Stab Wound Injury Model of the Adult Optic Tectum Using Zebrafish and Medaka for the Comparative Analysis of Regenerative Capacity
Summary
A mechanical brain injury model in the adult zebrafish is described to investigate the molecular mechanisms regulating their high regenerative capacity. The method explains to create a stab wound injury in the optic tectum of multiple species of small fish to evaluate the regenerative responses using fluorescent immunostaining.
Abstract
While zebrafish have a superior capacity to regenerate their central nervous system (CNS), medaka has a lower CNS regenerative capacity. A brain injury model was developed in the adult optic tectum of zebrafish and medaka and comparative histological and molecular analyses were performed to elucidate the molecular mechanisms regulating the high regenerative capacity of this tissue across these fish species. Here a stab wound injury model is presented for the adult optic tectum using a needle and histological analyses for proliferation and differentiation of the neural stem cells (NSCs). A needle was manually inserted into the central region of the optic tectum, and then the fish were intracardially perfused, and their brains were dissected. These tissues were then cryosectioned and evaluated using immunostaining against the appropriate NSC proliferation and differentiation markers. This tectum injury model provides robust and reproducible results in both zebrafish and medaka, allowing for comparing NSC responses after injury. This method is available for small teleosts, including zebrafish, medaka, and African killifish, and enables us to compare their regenerative capacity and investigate unique molecular mechanisms.
Introduction
Zebrafish (Danio rerio) have an increased ability to regenerate their central nervous system (CNS) compared to other mammals1,2,3. Recently, to better understand the molecular mechanisms underlying this increased regenerative capacity, comparative analyses of tissue regeneration using next-generation sequencing technology have been performed4,5,6. The brain structures in zebrafish and tetrapods are quite different7,8,9. This means that several brain injury models using small fish with similar brain structures and biological features have been developed to facilitate the investigation of the underlying molecular mechanisms contributing to this increased regenerative capacity.
In addition, medaka (Oryzias latipes) is a popular laboratory animal with a low capacity for heart and neuronal regeneration10,11,12,13 compared with zebrafish. Zebrafish and medaka have similar brain structures and niches for adult neural stem cells (NSCs)14,15,16,17. In zebrafish and medaka, the optic tectum includes two types of NSCs, neuroepithelial-like stem cells and radial glial cells (RGCs)15,18. A stab wound injury for the optic tectum of adult zebrafish was previously developed, and this model was used to investigate the molecular mechanisms regulating brain regeneration in these animals19,20,21,22,23. This young adult zebrafish stab wound injury model induced regenerative neurogenesis from RGCs19,24,25. This stab wound injury in the optic tectum is a robust and reproducible method13,19,20,21,22,23,24,25. When the same injury model was applied to adult medaka, the low neurogenic capacity of RGCs in medaka optic tectum was revealed via the comparative analysis of RGC proliferation and differentiation following injury13.
Stab wound injury models in the optic tectum have also been developed in mummichog models26, but details of the tectum injury have been not well documented when compared with telencephalic injury27. The stab wound injury in the optic tectum using zebrafish and medaka allows the investigation of the differential cellular responses and gene expression between species with differential regenerative capacity. This protocol describes how to perform a stab wound injury in the optic tectum using an injection needle. This method can be applied to small fish like zebrafish and medaka. The processes for sample preparation for histological analysis and cellular proliferation and differentiation analysis using fluorescent immunohistochemistry and cryosections are explained here.
Protocol
All experimental protocols were approved by the Institutional Animal Care and Use Committee at the National Institute of Advanced Industrial Science and Technology. Zebrafish and medaka were maintained according to standard procedures28.
1. Stab wound injury in the adult optic tectum
- Prepare a 0.4% (w/v) tricaine stock solution for anesthesia. For a 100 mL stock solution, dissolve 400 mg tricaine methanesulfonate (see Table of Materials) in 90 mL of distilled water and adjust the pH to 7.0 using 1 M Tris HCl buffer (pH 9.0). Once the pH is adjusted, add water up to 100 mL, then make appropriate aliquots and store them at -20 °C.
- To anesthetize the adult zebrafish or medaka, prepare a 0.02 % (w/v) tricaine solution by diluting the 0.4 % tricaine stock solution with fish facility water.
- Anesthetize the fish with 0.02% tricaine solution; they become anesthetized when they do not move or respond to touch (1-2 min).
