Measurement of Liver Stiffness Using Atomic Force Microscopy Coupled with Polarization Microscopy
Summary
We present a protocol to measure the elastic moduli of collagen-rich areas in normal and diseased liver using atomic force microscopy. The simultaneous use of polarization microscopy provides high spatial precision for localizing collagen-rich areas in the liver sections.
Abstract
Matrix stiffening has been recognized as one of the key drivers of the progression of liver fibrosis. It has profound effects on various aspects of cell behavior such as cell function, differentiation, and motility. However, as these processes are not homogeneous throughout the whole organ, it has become increasingly important to understand changes in the mechanical properties of tissues on the cellular level.
To be able to monitor the stiffening of collagen-rich areas within the liver lobes, this paper presents a protocol for measuring liver tissue elastic moduli with high spatial precision by atomic force microscopy (AFM). AFM is a sensitive method with the potential to characterize local mechanical properties, calculated as Young's (also referred to as elastic) modulus. AFM coupled with polarization microscopy can be used to specifically locate the areas of fibrosis development based on the birefringence of collagen fibers in tissues. Using the presented protocol, we characterized the stiffness of collagen-rich areas from fibrotic mouse livers and corresponding areas in the livers of control mice.
A prominent increase in the stiffness of collagen-positive areas was observed with fibrosis development. The presented protocol allows for a highly reproducible method of AFM measurement, due to the use of mildly fixed liver tissue, that can be used to further the understanding of disease-initiated changes in local tissue mechanical properties and their effect on the fate of neighboring cells.
Introduction
The liver is a vital organ for maintaining homeostasis in organisms1,2. Chronic liver diseases account for ~2 million deaths worldwide annually3. They originate most commonly as viral infections, autoimmune disorders, metabolic syndromes, or alcohol abuse-related diseases and are accompanied by progressive liver fibrosis. Liver injury elicits an inflammatory response, which leads to the activation of cells depositing extracellular matrix (ECM) in a wound-healing response. However, in the presence of a chronic insult, excess ECM forms unresolved scar tissue within the liver, leading to the development of liver fibrosis, cirrhosis, liver carcinoma, and, ultimately, to liver failure4.
Hepatocyte injury immediately results in increased liver stiffness5,6. This directly affects hepatocyte function, activates hepatic stellate cells (HSCs) and portal fibroblasts, and results in their transdifferentiation to collagen-depositing myofibroblasts7,8. The deposition of fibrous ECM further increases liver stiffness, creating a self-amplifying feedback loop of liver stiffening and matrix-producing cell activation.
Liver stiffness has, thus, become an important parameter in liver disease prognosis. The change in biomechanical tissue properties can be detected earlier than fibrosis can be diagnosed by histological analysis. Therefore, various techniques for liver stiffness measurement have been developed in both research and clinical applications. In clinical settings, transient elastography (TE)9,10,11,12,13 and magnetic resonance elastography (MRE)14,15,16,17,18 have been employed to non-invasively diagnose early stages of liver damage by examining gross liver stiffness19.
In TE, ultrasound waves of mild amplitude and low frequency (50 Hz) are propagated through the liver, and their velocity is measured, which is then used to calculate tissue elastic modulus13. However, this technique is not useful for patients with ascites, obesity, or lower intercostal spaces due to improper transmission of the ultrasound waves through the tissues surrounding the liver9.
MRE is based on magnetic resonance imaging modality and uses 20-200 Hz mechanical shear waves to target the liver. A specific magnetic resonance imaging sequence is then used to trace the waves inside the tissue and to calculate the tissue stiffness16. Stiffness values reported with both TE and MRE techniques correlate well with the degree of liver fibrosis obtained from biopsies of human liver samples ranked using histological METAVIR scores20 (Table 1). TE and MRE have also been adapted for the measurement of liver stiffness in rodent models for research purposes21,22,23. However, as both methods derive the stiffness values from the tissue's response to the propagating shear waves, the values obtained might not reflect the absolute mechanical stiffness of the tissue.
For a direct mechanical characterization of rodent livers, Barnes et al. developed a model-gel-tissue assay (MGT assay) involving the embedding of liver tissue in polyacrylamide gel24. This gel is compressed by a pulsed uniform force from which Young's modulus can be calculated. The MGT assay shows a good correlation with an indentation assay adapted for both normal and fibrotic livers24 (Table 1).
Table 1: Liver stiffness values at the bulk level. TE and MRE compared to direct ex vivo mechanical measurements of liver elastic moduli using indentation and MGT assays for livers from different sources. The relation between E and G is given by E = 2G (1 + v), where v is the Poisson's ratio of the sample; F0 to F4 represent the fibrosis score in the METAVIR scoring system, with F0 denoting low or no fibrosis and F4 cirrhotic livers. Abbreviations: TE = transient elastography; MRE = magnetic resonance elastography; MGT = model-gel-tissue; E = elastic (Young's) modulus; G = shear modulus. Please click here to download this Table.
One of the major drawbacks of generic liver stiffness measurements is that they do not provide cellular-level resolution of stiffness heterogeneity in the liver. During the progression of fibrosis, collagen-rich areas show higher rigidity compared to the surrounding parenchyma25,26. This stiffness gradient locally influences the resident cells and plays an important role in driving HSC heterogeneity27. Thus, changes in local mechanical properties during liver disease development need to be characterized on a microscopic level to better understand fibrosis progression.
AFM allows the mechanical properties of tissue to be measured with high resolution and high force sensitivity. AFM uses the tip of a cantilever to indent the surface of a sample with forces as low as several piconewtons, inducing a deformation at a microscopic or nanoscopic level based on the geometry and size of the tip employed. The force response of the sample to the applied strain is then measured as the deflection in the cantilever28. Force-displacement curves are collected from the approach and retraction of the cantilever, which can be fitted with appropriate contact mechanics models to evaluate the local stiffness of the sample29.
In addition to measuring the stiffness of a given area, AFM can also provide topographic information about specific features in the sample, such as the structure of collagen fibres30,31,32. Multiple studies have described the application of AFM to measure the stiffness of various healthy and diseased tissues, such as skin32,33, lung34,35, brain36, mammary37,38,39, cartilage40, or heart41,42,43,44 from both patient and mouse model samples. Furthermore, AFM has also been used in vitro to determine the stiffness of cells and extracellular protein scaffolds45,46,47.
The measurement of the mechanical properties of biological samples using AFM is nontrivial due to their softness and fragility. Thus, various studies have standardized different conditions and settings, which yield widely fluctuating values of Young's moduli (reviewed by Mckee et al.48). Similar to other soft tissues, liver Young's modulus values at different grades of liver fibrosis also show extensive variation (Table 2). The differences in Young's modulus values arise from differences in the mode of AFM operation, cantilever tip, sample preparation method, sample thickness, indentation depth and forces, liver tissue environment during measurement, and analysis method (Table 2).
