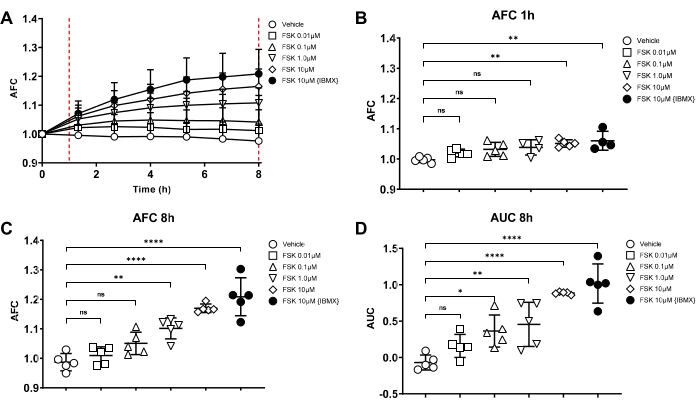
Figure 6: Rationale for the swelling assay length and analysis methods. Forskolin (FSK)-induced swelling (FIS) assay to test CFTR function on the primary nasal epithelial cells. Different dose of forskolin indicated in the figures was administrated into 21-day old organoids in the differentiation media; organoid swelling was immediately recorded with the automated imager for 8 h. After 8 h, swelling is shown in (A) (n = 5, non-CF subjects) using average fractional change (AFC). FSK dose-response is compared with AFC at 1 h (B) vs. at 8 h (C), which suggests that the 8 h assay can produce a more significant swelling difference among different FSK doses than those at 1 h. AFC (C) vs. the area under the curve, AUC (D) at 8 h are compared, indicating AUC can reflect a minor swelling difference than AFC. The X-axis in panels (B–D) represents the different treatment conditions corresponding with the symbols in the figure legend. All error bars in the figures indicate standard deviation. Please click here to view a larger version of this figure.
Table 1: All the components for making the expansion media. The detailed information about reagent stock concentration, stock storage, amount of stock for making a 500 mL media, and final concentration have been described. Please click here to download this Table.
Table 2: All the components for making differentiation media. The detailed information about reagent stock concentration, stock storage, amount of stock for making a 500 mL media, and final concentration have been described. Please click here to download this Table.
Supplementary File 1: An example protocol file specific to the imaging system is provided as a template for the automated imaging of organoids to monitor organoid differentiation. Please click here to download this File.
Supplementary File 2: An example protocol file containing settings specific for performing an FIS assay. Please click here to download this File.
Discussion
This manuscript provides detailed methodologies for comprehensive live and fixed imaging of the airway epithelial organoids derived from HNE brush biopsy. It describes functional assays that can determine CFTR activity in an individual. HNEs provide a minimally invasive, primary tissue for a variety of applications. The expansion techniques offered here can be used for modeling airways disease, including organoids. Organoids can be used for precision therapeutic approaches and to monitor the stability of gene or mRNA-based therapies over time, for precision trial design, and to aid in resolving inconclusive diagnoses39. The current research is on CF, but these models have applications for other diseases affecting epithelial function.
The initial expansion of HNE after biopsy is essential. It has been observed that cytology brushes yield larger initial cell numbers and better results than other biopsy tools14. From previous experiences, we have concluded that combining brushings from both nares in a single sample and processing that sample within 4 h yields the best results. Other investigators have used more extended time frames from biopsy to processing with success3. The appropriate initial collection of biopsies is vital to subsequent expansion and seeding as organoids. High-quality irradiated and inactivated fibroblasts for co-culture are required, which are grown and treated in-house in our laboratory but may also be purchased commercially. Investigators are advised that not all 3T3 fibroblasts are the same cell line and should be validated before use.