- Place the anesthetized fish on a Styrofoam tray (see Table of Materials) and fix the fish upright between two 30 G needles inserted vertically into the Styrofoam.
- Hold the fish body to prevent the fish head from moving and vertically insert a 30 G needle through the skull into the medial region of one of the optic tectum hemispheres (Figure 1A,B). The insertion depth is ~0.75 mm, almost equal to half the length of the 30 G bevel. This insertion depth induces brain injury on zebrafish and medaka because of the similar body size of these fish.
NOTE: Medaka has scales on the skull. Scratch and remove the scales using the 30 G needle to efficiently and accurately induce the stab wound injury (Figure 1C). - Transfer the injured fish into a fish tank with fresh fish facility water and move the tank to the fish breeding system once they have fully recovered from the anesthesia.
NOTE: Fish will recover and start freely moving within a few minutes. - For lineage analysis after the injury, prepare fresh fish facility water containing 5 mM bromodeoxyuridine (BrdU) (see Table of Materials) and then incubate the fish with this 5 mM BrdU solution for a predetermined amount of time13,19 (as determined by an experimental design). Then dissect these fish as per step 2 and evaluate BrdU incorporation and cell lineage identity as appropriate.
NOTE: This step is an optional step for cell lineage analysis. To detect BrdU signals, perform antigen retrieval as shown in step 4.3.
2. Brain dissection
- Prepare 0.02% tricaine solution, a Styrofoam tray for dissection, and a 10 mL syringe containing phosphate-buffered saline (1x PBS) with an extension tube and 30 G needles (see Table of Materials) for intracardial perfusion.
- Anesthetize the injured fish at selected time points.
- Place paper towels on the Styrofoam tray and place the anesthetized fish on the paper towels.
- Hold the fish body in place by vertically inserting two 30 G needles on either side of the anal fin (Figure 2A).
- Make a ventral incision from the anus to the heart (Figure 2B) and keep the heart visible by inserting another two 30 G needles on each side of this organ (Figure 2C).
NOTE: The heart is covered in the silvery epithelial layer of the hypodermis in both zebrafish and medaka (Figure 2C). - Gently remove the silver epithelial layer with the tip of the forceps (Figure 2D).
- Insert the 30 G needle into the ventricle, keeping the bevel up, and make an incision into the atrium using the forceps (Figure 2E). Carefully press down on the syringe to push the 1x PBS and confirm that the atrium flushed the blood.
NOTE: To prevent the needle from penetrating the ventricle, carefully insert the needle to the depth of the half bevel and then adjust the insertion depth. If the perfusion is well done, gills turn white (Figure 2F,G). - Stop the perfusion after the blood is drained and the gills turn white.
- Remove the needles fixing the fish body in place and fix the fish ventral side down (Figure 2H). Cut the spinal cord and remove the skull from the optic tectum and telencephalon (Figure 2I). Then, cut the optic nerves and dissect the brain carefully.
NOTE: Watch out for the optic nerves during the dissection because the nerves are connected to the tectum. When the optic nerve is pulled, the optic tectum can be detached from the brain. If the blood removal is not complete, the brain looks light pink (the right brain in Figure 2J). - Transfer the brains into 1.5 mL tubes with 1 mL of 4 % paraformaldehyde in 1x PBS and fix them overnight at 4 °C.
3. Preparation of frozen sections
- Wash the fixed brains three times in 1x PBS for 5 min each.
- To cryoprotect the brains, transfer them into 1.5 mL tubes with 1 mL of 30 % (w/v) sucrose in 1x PBS and incubate them overnight at 4 °C. Prepare the embedding compound (2:1 mixture of OCT compound and 30% sucrose, see Table of Materials) by combining 30 mL of OCT and 15 mL of 30 % sucrose. Store this at 4 °C.
NOTE: To remove the bubbles from the mixed compound, centrifuge the 50 mL tubes at 10,000 x g for 2 min at room temperature or place the tubes upright overnight. - Incubate an aluminum block overnight at -80 °C in order to cool the cryomold (see Table of Materials) immediately after the dissected brain is embedded in the cryomold with the embedding compound.
NOTE: Liquid nitrogen can also be used to cool the aluminum block. In that case, the aluminum block should be placed in a Styrofoam box filled with enough liquid nitrogen to cover the aluminum block just before embedding. After confirming the liquid nitrogen sublimation, place the cryomold on the precooled aluminum block. It is better to confirm how many cryomolds can be placed on the aluminum block at once. It is advisable to use liquid nitrogen or an additional precooled block to ensure enough space to cool all samples quickly. - Put the precooled aluminum block in a Styrofoam box. Use a Styrofoam box with a lid to prevent the aluminum block from warming.