Table 2: Liver stiffness values at the cellular level. Liver stiffness values obtained using AFM describe the mechanical properties of the liver at the cellular level. Abbreviations: AFM = atomic force microscopy; E = elastic (Young's) modulus; PFA = paraformaldehyde; PBS = phosphate-buffered saline. Please click here to download this Table.
This paper describes a protocol for the reproducible measurement of Young's moduli of collagen-rich fibrotic areas in liver tissue by AFM with a precise localization provided by the use of polarization microscopy. We administered carbon tetrachloride (CCl4) to induce collagen deposition in a centrilobular fashion49 in a mouse model, reliably mimicking crucial aspects of human liver fibrosis50. Polarized microscopic images enable the visualization of collagen in the liver due to the birefringence of collagen fibers51, which allows accurate positioning of the cantilever tip over the desired area of interest within the hepatic lobule52.
Protocol
All animal experiments were performed in accordance with an animal protocol approved by the Animal Care Committee of The Institute of Molecular Genetics and according to the EU Directive 2010/63/EU for animal experiments. An overall schematic diagram of the presented protocol is shown in Figure 1.
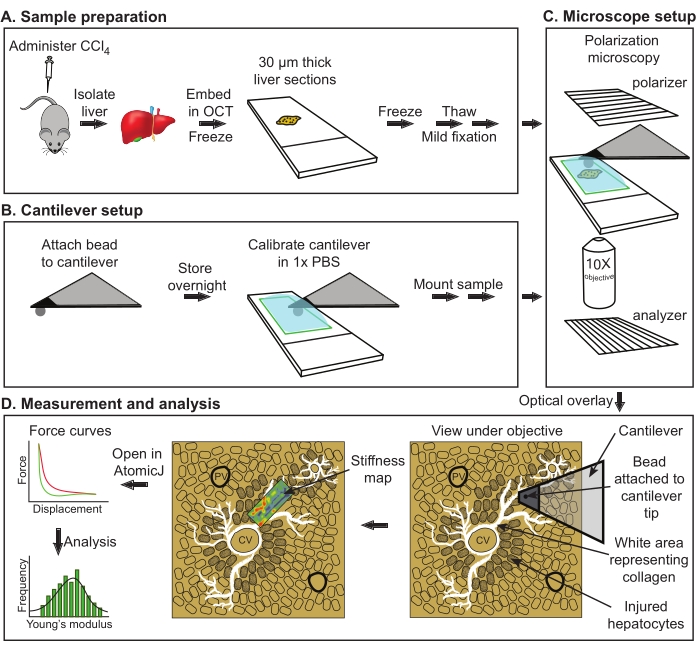
Figure 1: Overall schematic of AFM evaluation of Young's modulus from mouse livers. (A) Isolation of liver from control or treated mice followed by sectioning and storage at −80 °C (maximum storage, 2 weeks). (B) Attachment of the spherical bead to the cantilever with subsequent curing of glue overnight (left). Cantilever calibration followed by sample mounting (right). (C) Alignment of the polarizer and analyzer to visualize bright collagen structures followed by an overlay of the image in the camera with the measurement field under the AFM cantilever. (D) Acquisition of stiffness maps and analysis. Abbreviations: AFM = atomic force microscopy; PBS = phosphate-buffered saline; OCT = optimal cutting temperature compound; CCl4 = carbon tetrachloride. Please click here to view a larger version of this figure.
1. Sample preparation I
- Excise the liver from the opened abdomen of a mouse euthanized by cervical dislocation under anesthesia. Isolate the left lateral lobe and embed the lateral half of the lobe into optimal cutting temperature (OCT) compound by quick freezing on dry ice. Store the OCT-embedded tissue at −80 °C.
NOTE: Previous studies have shown that the stiffness values are similar between frozen and fresh tissues25,26. - Section 30 µm thick liver sections onto positively charged slides using a cryotome and store the slides at −80 °C until the day of AFM measurement no more than 2 weeks hence.
2. Setting up the instrument
- Attachment of a 5.7 µm bead to AFM cantilever tip (Supplemental Figure S1, steps 1-5)
NOTE: The attachment of the bead to the cantilever was also previously described by Norman et al.46.- Spread the suspension of 5.7 µm diameter melamine resin beads evenly on half of the area of a glass slide and air-dry to evaporate the solvent (Supplemental Figure S1, step 1).
- On the other half of the slide, using a 10 µL tip, make a thin line of premixed epoxy resin with a long working time (Supplemental Figure S1, step 1).
- Load the cantilever probe to the AFM head according to the manufacturer's instructions.
- Mount the slide and cover with the AFM head fitted with the cantilever probe.
NOTE: An SD-qp-BioT-TL-10 cantilever was used in the study. - Bring the premixed epoxy resin glue to the center of the slide and approach the glue with the cantilever with a low force (set a setpoint a 1 V to follow this protocol; Supplemental Figure S1, step 2).
NOTE: Before approaching the slide surface, ensure that the laser is aligned at the center of the cantilever tip, indicated by a high sum voltage by following steps 6-8 in Section 2, Part 3, Calibration of the spring constant of the AFM cantilever. - After the tip of the cantilever probe makes contact with the glue, move it just above the slide to remove excess glue.
- Now, retract the cantilever tip from the slide and move the slide to bring a single bead in the center (Supplemental Figure S1, step 3). Approach the bead again with the AFM cantilever probe with a higher force (setpoint 2 V) to attach the bead in the center of the cantilever probe and leave it for at least 10 s (Supplemental Figure S1, step 4).
- Extract the cantilever probe and keep it overnight at room temperature to harden the glue or follow the instructions for the epoxy resin (Supplemental Figure S1, step 5).
- Setting up the polarizer and analyzer
- To locate the areas of interest within the liver sections, set up the microscope with the polarizer and analyzer. Align their vibration azimuths at an angle between 0° and 90° to each other by rotating either one of them with respect to the other manually or in an automated way to minimize transmitted light and maximize extraordinary rays passing through the objective. Make sure that the collagen fibers appear bright against a dark background in the polarized image (Figure 2), which is reflected by a shift in the peak of the image histogram toward bright pixels.