For specimens not expected to have pathogenic bacterial or fungal culture, antibiotic treatment is limited to standard penicillin and streptomycin treatment for the experiments described in this manuscript. For those known to have chronic colonization of the upper airway, an antibiotic cocktail described above is utilized for only 3 days because antibiotics seem to slow epithelial expansion and yield poorer results in the experiments described here. Three days were selected to balance contamination risk with providing a similar expansion rate for both CF and non-CF specimens. For individuals with unusual pathogens, tailored antimicrobial treatment can rescue contaminated cultures if recognized early or initiated a priori. For initial biopsies that are unusually slow to grow, results will typically be poor for organoid studies. Investigators need to monitor both growth rate and morphology daily. The in-house culture media and reagents used are most helpful for functional swelling assays, but other commercially available media may benefit other applications12,25,40,41 depending on the experimental design. The type of ECM used can lead to different morphology and different results, and reproducible results are critical. All reagents used in this protocol are routinely tested before use for experiments. Despite this experience, some cultures will fail to expand or generate organoids for inexplicable reasons. Investigators are encouraged to consider these factors as they optimize this protocol for their applications.
A specific type of 15-well slide is used in this protocol optimized for optical imaging, utilizing minimal volumes and maximizing replicates while reducing the costs. These slides have a lower and upper chamber fixed on a polymer coverslip that minimizes menisci (which would otherwise impair imaging) and also media replacement without risk of dislodging the matrix and destroying the organoid cultures. These slides make bright-field imaging and live stain microscopy straightforward, with initial seeding, growth, and imaging in the same dish. Organoids will be lost during the collection, fixation, and staining process, so meticulous care must be taken during each step, and sufficient starting numbers must be obtained to ensure success. Growing organoids in culture inserts can help as these techniques are developed.
Imaging techniques that utilize common laboratory microscopes are included. However, the automated functional assays use an imaging system that is complex and requires a well-trained user. This protocol has been developed for users with a basic level of experience using this microscope and its software. It is recommended to first train the individuals on the primary use of the instrument and the software by representatives of the manufacturer; this same practice is followed in our lab. A minimum of 4 weeks of training was needed to use this microscope effectively for experiments.
As described above, this methodology has some limitations. Expertise in biopsy collection, quick processing time, and facility with feeder fibroblasts are needed to successfully expand and culture HNE organoids. This method was developed using specific reagents and equipment that may not be universally available. The methodology has proved useful for research in cystic fibrosis3,11,13 and may not be as applicable to other disease processes25. However, other methods and equipment may be used to develop similar strategies.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We gratefully acknowledge the contributions of all the participants who donated HNE brush biopsies to develop this protocol. We thank Latona Kersh and Children's Research Unit staff for coordinating study volunteer recruitment and sample collections. We thank Lily Deng, Johnathan Bailey, and Stephen Mackay, former trainees in our laboratory, for technical assistance. We thank Zhong Liu and Rui Zhao for their technical help. Steven M. Rowe, Director of the CF Research Center at UAB, provides leadership and resources, without which this work would not be possible. We also would like to thank Sarah Guadiana at Biotek for assistance with instrument training, Robert Grabski for confocal microscopy assistance at the UAB High-Resolution Imaging Facility, and Dezhi Wang for histological assistance at the UAB Histology Core. This work was supported by the National Institutes of Health (NIH.) Grant K23HL143167 (to JSG), Cystic Fibrosis Foundation (CFF) Grant GUIMBE18A0-Q (to JSG), the Gregory Fleming James Cystic Fibrosis Center [NIH Grants R35HL135816 and DK072482 and the CFF University of Alabama at Birmingham (UAB) Research and Development Program (Rowe19RO)], and the UAB Center for Clinical and Translational Science (NIH Grant UL1TR001417).