- For coronal sections, embed the brain in a cryomold and adjust the orientation using forceps under the microscope (Figure 3A).
- Place the cryomold on the precooled aluminum block in the Styrofoam box and allow the OCT compound to freeze (Figure 3B). When the OCT compound starts to freeze, it becomes white.
- Apply a small circle of OCT compound on a specimen disc, and mount the cryoblock to attach the cryoblock to the specimen disc.
- Place the specimen disc in the cryostat and freeze the OCT compound completely.
- Place the specimen disc on the specimen head in the cryostat.
- Set the orientation of the cryoblock and trim the OCT to remove any extra regions.
NOTE: Adjust the orientation of the cutting plane while cutting the telencephalon. The blade is equipped with a cryostat. It is possible to adjust the orientation of the cutting plane by changing the angle of the specimen head. - Cut 14 µm thick serial sections through the whole optic tectum using a cryostat. Store the slides at -25 °C.
NOTE: Long-term storage should be at -80 °C.
4. Fluorescent immunostaining
- Dry glass slides in a slide rack for 30 min at room temperature (RT). Wash slides in 1x PBS for 30 min at RT. If immunostaining with anti-PCNA or BrdU antibodies is to be performed, proceed to step 4.2 or step 4.3.
- Prepare 500 mL of 10 mM sodium citrate in a 500 mL beaker and warm the citrate buffer to 85 °C on a hot plate magnetic stirrer, then place the slides in a slide staining rack and incubate the rack in the citrate buffer at 85 °C for 30 min. To avoid boiling, keep the temperature around 85 °C. After antigen retrieval, wash the slides three times in 0.1 % Triton in 1x PBS (PBSTr), with each wash taking 5 min at RT.
NOTE: This step is an optional step for proliferating cell nuclear antigen (PCNA) retrieval. - Prepare a 2 N HCl solution by diluting 12 N HCl in distilled water. Using a liquid blocker (see Table of Materials), draw a line as a hydrophobic barrier to keep the 2 N HCl solution around the sections.
- Place the slides on immunostaining trays and apply 200-300 µL of the 2N HCl solution. Incubate the trays at 37 °C for 30 min. After antigen retrieval, wash three times in 0.1 % PBSTr with each wash taking 5 min at RT.
NOTE: This step is an optional step for BrdU antigen retrieval.
- Place the slides on immunostaining trays and apply 200-300 µL of the 2N HCl solution. Incubate the trays at 37 °C for 30 min. After antigen retrieval, wash three times in 0.1 % PBSTr with each wash taking 5 min at RT.
- Absorb and remove the remaining solution using paper towels. Then, use a liquid blocker to draw a hydrophobic barrier to keep the antibody solution around the sections as necessary.
- Place the slides on trays and apply 200-300 µL of blocking solution, 3% (v/v) horse serum in PBSTr) to every slide, and incubate the slides for 1 h at RT.
- Wash the sections with PBSTr for 5 min at RT and repeat three times.
- Prepare the primary antibodies (see Table of Materials) using the blocking solution (200-300 µL of antibody solution for each slide). Remove the remaining PBSTr with paper towels and place the slides on the immunostaining trays. Apply the primary antibody solution to every slide and incubate trays overnight at 4 °C.
- Wash the sections with PBSTr for 5 min at RT and repeat three times.
- Prepare fluorescent secondary antibody solution (see Table of Materials) using the blocking buffer (1:500). Remove the remaining PBSTr with paper towels and place the slides on the immunostaining trays. Apply the secondary antibody solution to every slide and shade trays with aluminum foil. Incubate for 1-2 h at RT.
- Wash the sections with PBSTr for 5 min at RT three times without exposing them to light.
- Prepare Hoechst solution (1:500 dilution in 1x PBS, see Table of Materials) for nuclear staining. Remove the remaining PBSTr with paper towels and place the slides on the immunostaining trays. Apply the Hoechst solution to every slide and cover the trays with aluminum foil. Incubate for 30 min at RT.
- Wash the sections with 1x PBS for 10 min at RT without exposing them to light.
- Absorb and remove the remaining 1x PBS using paper towels and mount the sections on slides using a water-soluble mounting medium (see Table of Materials) for fluorescent immunostaining. Slowly set a 24 x 45 mm coverslip onto the sections. Store at 4 °C, without exposing the slides to light.