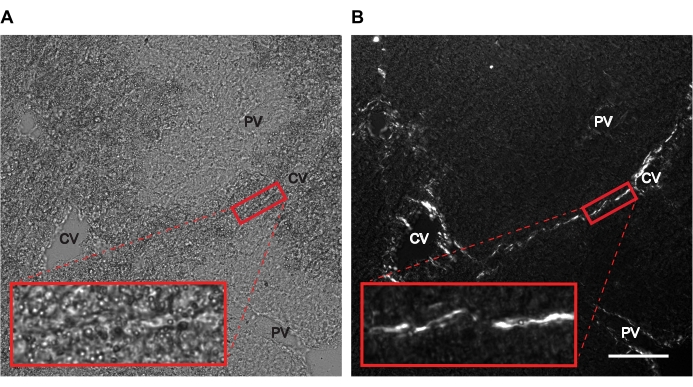
Figure 2: Representative microscopy images show pronounced visualization of collagen fibers in polarized microscopy as compared to brightfield images. Liver sections from mice treated with CCl4 for 3 weeks were subjected to (A) brightfield and (B) polarized microscopy. Birefringent collagen fibers are clearly visible in white in polarized images compared to brightfield images. The red box represents collagen-rich area used for the AFM measurement. Insets show zoomed-in views of the area in the red box. Scale bar = 100 µm. Abbreviations: AFM = atomic force microscopy; CCl4 = carbon tetrachloride; CV = central vein; PV = portal vein. Please click here to view a larger version of this figure.
NOTE: For this protocol, the AFM head can be installed on any suitable inverted microscope with the possibility to insert a polarizer and an analyzer. The system must be placed in an isolation unit to reduce the background noise.
- Calibration of the spring constant of the AFM cantilever
- Load the cantilever probe (prepared according to steps 1-8 in Section 2, Part 1, Attachment of a 5.7 μm bead to AFM cantilever tip) to the AFM head according to the manufacturer's instructions.
- Clean the cantilever with 70% ethanol to prevent contamination of the cantilever during measurement. Wash extensively with distilled water to remove residual ethanol from the tip.
- Enable contact mode and select force mapping as the method of measurement.
- Open all the relevant windows (Z Stepper Motors, Motorized Stage Control, Data Viewer, Force Scan Map Oscilloscope, Laser Alignment, and Camera window) in the software tabs by clicking on the corresponding buttons.
- Mount a clean glass slide containing 1.2 mL of 1x phosphate-buffered saline (PBS) in an area of approximately 2 cm x 4 cm delineated with a hydrophobic marker pen. Mount the AFM head on the microscope stage and ensure that the cantilever is fully immersed in PBS.
- Focus the objective on the cantilever tip. Reduce the intensity of transmitted light on the microscope to get a better view of the laser position on the monitor.
- Target the laser to the translucent end of the cantilever (under which the spherical bead is attached) and align the mirror using the knobs on the AFM head to maximize the total intensity of the laser beam on the detector (depicted by the sum in the Laser Alignment window).
- Align the photodiode detector using the knobs present on the AFM head to position the laser at its center.
- Let the cantilever stabilize for 15 min before proceeding with the calibration.
- Open the Calibration manager and insert the type of cantilever, cantilever dimensions, and environmental conditions. Calibrate the spring constant and sensitivity (also known as inverse optical lever sensitivity, InvOLS) by clicking on Calibrate. Confirm the accuracy of the spring constant obtained after calibration with the manufacturer's declaration. Calibrate the cantilever in contact-free mode (calibration is performed by the software using the thermal theorem derived by Sader et al.58).
NOTE: The spring constant of the cantilever represents the stiffness of the cantilever and is given by the resistance force of the cantilever as it deforms per the deformation length in terms of N/m. The sensitivity of the cantilever depicts the value of photodiode response (in volts) in response to the deflection of the cantilever (in nanometers) and is usually presented in terms of nm/V59. In a contact-free mode, the thermal noise spectrum is recorded and fitted with the hydrodynamic function60 by the AFM control software automatically after calibration. The fitting provides the calibration parameters, namely the spring constant and the InvOLS. The spring constant of the SD-qp-BioT-TL-10 cantilever used in this study was 0.09 N/m, as declared by the manufacturer.
3. Sample preparation II
- Thaw the frozen sections (stored at −80 °C, as described in Section 1, Sample preparation I) at room temperature for 2 min.
- Fix the sections with ice-cold 4% paraformaldehyde (PFA) in 1x PBS for 10 min at 4 °C followed by extensive washing (5x) with 1x PBS. Wipe residual PBS around the section with a tissue and mark a boundary of approximately 2 cm x 4 cm around the liver section with a hydrophobic marker pen. Cover the sample with 1x PBS (ensure the demarcated area contains ~1.2 mL of 1x PBS). Use the sample for AFM measurements.
NOTE: Since PFA is a hazardous chemical, it must be handled with care to prevent contact with skin or eyes. To avoid its toxic fumes, all procedures involving PFA must be carried out in a certified chemical fume hood or other approved ventilated area.
4. Measurement
- Enable autosave by going to Setup tab and making a tick on Autosave. Specify the file name and directory for saving the measurement files by going to the Setup tab and then clicking on Saving Settings.
- Load the sample (prepared according to Section 3, Sample preparation II). Approach the tissue surface with the AFM cantilever by switching on the Laser and clicking on the Approach key. After the tip is in contact with the tissue surface, retract the cantilever tip such that the tip remains in the focus of the objective (by clicking on the Retraction key, which retracts the cantilever to the top end of the piezo range). Realign the detector if the laser position is moved away from its center on the Laser Alignment window.
- Switch off the Laser and overlay the optical field of the microscope with AFM measurement maps by clicking on the Accessories tab and then selecting Direct Overlay Optical Calibration. In the succeeding window, click on Next to take a series of images of the cantilever scanning a specific area. Click Next again to go to the next window.
- In the first image, click on the center of the tip of the cantilever manually to depict the tip position in the software. The circle depicting the tip position can be manipulated in size by indicating its Radius to increase accuracy. Click Calibrate to automatically detect the cantilever tip position in all images. Confirm the accuracy of tip detection by going through the images.
- Click Next and then Finish to finish the optical overlay and save the series of images captured during optical calibration.
- Retract the cantilever further to avoid its collision with higher surfaces on the slide. Move the stage to position a collagen-rich area inside the green box visible in the Data Viewer tab using the polarized image. Select the collagen-rich area by making a rectangle around it using a long press on the left mouse button inside the specified green box. Define the dimensions, orientation, and resolution of the selected area under the Grid tab on the left-hand side. Click on Confirm New Scan Region to set the selected area as area of measurement.
- Keep IGain and PGain-parameters of the feedback loop-at default values, if no major instabilities are presented in the form of system oversensitivity. To follow this protocol, set IGain at 50 Hz and PGain at 0.001.
- Set the Setpoint at 1 nN.