Materials
| Nasal brush | Medical Packaging CYB1 | CYB-1 | Length: 8 inches, width approximately 7 mm |
| Large-Orifice Pipette Tips | ThermoFisher Scientific | 02-707-141 | Large bore pipette tips |
| Accutase | ThermoFisher Scientific | A1110501 | Cell detachment solution |
| 0.05% trypsin -EDTA | Gibco | 25300-054 | |
| Trypsin inhibitor from soybean | Sigma | T6522 | Working solution: 1mg/mL in 1XDPBS |
| Matrigel matrix | Corning | 356255 | Extracellular matrix (EM) |
| µ-Slide Angiogenesis | Ibidi | 81506 | 15-well slide |
| 24-Well Transwell | Corning | 7200154 | Culture insert |
| Chambered Coverglass | ThermoFisher Scientific | 155409 | 8-well glass-bottom chamber slides |
| Cell-Tak Cell and Tissue Adhesive | ThermoFisher Scientific | 354240 | Cell adhesive |
| Paraformaldehyde | Electron Microscopy Sciences | 50980487 | |
| Triton X-100 | Alfa Aesar | A16046 | |
| BSA | ThermoFisher Scientific | BP1600-100 | |
| NucBlue | ThermoFisher Scientific | R37605 | DAPI |
| Eclipse Ts2-FL (Inverted Routine Microscope) | Nikon | Inverted epi-fluorescence microscope or bright-field microscope | |
| Nikon A1R-HD25 | Nikon | Confocal microscope | |
| NIS Elements- Basic Research | Nikon | manual imaging analysis software | |
| Histogel | ThermoFisher Scientific | HG-4000-012 | |
| Disposable Base Molds | ThermoFisher Scientific | 41-740 | |
| Lionheart FX | BioTek | BTLFX | Automated image system |
| Lionheart Cover | BioTek | BT1450009 | Environmental Control Lid |
| Humidity Chamber | BioTek | BT1450006 | Stage insert (environmental chamber) |
| Gas Controller for CO2 and O2 | BioTek | BT1210013 | Gas controller |
| Microplate/Slide Stage Insert | BioTek | BT1450527 | Slide holder |
| Gen5 Imaging Prime Software | BioTek | BTGEN5IPRIM | Automated imaging analysis software |
| 4x Phase Contrast Objective | BioTek | BT1320515 | |
| 10x Phase Contrast Objective | BioTek | BT1320516 | |
| LED Cube | BioTek | BT1225007 | |
| Filter Cube (DAPI) | BioTek | BT1225100 | DAPI |
| CFTRinh-172 | Selleck Chemicals | S7139 | |
| Forskolin | Sigma | F6886 | |
| IBMX | Sigma | I5879 | |
| Expansion Media | |||
| DMEM | ThermoFisher Scientific | 11965 | |
| F12 Nutrient mix | ThermoFisher Scientific | 11765 | |
| Fetal Bovine Serum | ThermoFisher Scientific | 16140-071 | |
| Penicillin/Streptomycin | ThermoFisher Scientific | 15-140-122 | |
| Cholera Toxin | Sigma | C8052 | |
| Epidermal Growth Factor (EGF) | ThermoFisher Scientific | PHG0314 | |
| Hydrocortisone (HC) | Sigma | H0888 | |
| Insulin | Sigma | I9278 | |
| Adenine | Sigma | A2786 | |
| Y-27632 | Stemgent | 04-0012-02 | |
| Antibiotic Media | |||
| Ceftazidime | Alfa Aesar | J66460-03 | |
| Tobramycin | Alfa Aesar | J67340 | |
| Vancomycin | Alfa Aesar | J67251 | |
| Amphotericin B | Sigma | A2942 | |
| Differentiation Media | |||
| DMEM/F-12 (1:1) | ThermoFisher Scientific | 11330-32 | |
| Ultroser-G | Pall | 15950-017 | |
| Fetal Clone II | Hyclone | SH30066.03 | |
| Bovine Brain Extract | Lonza | CC-4098 | |
| Insulin | Sigma | I-9278 | |
| Hydrocortisone | Sigma | H-0888 | |
| Triiodothyronine | Sigma | T-6397 | |
| Transferrin | Sigma | T-0665 | |
| Ethanolamine | Sigma | E-0135 | |
| Epinephrine | Sigma | E-4250 | |
| O-Phosphorylethanolamine | Sigma | P-0503 | |
| Retinoic Acid | Sigma | R-2625 | |
| Primary antibodies | |||
| Human CFTR antibody | R&D Systems | MAB1660 | Dilution: 100x |
| ZO-1 antibody | Thermo Fisher | MA3-39100-A647 | Dilution: 1000x |
| Anti-MUC5B antibody | Sigma | HPA008246 | Dilution: 100x |
| Anti-acetylated tubulin | Sigma | T7451 | Dilution: 100x |
| Anti-beta IV Tubulin antibody | Abcam | Ab11315 | Dilution: 100x |
| Secondary antibodies | |||
| Donkey anti-Mouse IgG (H+L), Alexa Fluor 488 | Invitrogen | A21202 | Dilution: 2000x |
| Donkey anti-Rabbit IgG (H+L), Alexa Fluor 594 | Invitrogen | A21207 | Dilution: 2000x |
Riferimenti
- Brewington, J. J., et al. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight. 3 (13), (2018).