- Observe and image the sections under fluorescent microscopy or confocal microscopy.
Representative Results
Stab wound injury in the optic tectum using needle insertion into the right hemisphere (Figure 1, Figure 4A, and Figure 5A) induces various cellular responses, including radial glial cell (RGC) proliferation and the generation of newborn neurons. Similarly, aged populations of zebrafish and medaka were used to counteract any aging effects in the regenerative response. Then fluorescent immunostaining was performed on the frozen sections, and the RGC proliferation and differentiation were analyzed after the tectum injury in the zebrafish and medaka (Figure 4-5)13.
Antibodies against a proliferating cell marker were used, proliferating cell antigen (PCNA), and an RGC marker, brain lipid-binding protein (BLBP), available in zebrafish and medaka to evaluate the RGC proliferation in these tissues13,19. As previously described, most of the RGCs were quiescent (PCNA negative) in the contralateral uninjured hemisphere (Figure 4B)13. Still, RGC proliferation was induced at 2 days post-injury (dpi) in the medaka tectum (Figure 4C,D)13. Induction of RGC proliferation after the injury is a common feature of both the zebrafish and medaka regenerative responses13,19.
BrdU labeling is a simple method used to evaluate cell lineage and analyze RGC differentiation after brain injury (Figure 5A)13. Immunostaining with antibodies for a pan-neuronal marker, HuC, and BrdU was previously used to compare RGC differentiation in the injured tectum of zebrafish and medaka (Figure 5B,C)13. If these antibodies are available in the target species, comparative analyses can be performed.
If the injury is appropriately induced, the injury site is located in the central-dorsal region in the optic tectum (Figure 4C). Nuclear staining and hematoxylin and eosin staining can then be used to confirm the injury site13,19,20,21,22,23,24,25. After the injury, a disturbed periventricular gray zone with nuclear staining can be observed (Figure 4C). If the injury is located in the medial dorsal region, RGC proliferation is not significantly increased25.
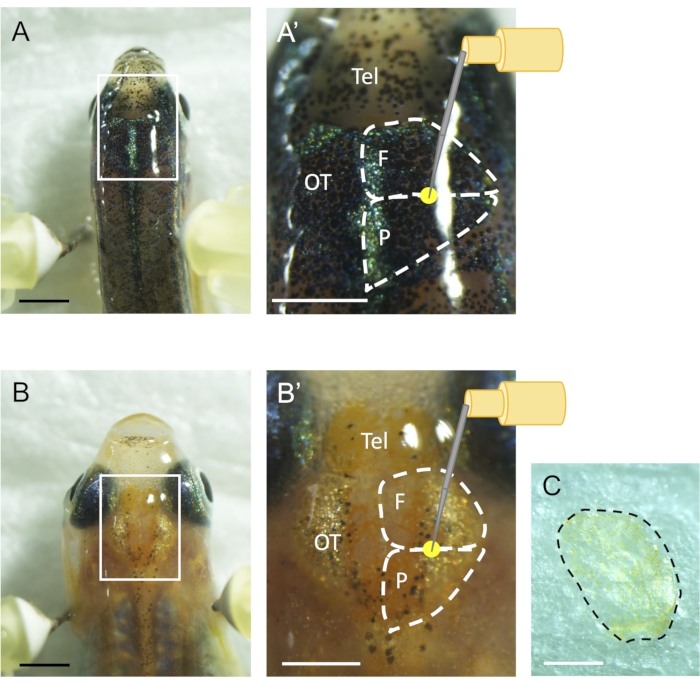
Figure 1: Stab wound injury in adult optic tectum using a needle. (A) Dorsal view of an adult zebrafish. Zebrafish is kept upright between two bent needles inserted vertically into a Styrofoam. (A') Magnified image of the boxed area in (A). 30 G needle is inserted into the medial region of the border between two skulls called os frontale and os parietale on the optic tectum. The yellow circle indicates the injury site, and white dashed lines indicate the two skulls on the optic tectum. (B) Dorsal view of adult medaka. (B') A magnified image of the boxed area in (B). 30 G needle is inserted into the border on the optic tectum. The yellow circle indicates the injury site, and white dashed lines indicate two skulls on the optic tectum. (C) Medaka has scales on the skull. Skull scales are to be removed before the stab wound injury. The dashed line indicates the scale on the optic tectum. Scale bar: 2 mm in A-B, 1 mm in A', B' and C. Telencephalon (Tel), optic tectum (OT), os frontale (F), and os parietale (P). Please click here to view a larger version of this figure.