NOTE: Setpoint is the force of the tip-surface interaction during the stationary state. For most soft samples (cells, gels, and tissues), Setpoint values in the range of 0.5-2.0 nN are appropriate. - Select the Relative Setpoint Value according to the mechanical properties of the studied material and the stiffness of the cantilever. For this protocol, set the value at 5 nN.
NOTE: The relative setpoint represents the maximal force of interaction, when the peak of the force-distance curve is reached and the tip movement returns to the baseline. For soft materials (1-50 kPa), this parameter is set in units of nanonewtons (nN). Moreover, for a soft cantilever (with a spring constant from 0.05 N/m to 0.35 N/m), the maximum force that can be applied is approximately 50 nN. Adjust the Setpoint and Relative Setpoint Values accordingly.- Ensure that the given setpoint value does not lead to an indentation depth larger than the radius of the spherical indenter, otherwise the indenting surface cannot be properly defined in the calculation of Young's modulus. Calculate the average value of the indentation depth after the first run of the indentations; see Section 5, Data analysis, to learn how to process the recorded data. Adjust the Setpoint Value if needed.
- Set Adjust Baseline as 5.
NOTE: Here, the Baseline refers to the degree of polynomial used for fitting the approach and retract curves. Setting the baseline to 5 fits the curves to high resolution and thus ensures capturing of background noise during measurements. - Select the length of the cantilever movement in the Z-axis (Z-length) according to the surface topography of the sample. To follow this protocol, set the Z-length at 15 µm.
NOTE: A high value (e.g. 15 μm) reduces measurement sensitivity but is usually required for samples with a highly irregular surface such as fibrotic liver tissue. - Set Z movement a Constant Duration.
NOTE: Z movement can also be set to Constant speed to view data referring to Z movement (Extend Speed and Extend Time) in a different mode. - Set the Extend Time a 1 s. Set Extension Delay and Retraction Delay as 0.
NOTE: Use extend speeds of >5.0 µm/s for very soft materials61, as lower indentation speeds will result in a more viscous and less elastic response from soft surfaces. Extension Delay and Retraction Delay are parameters that can be used for studying specific interaction between the cantilever tip and substrate (e.g. interactions of a proteins immobilized on the surface of the cantilever with proteins immobilized on the slide surface). - Set Sample Rate as 5000 Hz.
NOTE: The Sample Rate refers to the frequency of points that are recorded on a complete approach-retract curve. Set this to a high value (e.g. 5000 Hz) to avoid missing certain regions of the curves due to their extremely fast transition. - Mark the Z Closed Loop with a Tick to enable a feedback loop system, which ensures contant distance between sample surface and the cantilever tip.
- Disengage the motorized stage by deselecting Engage and Click on Approach to approach the sample with the cantilever. Click on Start Scanning to start collecting force-distance curves in the area set in step 6; Section 4, Measurement.
5. Data analysis
- Analyze the acquired data using the open-source software "AtomicJ" (which can be downloaded from https://sourceforge.net/projects/jrobust/).
NOTE: It supports files collected from Agilent Technologies, JPK Instruments, or Bruker atomic force microscopes. - Load the force curves into the program by clicking on the process force curves and maps icon in AtomicJ (Supplemental Figure S2A, x). In the processing assistant, add the maps to be analyzed by clicking on the Add button (Supplemental Figure S2A, y). Click on Next after the maps are loaded (Supplemental Figure S2A, z).
- Specify the processing settings in the next window according to the steps described below (corresponding to the steps in Supplemental Figure S2B, steps 1-11):
- Estimate the contact point between the sample and the cantilever manually by calculation or automatically using a set of fitting curve parameters. To follow this protocol, use the automatic estimation of the contact point.
- Determine the contact point between the cantilever and the sample by the Classical focused grid method.
- Select the estimation method to yield the best determination of the contact point based on the quality of the measured force curves, which needs to be empirically determined during the optimization of data analysis. To follow this protocol, use the Model independent method.
- Fit the force indentation curve using a Classical model (use Classical L2 for model fit to follow this protocol).
NOTE: Model fit and Contact estimator are parameters that govern how the curves are fitted by the software and how the contact point is established in the fitted curve, respectively. For these measurements, the Classical option was used, which uses least squares regression. This processes each low-force point on the curve as a trial contact point and fits a polynomial to the region before the trial contact point. It then fits the appropriate contact model to the collected force indentation data. The point that gives the lowest sum of squares is assumed as the contact point. Other methods can be used based on the quality of force curves obtained62,63. - Set the fit of the model to the Withdraw curve.
- Set Poisson's ratio as 0.45, as recommended for soft tissues such as liver24.
- Set the fit of the curve using a baseline degree of 3 and an in-contact degree of 1. Change the degree of polynomial fit based on the scale of deviations of the curves from the model.
- Select the model used to fit the withdraw curves. Use the Sneddon model, which calculates Young's modulus based on equation (1) and equation (2):
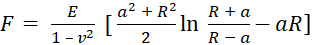 (1)
(1)
δ =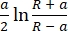 (2)
(2)
where F is force, E is Young's modulus, v is the sample's Poisson's ratio, δ is the depth of indentation, a is the contact radius, and R is the sphere radius62,63. - Fill in the radius of the spherical tip in micrometers (2.9 µm in this protocol).
- Load the Spring constant and InvOLS from the data files by enabling read-in (check the boxes).
- Click on Finish.
NOTE: The analyzed data are presented in the form of maps of vertical deflection, height, adhesion, contact force, deformation, adhesion force, R2 values, slope, Young's modulus, transition indentation, transition force, and contact position calculated for each force curve (Supplemental Figure S3, upper left). Two additional windows display force curves and raw values (Supplemental Figure S3, upper right and lower panels, respectively).
- Exclude force curves where the cantilever approached the surface of the liver section incorrectly. To identify these, look for curves with high noise, aberrant shapes, and/or incomplete approach, as demonstrated in Figure 3 and discussed in detail elsewhere46.