- Brewington, J. J., et al. Generation of human nasal epithelial cell spheroids for individualized cystic fibrosis transmembrane conductance regulator study. Journal of Visualized Experiments: JoVE. (134), e57492 (2018).
- Brewington, J. J., et al. Detection of CFTR function and modulation in primary human nasal cell spheroids. Journal of Cystic Fibrosis. 17 (1), 26-33 (2017).
- Bridges, M. A., Walker, D. C., Davidson, A. G. Cystic fibrosis and control nasal epithelial cells harvested by a brushing procedure. In Vitro Cellular & Developmental Biology. 27 (9), 684-686 (1991).
- Bridges, M. A., Walker, D. C., Harris, R. A., Wilson, B. R., Davidson, A. G. Cultured human nasal epithelial multicellular spheroids: polar cyst-like model tissues. Biochemistry and Cell Biology. 69 (2-3), 102-108 (1991).
- Collie, G., Buchwald, M., Harper, P., Riordan, J. R. Culture of sweat gland epithelial cells from normal individuals and patients with cystic fibrosis. In Vitro Cellular & Developmental Biology. 21 (10), 597-602 (1985).
- Conger, B. T., et al. Comparison of cystic fibrosis transmembrane conductance regulator (CFTR) and ciliary beat frequency activation by the CFTR Modulators Genistein, VRT-532, and UCCF-152 in primary sinonasal epithelial cultures. JAMA Otolaryngology-Head & Neck Surgery. 139 (8), 822-827 (2013).
- de Courcey, F., et al. Development of primary human nasal epithelial cell cultures for the study of cystic fibrosis pathophysiology. American Journal of Physiology-Cell Physiology. 303 (11), 1173-1179 (2012).
- Gruenert, D. C., Basbaum, C. B., Widdicombe, J. H. Long-term culture of normal and cystic fibrosis epithelial cells grown under serum-free conditions. In Vitro Cellular & Developmental Biology. 26 (4), 411-418 (1990).
- Mosler, K., et al. Feasibility of nasal epithelial brushing for the study of airway epithelial functions in CF infants. Journal of Cystic Fibrosis. 7 (1), 44-53 (2008).
- Anderson, J. D., Liu, Z., Odom, L. V., Kersh, L., Guimbellot, J. S. CFTR function and clinical response to modulators parallel nasal epithelial organoid swelling. The American Journal of Physiology – Lung Cellular and Molecular Physiology. 321 (1), 119-129 (2021).
- Guimbellot, J. S., et al. Nasospheroids permit measurements of CFTR-dependent fluid transport. JCI Insight. 2 (22), (2017).
- Liu, Z., et al. Human nasal epithelial organoids for therapeutic development in cystic fibrosis. Genes (Basel). 11 (6), (2020).
- Muller, L., Brighton, L. E., Carson, J. L., Fischer, W. A., Jaspers, I. Culturing of human nasal epithelial cells at the air liquid interface. Journal of Visualized Experiments: JoVE. , (2013).
- Dekkers, J. F., vander Ent, C. K., Beekman, J. M. Novel opportunities for CFTR-targeting drug development using organoids. Rare Diseases. 1, 27112 (2013).
- Dekkers, J. F., et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature Medicine. 19 (7), 939-945 (2013).
- Okiyoneda, T., et al. Mechanism-based corrector combination restores DeltaF508-CFTR folding and function. Nature Chemical Biology. 9 (7), 444-454 (2013).
- Schwank, G., et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13 (6), 653-658 (2013).