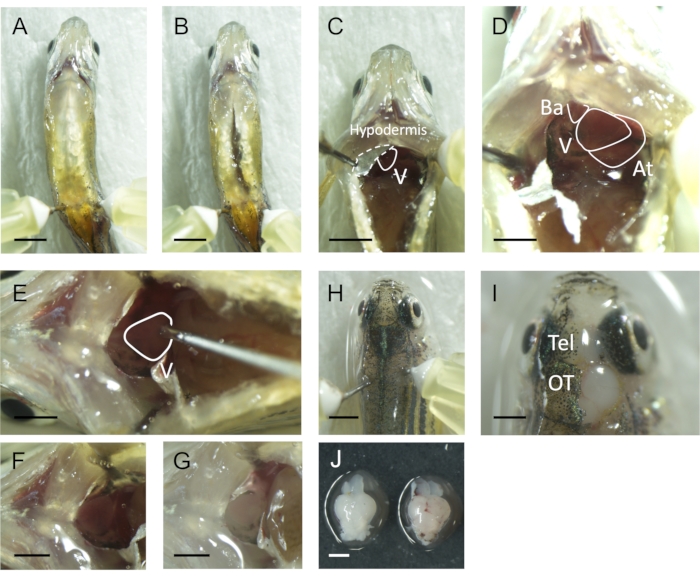
Figure 2: Intracardiac perfusion in small adult fish. (A) Ventral view of a zebrafish fixed on Styrofoam using bent needles ready for intracardiac perfusion and brain dissection. (B) A ventral incision is made from the origin of the anal fins to the chest. (C) The heart is behind a silvery epithelial layer called the hypodermis in both zebrafish and medaka. Another fixation using a bent needle beside the silver epithelial layer allows for easier access. The solid white line indicates the ventricle (V), and the dotted line indicates the hypodermis. (D) The silver epithelial layer is removed before the intracardial perfusion of 1x PBS. (E-G) Canula is inserted into the ventricle for intracardiac perfusion. Gills before (F) and after (G) the intracardiac perfusion. If the blood removal is not complete, the gills remain red. (H-J) Brain dissection after PBS perfusion to remove the blood from the tissues. Remove skulls on the optic tectum and telencephalon as shown in (I). If the blood removal is not complete, the brain looks light pink (the right brain in (J)). Scale bar: 2 mm in A-C and H, 1 mm in D-G and I-J. The olfactory bulb (OB), telencephalon (Tel), optic tectum (OT), bulbus arteriosus (Ba), ventricle (V), and atrium (At). Please click here to view a larger version of this figure.
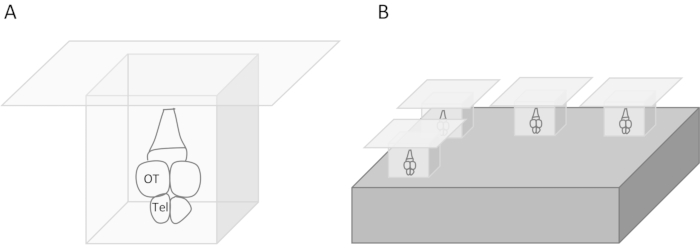
Figure 3: Brain embedding for frozen sections. (A) The brain is embedded in a cryomold with an embedding compound. Anterior is down. (B) Cryomolds are cooled on a precooled aluminum block. Telencephalon (Tel), optic tectum (OT). Please click here to view a larger version of this figure.