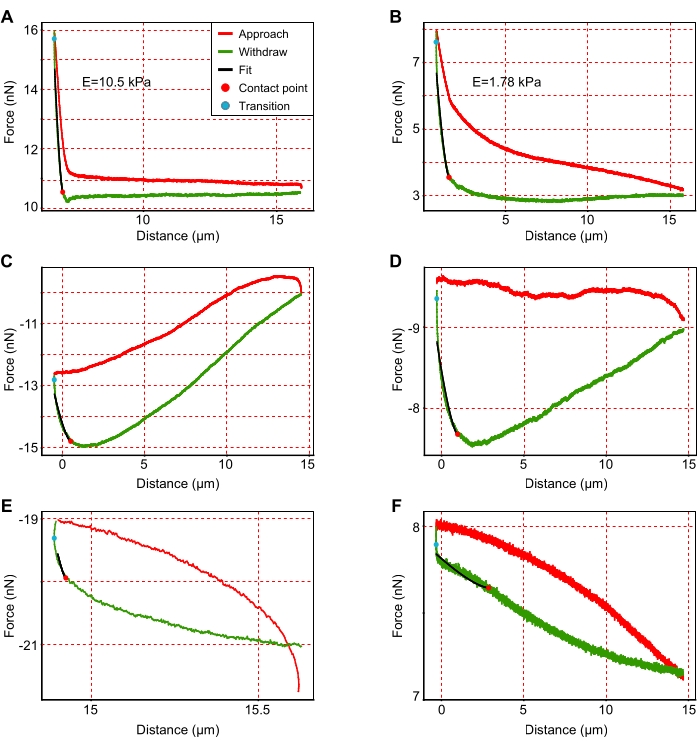
Figure 3: Examples of representative force-displacement curves. (A,B) Representative interpretable force curves for stiffer (A; E = 10.5 kPa) and softer (B; E = 1.78 kPa) areas that are suitable for analysis. (C–F) Representative uninterpretable graphs that need to be excluded from the analysis due to (C–E) incorrect approach or (F) higher noise. As indicated in the legend provided in (A), red curves show the approach of the cantilever, and green curves show the retraction of the cantilever. Black lines show the fitting of the withdrawal curve of the cantilever. The slope of the black lines corresponds to Young's modulus. The red and blue points correspond to the contact point and the transition point, respectively. The contact point is the last point of contact between the cantilever and substrate during retraction, while the transition point describes the transition of the cantilever from approach to retraction. Please click here to view a larger version of this figure.
Representative Results
Mildly fixed, 30 µm thick liver sections obtained from control mice and from mice with mild or advanced fibrosis (induced by injection of CCl4 for 3 weeks or 6 weeks, respectively49) were probed with AFM as described in this protocol. Collagen fibers close to central veins were selected for the measurement of stiffness maps. Areas close to the central veins, which correspond to the areas where collagen fibers in CCl4-treated animals usually form, were analyzed in control livers (Figure 4A). The distribution of Young's moduli was reproducible across different control livers and collagen-rich areas within a single liver section (Figure 4B: left violin plot).
In CCl4-treated animals, stiffness maps corresponding to the pericentral areas of collagen deposits showed significantly higher values of Young's moduli compared to equivalent areas in control mice (Figure 4B: 1.9 kPa vs. 2.6 kPa median Young's modulus values for control vs. 3 week CCl4-treated mouse; p = 0.07; 1.9 kPa vs. 5.1 kPa median Young's modulus values for control vs. 6 weeks CCl4-treated mouse; p = 0.02). Moreover, there was a significant increase in the values of Young's moduli with longer CCl4 treatment (Figure 4B; 2.6 kPa vs. 5.1 kPa median Young's modulus values for 3 week vs. 6 week CCl4-treated mouse; p = 0.04). This shows a gradual stiffening of collagen deposits with fibrosis progression and that AFM measurements reflect the fibrogenesis.
To evaluate the effect of prolonged storage of OCT-embedded liver sections on the mechanical properties of collagen fibers, we measured the stiffness of collagen fibers in sections of CCl4-treated mice, which were stored at −80 °C for 2 weeks or 3 months on the slide after cutting (Figure 5). AFM measurements showed significantly lower values of Young's moduli in collagen-rich areas for sections stored for 3 months compared to those obtained from sections measured within 2 weeks of sample sectioning (Figure 5; 4.7 kPa vs. 3.6 kPa median Young's modulus values for 2 weeks vs. 3 months storage; p < 0.001). Thus, it is important to measure the mechanical properties of the liver tissue shortly after the sections are prepared from the OCT-embedded liver lobes.
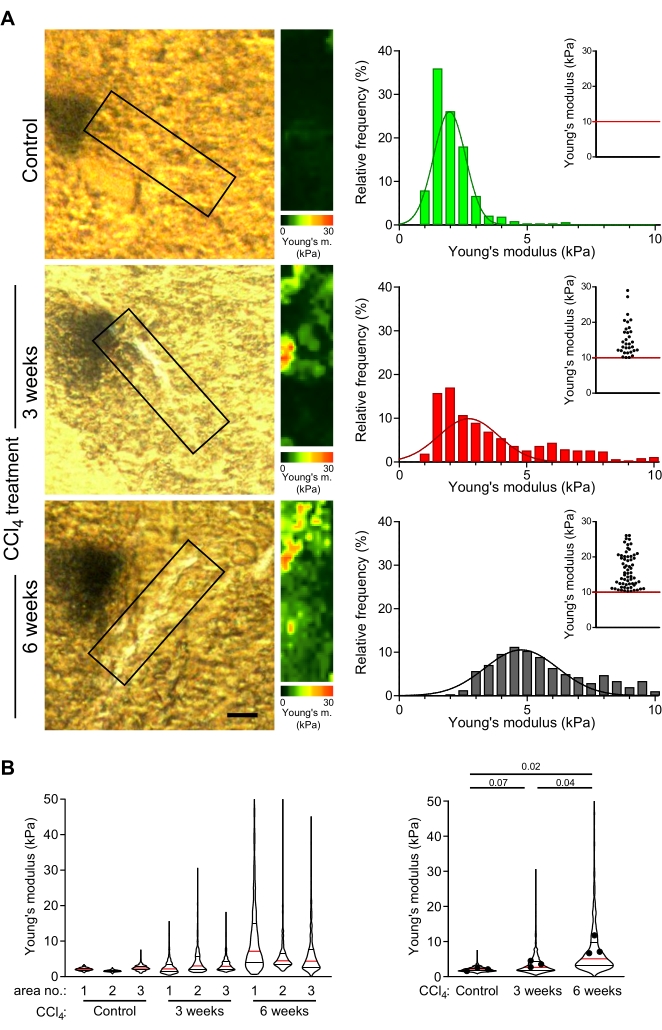
Figure 4: AFM measurements reveal progressive stiffening of collagen-rich areas correlating with prolonged CCl4 treatment. (A) Liver sections from control mice and mice treated with CCl4 for 3 weeks or 6 weeks were used to measure the mechanical properties of collagen-rich areas. The boxed areas of liver sections shown in the polarized microscopy images (left) are collagen-rich scan areas (or corresponding regions in the control liver) selected for AFM measurements (30 µm x 100 µm, 10 pixels x 36 pixels). The Young's modulus maps with color scales corresponding to these boxed areas are shown on the right, including histograms of Young's modulus values from these maps; inset scatter plots show values >10 kPa for each condition. The stiffening of the liver is visualized as a gradual rightward shift in the histogram distribution and a higher frequency of points in the inset scatter plot. Scale bar = 20 µm. (B) Violin plots show the distribution of elastic moduli from three areas measured for each condition (left) and summarized elastic moduli values from all three maps (right). Violin plots show the median (red line), 25th percentile and 75th percentile (black lines); dots represent the mean values of individual maps from areas 1-3. The presented p-values were calculated using a Student's t-test performed on means. Abbreviations: AFM = atomic force microscopy; CCl4 = carbon tetrachloride. Please click here to view a larger version of this figure.