- Geurts, M. H., et al. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell Stem Cell. 26 (4), 503-510 (2020).
- Bove, P. F., et al. Breaking the in vitro alveolar type II cell proliferation barrier while retaining ion transport properties. American Journal of Respiratory Cell and Molecular Biology. 50 (4), 767-776 (2014).
- Chapman, S., Liu, X., Meyers, C., Schlegel, R., McBride, A. A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. Journal of Clinical Investigation. 120 (7), 2619-2626 (2010).
- Liu, X., et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. The American Journal of Pathology. 180 (2), 599-607 (2012).
- Palechor-Ceron, N., et al. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. The American Journal of Pathology. 183 (6), 1862-1870 (2013).
- Scudieri, P., et al. Ionocytes and CFTR chloride channel expression in normal and cystic fibrosis nasal and bronchial epithelial cells. Cells. 9 (9), (2020).
- Marthin, J. K., Stevens, E. M., Larsen, L. A., Christensen, S. T., Nielsen, K. G. Patient-specific three-dimensional explant spheroids derived from human nasal airway epithelium: a simple methodological approach for ex vivo studies of primary ciliary dyskinesia. Cilia. 6, 3 (2017).
- Blouquit, S., et al. Ion and fluid transport properties of small airways in cystic fibrosis. American Journal of Respiratory Cell and Molecular Biology. 174 (3), 299-305 (2006).
- Birket, S. E., et al. Combination therapy with cystic fibrosis transmembrane conductance regulator modulators augment the airway functional microanatomy. The American Journal of Physiology – Lung Cellular and Molecular Physiology. 310 (10), 928-939 (2016).
- Birket, S. E., et al. A functional anatomic defect of the cystic fibrosis airway. American Journal of Respiratory and Critical Care Medicine. 190 (4), 421-432 (2014).
- Chu, K. K., et al. Particle-tracking microrheology using micro-optical coherence tomography. Biophysical Journal. 111 (5), 1053-1063 (2016).
- Chu, K. K., et al. et al. In vivo imaging of airway cilia and mucus clearance with micro-optical coherence tomography. Biomedical Optics Express. 7 (7), 2494-2505 (2016).
- Liu, L., et al. Method for quantitative study of airway functional microanatomy using micro-optical coherence tomography. PLoS One. 8 (1), 54473 (2013).
- Tuggle, K. L., et al. Characterization of defects in ion transport and tissue development in cystic fibrosis transmembrane conductance regulator (CFTR)-knockout rats. PLoS One. 9 (3), 91253 (2014).
- McCravy, M. S., et al. Personalised medicine for non-classic cystic fibrosis resulting from rare CFTR mutations. European Respiratory Journal. 56 (1), 2000062 (2020).
- Mutyam, V., et al. Therapeutic benefit observed with the CFTR potentiator, ivacaftor, in a CF patient homozygous for the W1282X CFTR nonsense mutation. Journal of Cystic Fibrosis. 16 (1), 24-29 (2017).
- Corning Inc. . CORNING CELL-TAK CELL AND TISSUE ADHESIVE. , (2013).
- Anderson, J. D., Liu, Z., Odom, L. V., Kersh, L., Guimbellot, J. S. CFTR function and clinical response to modulators parallel nasal epithelial organoid swelling. The American Journal of Physiology – Lung Cellular and Molecular Physiology. 321 (1), 119-129 (2021).
- Biotek Instruments, Incorporated. . Lionheart FX Live Cell Imager Operator’s Manual. , (2016).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 9 (7), 671-675 (2012).
- Simmonds, N. J. Is it cystic fibrosis? The challenges of diagnosing cystic fibrosis. Paediatric Respiratory Reviews. 31, 6-8 (2019).
- McGarry, M. E., et al. In vivo and in vitro ivacaftor response in cystic fibrosis patients with residual CFTR function: N-of-1 studies. Pediatric Pulmonology. 52 (4), 472-479 (2017).
- Garratt, L. W., et al. Determinants of culture success in an airway epithelium sampling program of young children with cystic fibrosis. Experimental Lung Research. 40 (9), 447-459 (2014).