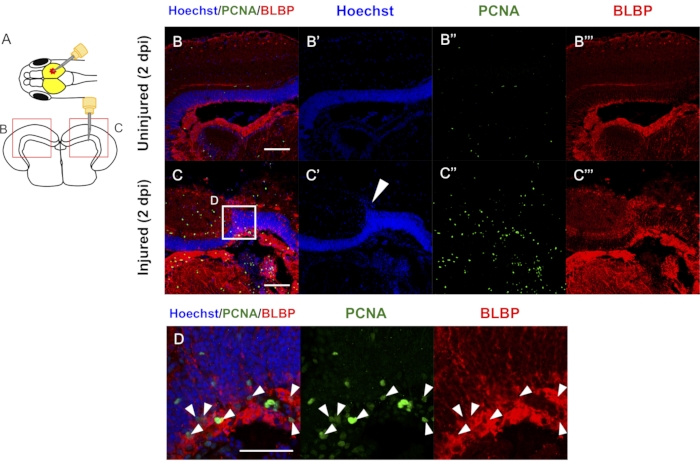
Figure 4: Representative results of fluorescent immunostaining against RGC proliferation after tectum injury in adult medaka. (A) Schematic view of the stab wound injury to the right hemisphere of the optic tectum and coronal section. (B-C) Representative results of proliferative RGCs (PCNA + BLBP + cells) in the contralateral uninjured (B) and injured (C) side at 2 days post-injury. White arrowhead in C' indicates a disturbed periventricular gray zone by the stab wound injury. (D) Magnified images of the boxed area in (C). White arrowheads in (D) indicate PCNA + BLBP + cells. Scale bar: 50 µm in B-D. Adapted with permission from Reference13. Please click here to view a larger version of this figure.
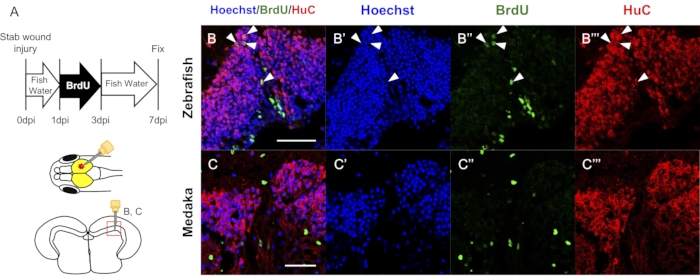
Figure 5: Representative results of the fluorescent immunostaining for the generation of newborn neurons after tectum injury. (A) Schematic view of bromodeoxyuridine (BrdU) treatment and the stab wound injury in the optic tectum and coronal section. (B-C) Representative results of newborn neurons (BrdU + HuC + cells) at 7 days post-injury in the injured zebrafish (B) and medaka (C). Scale bar: 50 µm in B-C. Adapted with permission from Reference13. Please click here to view a larger version of this figure.
Discussion
Here a set of methods is described which can be used to induce stab wound injuries in the optic tectum utilizing a needle to facilitate the evaluation of RGC proliferation and differentiation after brain injury. Needle-mediated stab wounds are a simple, efficiently implemented method that can be applied to many experimental samples using a standard set of tools. Stab wound injury models for several regions of the zebrafish brain have been developed3,19,29. The optic tectum is one of the most largest parts of the brain and is easy to manipulate. Moreover, most RGCs in the optic tectum are quiescent under physiological conditions when compared to the telencephalon, making it easier to observe RGC proliferation and differentiation depending on the injury3,19.
One of the critical steps and limitations in stab wound injuries is manual needle insertion; a consistent injury is necessary for creating reproducible results and facilitating comparative analysis. The precise location and depth of insertion are crucial and help create reproducible injuries in experiments. This paper provides clear guidelines for making similar injuries each time. Moreover, the proliferation of RGC after the injury is essential for the neurogenesis of the injured tectum. In injured zebrafish and medaka, RGC proliferation increases at 1 dpi and returns to basal levels, the same as in the contralateral uninjured hemisphere, at 7 dpi13,19.
Stab wound injury is one of the mechanical injury methods that induce non-specific cell ablation. In contrast, transgenic approaches to cell-specific ablation such as nitroreductase/metronidazole system have also been developed30,31,32. These ablation models should be selected based on the experimental purpose. Non-specific ablation is suitable for brain injuries such as stab wounding and ischemic stroke. In contrast, cell-specific ablation might be more appropriate for evaluating the degeneration of specific cells associated with neurodegenerative diseases such as Parkinson's disease.
Recently, ischemic injury models using zebrafish have been developed33, but these models need transgenic lines and fluorescent microscopy to monitor blood flow. Therefore, these models are challenging to apply to species with poor genetic approaches, and their throughput is lower than the stab wound injury model.
As mentioned above, stab wound injury in the optic tectum is simple and easily applied in other small fish models such as African killifish and mummichog with common tools26. Furthermore, comparative analysis between species has been well investigated using sequencing technology. Therefore, this simple method remains essential when studying the regenerative capacity of NSCs in zebrafish using comparative analysis of cellular responses and gene expression.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 18K14824 and 21K15195 and an internal grant of AIST, Japan.