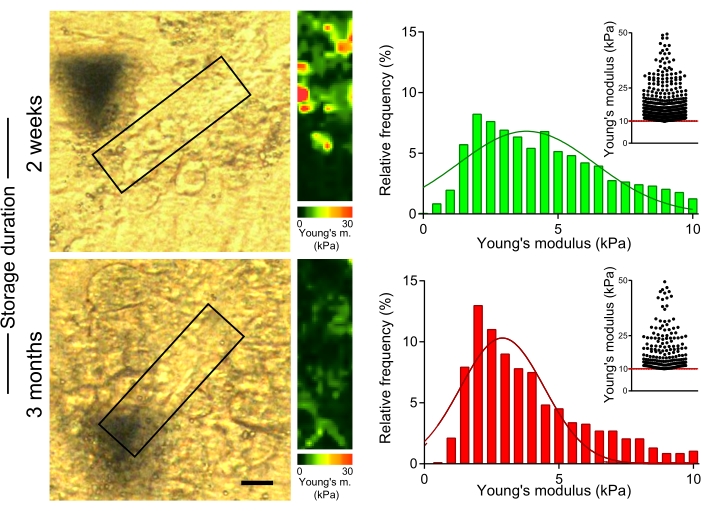
Figure 5: Extended storage of liver sections leads to a decrease in stiffness of collagen-rich areas. Liver sections (prepared from mice treated with CCl4 for 2 weeks) stored at −80 °C for 2 weeks or 3 months were used to measure Young's modulus. Polarized microscopy images (left) with boxes indicating the collagen-rich areas used for AFM measurement (30 µm x 100 µm, 10 pixels x 36 pixels). Corresponding Young's modulus maps with color scales (right). Histograms show Young's modulus values collected from 4-6 areas in each sample; inset scatter graphs show values >10 kPa for each condition. Scale bar = 20 µm. Abbreviations: AFM = atomic force microscopy; CCl4 = carbon tetrachloride. Please click here to view a larger version of this figure.
Supplemental Figure S1: Method for modifying the cantilever with a melamine resin micro bead. (A) Drawn schematic illustrates the attachment of a spherical bead to the tip of the cantilever. For a step-wise description, see Section 2, Part 1, Attachment of a 5.7 µm bead to AFM cantilever tip. (B) Microscopy image of a spherical 5.7 µm bead attached to the cantilever tip shown from top (left) and lateral view (right). Scale bars = 20 µm. Abbreviations: AFM = atomic force microscopy; RT = room temperature. Please click here to download this File.
Supplemental Figure S2: Data analysis in AtomicJ. (A) The sequence of steps to be followed for opening stiffness maps in AtomicJ. A single left-click on the process force curves and maps (x) opens the processing assistant. Files can be loaded to the processing assistant by clicking on add (y) and selecting the required files. Click on next (z) to proceed to the next step. (B) Parameters for curve fitting, appropriate contact mechanics model, and AFM settings used during measurement. Steps 1-11 refer to corresponding sub-points detailed in protocol step 3, Section 5, Data analysis. Please click here to download this File.
Supplemental Figure S3: Outline of analyzed data in AtomicJ. The preview of analyzed data shows stiffness maps (upper left window), force curves (upper right window), and raw data (lower window). Abbreviation: AFM = atomic force microscopy. Please click here to download this File.
Discussion
The presented protocol provides a step-by-step reproducible method for AFM measurement of normal and fibrotic mouse liver tissue. Coupled polarization microscopy provides high spatial precision and enables visualization of collagen fibers due to their birefringence. Further, a detailed description of the analysis of the obtained force curves is provided. AFM stiffness measurement can be performed on the cellular level, which allows local changes in liver tissue mechanical properties due to developing fibrotic disease to be monitored. Liver fibrosis is not a homogeneous process affecting the entire organ. On the contrary, areas of collagen-rich fibrotic septa are interspersed with areas of low or no collagen deposits. Thus, stiffness changes are specific to the local microenvironment and only affect cells locally in contact with areas damaged by injury. This microscale of stiffness heterogeneity is also apparent in the details of AFM Young's modulus maps, where points of high stiffness border the areas of almost normal stiffness. This variation shows that even collagen scar tissue area is not mechanically homogeneous and requires AFM measurement to be characterized on a cellular level (Figure 4).
The presented protocol allows the measurements of liver stiffness by AFM independently of the liver collection, as the whole liver lobes embedded in OCT can be stored for a prolonged period at −80 °C. However, once the tissue is sectioned, we recommend measuring the samples within ~2 weeks as we have observed a gradual softening of tissue sections stored for longer periods of time (Figure 5).
The AFM equipped with polarization microscopy allows for precisely locating the area of interest within the liver lobule structure. However, it also has some limitations that need to be considered when interpreting the results. The stiffness values obtained here were measured at room temperature. We assume that the effects of temperature on the mechanical properties of soft tissue will be small; however, this might be one of the reasons for differences between the reported in vivo values of mechanical properties of liver tissues and the values in this study.
Furthermore, this protocol allows AFM analysis of liver tissue for up to 3 h, which requires mild fixation of the tissue. The mild fixation of tissue sections, as well as the freeze-thaw cycle, will most likely affect the absolute values of Young's modulus. Thus, the reported values of Young's moduli might differ from in vivo values. Further studies are needed to optimize the protocol for the measurement of absolute values of Young's modulus from liver sections, which may be achieved by a different method for fixation of liver tissue64.
Nonetheless, we observed increasing stiffness of collagen-rich areas in the livers of mice treated with CCl4 for 3 weeks compared to 6 weeks. Such changes correspond to fibrosis progression during prolonged injury (Figure 4) and show that relative differences can be probed between different treatments using the presented protocol. This is in agreement with the observations of Calò et al., who showed that mildly fixed liver sections show similar differences in stiffness values between collagen-rich and collagen-lacking areas as in fresh tissue25.
We used the SD-qp-BioT-TL-10 cantilever (theoretical spring constant ~0.09 N/m) modified with a 5.7 µm diameter spherical tip to minimize mechanical disruption of the liver tissue during measurements. A 5.7 µm bead enabled sufficient indentation of the sample to probe its stiffness while preserving its integrity. A bead with a smaller diameter can be used, after several optimizations, for gaining higher resolution in the stiffness maps but might lead to further overestimation of the Young's modulus values (for more details, see Crichton et al.65). Using the specified cantilever-bead ensemble, we were able to characterize sample stiffness in a broad range, from tens of units of Pa to ~100 kPa.
Sneddon's model was used to derive Young's modulus from force curves, as it allows analysis of deep indentations with colloidal probes62. Sneddon's model, unlike Hertz's model, does not suffer from the constraint that the contact radius must be much smaller than the sphere radius. It further assumes that the sample thickness is several times greater than the indentation depth30,66. In the present study, the indentation was ~2 µm with a bead size of 5.7 µm and a sample thickness of 30 µm in collagen-rich areas; thus, Sneddon's model was appropriate. Other models63 considering the adhesion force between tip and substrate can be used for different types of tissues.
Analysis in AtomicJ implements corrections for the finite thickness of the samples to minimize the contribution of a substrate while deriving Young's modulus62,67. In the analysis of the obtained force curves, we used a single Poisson's ratio of 0.45, which has been previously recommended for soft tissue organs24. This approximation does not have a significant effect on calculated values of Young's modulus, as the change in the value of Poisson's ratio from 0.4 to 0.5 results only in a 0.893x decrease in Young's modulus values calculated according to the Sneddon's equation. Given the multi-fold differences in Young's modulus between the different durations of CCl4 treatments, the errors produced by approximating Poisson's ratio are only marginal.
We used withdraw curves to calculate stiffness values, as we were interested in the elastic response of the tissue to the load provided by the cantilever rather than in the plastic response to indentation68. Due to the viscoelastic response of the soft tissue, fitting withdraw curves might overestimate Young's modulus, which should be kept in mind. Furthermore, we have observed that data analysis with approach curves yields similar trends in stiffness values between fibrotic and control areas, though the absolute values are correspondingly lower (data not shown).
While optimizing the protocol, we identified several steps critical for the reproducibility of the measurements. First, it is important to ensure that the bead is approximately in the center of the translucent tip while attaching to the cantilever. This prevents possible mechanical imbalance during indentation. Second, during liver fixation with PFA, it is necessary to strictly follow the time limits for thawing and fixation. Changing the timing of this step might severely affect the mechanical properties of tissue sections. Third, the cantilever must be repeatedly calibrated with continuous monitoring and input of concurrent temperature values to avoid any artifacts occurring in stiffness values due to temperature fluctuations. Last, a single liver section should not be measured for longer than 3 h from preparation, as overlaid PBS may evaporate over longer periods. Readers can refer to the troubleshooting table (Table 3) for solving problems encountered during the AFM measurement, also discussed in length in Norman et al.46.
Table 3: Troubleshooting guide. Please click here to download this Table.
The presented protocol allows for reproducible AFM probing of liver tissue. It has the potential to reveal information on the development and eventual regression of fibrotic liver disease on a microscopic level and can contribute to the development of therapies targeting fibrotic scar regions formed during the progression of chronic liver disease.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Grant Agency of the Czech Republic (18-02699S), the Institutional Research Project of the Czech Academy of Sciences (RVO 68378050), and MEYS CR project NICR EXCELES (LX22NPO05102). CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2018127 and European Regional Development Fund-Project "UP CIISB" (No. CZ.02.1.01/0.0/0.0/18_046/0015974) financially supported the measurements at the CF Nanobiotechnology, CEITEC MU. We also acknowledge the Light Microscopy Core Facility, IMG CAS, Prague, Czech Republic, supported by MEYS (LM2018129, CZ.02.1.01/0.0/0.0/18_046/0016045) and RVO: 68378050-KAV-NPUI, for their support with the microscopy imaging presented herein.
Materials
| AFM head | Bruker | JPK nanowizard 3 | |
| Cameras | Andor | Zyla 5.5 USB (sCMOS, water cooled) | |
| The Imagingsource | S/N:12310015 | ||
| Cantilever | SD-qp-BioT-TL-10, Nanosensors | S/N:73750F05 | |
| Cryotome | Leica | CM1950 | |
| Epoxy resin glue (Long working time ) | Bison epoxy universal | ||
| Melamine beads; diameter, 5.7 um | Microparticles, GmbH | MF-R-5.7 | |
| Microscope | Olympus | IX81 | |
| Hydrophobic slide marker | SuperHT | PAP PEN | |
| Software | JPK nanowizard v6.1.151 | ||
| AtomicJ v2.3.1 | |||
| Superfrost slides | Thermoscientific | ref no. J1800AMNZ | |
| System | Ubuntu 14.04.5 LTS | ||
| Vibration isolation control unit | Tablestable | AVI-200-S |
Riferimenti
- vanden Berghe, G. The role of the liver in metabolic homeostasis: Implications for inborn errors of metabolism. Journal of Inherited Metabolic Dis. 14 (4), 407-420 (1991).
- Stanger, B. Z. Cellular homeostasis and repair in the mammalian liver. Annual Reviews of Physiology. 77, 179-200 (2015).
- Asrani, S. K., Devarbhavi, H., Eaton, J., Kamath, P. S. Burden of liver diseases in the world. Journal of Hepatology. 70 (1), 151-171 (2019).
- Hernandez-Gea, V., Friedman, S. L. Pathogenesis of liver fibrosis. Annual Review of Pathology. 6, 425-456 (2011).
- Georges, P. C., et al. Increased stiffness of the rat liver precedes matrix deposition: Implications for fibrosis. American Journal of Physiology. Gastrointestinal and Liver Physiology. 293 (6), 1147-1154 (2007).
- Perepelyuk, M., et al. Hepatic stellate cells and portal fibroblasts are the major cellular sources of collagens and lysyl oxidases in normal liver and early after injury. American Journal of Physiology. Gastrointestinal and Liver Physiology. 304 (6), 605-614 (2013).
- Wells, R. G. The role of matrix stiffness in regulating cell behavior. Hepatology. 47 (5), 1394-1400 (2008).
- Olsen, A. L., et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. American Journal of Physiology. Gastrointestinal and Liver Physiology. 301 (1), 110-118 (2011).
- Sandrin, L., et al. Transient elastography: A new noninvasive method for assessment of hepatic fibrosis. Ultrasound in Medicine & Biology. 29 (12), 1705-1713 (2003).
- Ling, W., et al. Effects of vascularity and differentiation of hepatocellular carcinoma on tumor and liver stiffness: In vivo and in vitro studies. Ultrasound in Medicine & Biology. 40 (4), 739-746 (2014).