Materials
| 10 mL syringe | TERUMO | SS-10ESZ | |
| 1M Tris-HCl (pH 9.0) | NIPPON GENE | 314-90381 | |
| 30 G needle | Dentronics | HS-2739A | |
| 4% Paraformaldehyde Phosphate Buffer Solution | Wako | 163-20145 | |
| Aluminum block | 115 x 80 x 37 mm (W x D x H) is enough size to freeze 6 cryomolds | ||
| Anti-BLBP | Millipore | ABN14 | 1:500 |
| Anti-BrdU | Abcam | ab1893 | 1:500 |
| Anti-HuC | Invitrogen | A21271 | 1:100 |
| Anti-PCNA | Santa Cruz Biotechnology | sc-56 | 1:200 |
| Brmodeoxyuridine | Wako | 023-15563 | |
| Confocal microscope C1 plus | Nikon | ||
| Cryomold | Sakura Finetek Japan | 4565 | 10 x 10 x 5 mm (W x D x H) |
| Cryostat | Leica | CM1960 | |
| Danio rerio WT strains RW | |||
| Extension tube | TERUMO | SF-ET3520 | |
| Fluoromount (TM) Aqueous Mounting Medium, for use with fluorescent dye-stained tissues | SIGMA-ALDRICH | F4680-25ML | |
| Forceps | DUMONT | 11252-20 | |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 | Invitrogen | A32723 | |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Invitrogen | A11035 | |
| Hoechst 33342 solution | Dojindo | 23491-52-3 | |
| Hydrochloric Acid | Wako | 080-01066 | |
| Incubation Chamber for 10 slides Dark Orange | COSMO BIO CO., LTD. | 10DO | |
| MAS coat sliding glass | Matsunami glass | MAS-01 | |
| Micro cover glass | Matsunami glass | C024451 | |
| Microscopy | Nikon | SMZ745T | |
| Normal horse serum blocking solution | VECTOR LABRATORIES | S-2000-20 | |
| O.C.T Compound | Sakura Finetek Japan | 83-1824 | |
| Oryzias latipes WT strains Cab | |||
| PAP Pen Super-Liquid Blocker | DAIDO SANGYO | PAP-S | |
| Phosphate Buffered Saline (PBS) Tablets, pH 7.4 | TaKaRa | T9181 | |
| Styrofoam tray | 100 x 100 x 10 mm (W x D x H) styrofoam sheet is available as tray | ||
| Sucrose | Wako | 196-00015 | 30 % (w/v) Sucrose in PBS |
| Tricaine (MS-222) | nacarai tesque | 14805-24 | |
| Trisodium Citrate Dihydrate | Wako | 191-01785 | |
| Triton X-100 | Wako | 04605-250 |
Riferimenti
- Becker, T., Wullimann, M. F., Becker, C. G., Bernhardt, R. R., Schachner, M. Axonal regrowth after spinal cord transection in adult zebrafish. Journal of Comparative Neurology. 377 (4), 577-595 (1997).
- Raymond, P. A., Barthel, L. K., Bernardos, R. L., Perkowski, J. J. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Developmental Biology. 6, 36 (2006).
- März, M., Schmidt, R., Rastegar, S., Strähle, U. Regenerative response following stab injury in the adult zebrafish telencephalon. Developmental Dynamics. 240 (9), 2221-2231 (2011).
- Kang, J., et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 532 (7598), 201-206 (2016).
- Simões, F. C., et al. Macrophages directly contribute collagen to scar formation during zebrafish heart regeneration and mouse heart repair. Nature Communications. 11 (1), 600 (2020).
- Hoang, T., et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science. 370 (6519), (2020).
- Alunni, A., Bally-Cuif, L. A comparative view of regenerative neurogenesis in vertebrates. Development. 143 (5), 741-753 (2016).
- Diotel, N., Lübke, L., Strähle, U., Rastegar, S. Common and distinct features of adult neurogenesis and regeneration in the telencephalon of zebrafish and mammals. Frontiers in Neuroscience. 14, 568930 (2020).
- Labusch, M., Mancini, L., Morizet, D., Bally-Cuif, L. Conserved and divergent features of adult neurogenesis in zebrafish. Frontiers in Cell and Developmental Biology. 8, 525 (2020).
- Ito, K., et al. Differential reparative phenotypes between zebrafish and medaka after cardiac injury. Developmental Dynamics. 243 (9), 1106-1115 (2014).
- Lai, S. L., et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. eLife. 6, 25605 (2017).
- Lust, K., Wittbrodt, J. Activating the regenerative potential of Müller glia cells in a regeneration-deficient retina. eLife. 7, 32319 (2018).
- Shimizu, Y., Kawasaki, T. Differential regenerative capacity of the optic tectum of adult medaka and zebrafish. Frontiers in Cell and Developmental Biology. 9, 686755 (2021).
- Adolf, B., et al. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Biologia dello sviluppo. 295 (1), 278-293 (2006).
- Grandel, H., Kaslin, J., Ganz, J., Wenzel, I., Brand, M. Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Biologia dello sviluppo. 295 (1), 263-277 (2006).
- Alunni, A., et al. Evidence for neural stem cells in the medaka optic tectum proliferation zones. Developmental Neurobiology. 70 (10), 693-713 (2010).
- Kuroyanagi, Y., et al. Proliferation zones in adult medaka (Oryzias latipes) brain. Brain Research. 1323, 33-40 (2010).
- Ito, Y., Tanaka, H., Okamoto, H., Ohshima, T. Characterization of neural stem cells and their progeny in the adult zebrafish optic tectum. Biologia dello sviluppo. 342 (1), 26-38 (2010).
- Shimizu, Y., Ueda, Y., Ohshima, T. Wnt signaling regulates proliferation and differentiation of radial glia in regenerative processes after stab injury in the optic tectum of adult zebrafish. Glia. 66 (7), 1382-1394 (2018).
- Ueda, Y., Shimizu, Y., Shimizu, N., Ishitani, T., Ohshima, T. Involvement of sonic hedgehog and notch signaling in regenerative neurogenesis in adult zebrafish optic tectum after stab injury. Journal of Comparative Neurology. 526 (15), 2360-2372 (2018).
- Kiyooka, M., Shimizu, Y., Ohshima, T. Histone deacetylase inhibition promotes regenerative neurogenesis after stab wound injury in the adult zebrafish optic tectum. Biochemical and Biophysical Research Communications. 529 (2), 366-371 (2020).
- Shimizu, Y., Kawasaki, T. Histone acetyltransferase EP300 regulates the proliferation and differentiation of neural stem cells during adult neurogenesis and regenerative neurogenesis in the zebrafish optic tectum. Neuroscience Letters. 756, 135978 (2021).
- Shimizu, Y., Kiyooka, M., Ohshima, T. Transcriptome analyses reveal IL6/Stat3 signaling involvement in radial glia proliferation after stab wound injury in the adult zebrafish optic tectum. Frontiers in Cell and Developmental Biology. 9, 668408 (2021).
- Lindsey, B. W., et al. Midbrain tectal stem cells display diverse regenerative capacities in zebrafish. Scientific Reports. 9 (1), 4420 (2019).
- Yu, S., He, J. Stochastic cell-cycle entry and cell-state-dependent fate outputs of injury-reactivated tectal radial glia in zebrafish. eLife. 8, 48660 (2019).
- Bisese, E. C., et al. The acute transcriptome response of the midbrain/diencephalon to injury in the adult mummichog (Fundulus heteroclitus). Molecular Brain. 12 (1), 119 (2019).
- Schmidt, R., Beil, T., Strähle, U., Rastegar, S. Stab wound injury of the zebrafish adult telencephalon: a method to investigate vertebrate brain neurogenesis and regeneration. Journal of Visualized Experiments. (4), e51753 (2014).
- Westerfield, M. . The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 5th ed. , (2007).
- Kaslin, J., Kroehne, V., Ganz, J., Hans, S., Brand, M. Distinct roles of neuroepithelial-like and radial glia-like progenitor cells in cerebellar regeneration. Development. 144 (8), 1462-1471 (2017).
- Curado, S., et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Developmental Dynamics. 236 (4), 1025-1035 (2007).
- Shimizu, Y., Ito, Y., Tanaka, H., Ohshima, T. Radial glial cell-specific ablation in the adult zebrafish brain. Genesis. 53 (7), 431-439 (2015).
- Godoy, R., et al. Dopaminergic neurons regenerate following chemogenetic ablation in the olfactory bulb of adult zebrafish (Danio rerio). Scientific Reports. 10 (1), 12825 (2020).
- Sawahata, M., Izumi, Y., Akaike, A., Kume, T. In vivo brain ischemia-reperfusion model induced by hypoxia-reoxygenation using zebrafish larvae. Brain Research Bulletin. 173, 45-52 (2021).