- Wong, V. W. S., et al. Diagnosis of fibrosis and cirrhosis using liver stiffness measurement in nonalcoholic fatty liver disease. Hepatology. 51 (2), 454-462 (2010).
- Cha, S. W., et al. Nondiseased liver stiffness measured by shearwave elastography a pilot study. Journal of Ultrasound in Medicine. 33 (1), 53-60 (2014).
- Castera, L., Forns, X., Alberti, A. Non-invasive evaluation of liver fibrosis using transient elastography. Journal of Hepatology. 48 (5), 835-847 (2008).
- Chang, W., et al. Liver fibrosis staging with MR elastography: Comparison of diagnostic performance between patients with chronic hepatitis B and those with other etiologic causes. Radiology. 280 (1), 88-97 (2016).
- Venkatesh, S. K., Yin, M., Ehman, R. L. Magnetic resonance elastography of liver: Clinical applications. Journal of Computer Assisted Tomography. 37 (6), 887-896 (2013).
- Venkatesh, S. K., Yin, M., Ehman, R. L. Magnetic resonance elastography of liver: Technique, analysis and clinical applications. Journal of Magnetic Resonance Imaging. 37 (3), 544-555 (2013).
- Venkatesh, S. K., Wang, G., Teo, L. L. S., Ang, B. W. L. Magnetic resonance elastography of liver in healthy Asians: Normal liver stiffness quantification and reproducibility assessment. Journal of Magnetic Resonance Imaging. 39 (1), 1-8 (2014).
- Lee, D. H., Lee, J. M., Han, J. K., Choi, B. I. MR elastography of healthy liver parenchyma: Normal value and reliability of the liver stiffness value measurement. Journal of Magnetic Resonance Imaging. 38 (5), 1215-1223 (2013).
- Mueller, S. Liver stiffness: A novel parameter for the diagnosis of liver disease. Hepatic Medicine. Evidence and Research. 2, 49-67 (2010).
- Goodman, Z. D. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. Journal of Hepatology. 47 (4), 598-607 (2007).
- Salameh, N., et al. Hepatic viscoelastic parameters measured with MR elastography: Correlations with quantitative analysis of liver fibrosis in the rat. Journal of Magnetic Resonance Imaging. 26 (4), 956-962 (2007).
- Yin, M., et al. Quantitative assessment of hepatic fibrosis in an animal model with magnetic resonance elastography. Magnetic Resonance in Medicine. 58 (2), 346-353 (2007).
- Bastard, C., et al. Transient micro-elastography: A novel non-invasive approach to measure liver stiffness in mice. World Journal of Gastroenterology. 17 (8), 968-975 (2011).
- Barnes, S. L., Lyshchik, A., Washington, M. K., Gore, J. C., Miga, M. I. Development of a mechanical testing assay for fibrotic murine liver. Medical Physics. 34 (11), 4439-4450 (2007).
- Calò, A., et al. Spatial mapping of the collagen distribution in human and mouse tissues by force volume atomic force microscopy. Scientific Reports. 10, 15664 (2020).
- Desai, S. S., et al. Physiological ranges of matrix rigidity modulate primary mouse hepatocyte function in part through hepatocyte nuclear factor 4 alpha. Hepatology. 64 (1), 261-275 (2016).
- Kostallari, E., et al. Stiffness is associated with hepatic stellate cell heterogeneity during liver fibrosis. American Journal of Physiology. Gastrointestinal and Liver Physiology. 322 (2), 234-246 (2022).
- Binnig, G., Qaute, C. F., Gerber, C. Atomic force microscope. Physical Review Letters. 56 (9), (1986).
- Johnson, K. L. . Contact Mechanics. , (1985).
- Asgari, M., Latifi, N., Giovanniello, F., Espinosa, H. D., Amabili, M. Revealing layer-specific ultrastructure and nanomechanics of fibrillar collagen in human aorta via atomic force microscopy testing: Implications on tissue mechanics at macroscopic scale. Advanced NanoBiomed Research. 2 (5), 2100159 (2022).
- Amabili, M., et al. Microstructural and mechanical characterization of the layers of human descending thoracic aortas. Acta Biomaterialia. 134, 401-421 (2021).
- Grant, C. A., Twigg, P. C., Tobin, D. J. Static and dynamic nanomechanical properties of human skin tissue using atomic force microscopy: Effect of scarring in the upper dermis. Acta Biomaterialia. 8 (11), 4123-4129 (2012).
- Geerligs, M., et al. In vitro indentation to determine the mechanical properties of epidermis. Journal of Biomechanics. 44 (6), 1176-1181 (2011).
- Liu, F., Tschumperlin, D. J. Micro-mechanical characterization of lung tissue using atomic force microscopy. Journal of Visualized Experiments. (54), e2911 (2011).
- Sicard, D., Fredenburgh, L. E., Tschumperlin, D. J. Measured pulmonary arterial tissue stiffness is highly sensitive to AFM indenter dimensions. Journal of the Mechanical Behavior of Biomedical Materials. 74, 118-127 (2017).
- Babu, P. K. V., Radmacher, M. Mechanics of brain tissues studied by atomic force microscopy: A perspective. Frontiers in Neuroscience. 13, 600 (2019).
- del Mar Vivanco, M. . Mammary Stem Cells: Methods and protocols. , (2015).
- Zanetti-Dällenbach, R., et al. Length scale matters: Real-time elastography versus nanomechanical profiling by atomic force microscopy for the diagnosis of breast lesions. Biomed Research International. 2018, 3840597 (2018).
- Lopez, J. I., Kang, I., You, W. -. K., McDonald, D. M., Weaver, V. M. In situ force mapping of mammary gland transformation. Integrative Biology. 3 (9), 910-921 (2011).
- Stolz, M., et al. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nature Nanotechnol. 4 (3), 186-192 (2009).
- Borin, D., Pecorari, I., Pena, B., Sbaizero, O. Novel insights into cardiomyocytes provided by atomic force microscopy. Seminars in Cell & Developmental Biology. 73, 4-12 (2018).
- Lachaize, V., et al. Atomic Force Microscopy: An innovative technology to explore cardiomyocyte cell surface in cardiac physio/pathophysiology. Letters in Applied NanoBioScience. 4 (4), 321-324 (2015).
- Lieber, S. C., et al. Aging increases stiffness of cardiac myocytes measured by atomic force microscopy nanoindentation. American Journal of Physiology. Heart and Circulatory Physiology. 287 (2), 645-651 (2004).
- Guedes, A. F., et al. Atomic force microscopy as a tool to evaluate the risk of cardiovascular diseases in patients. Nature Nanotechnology. 11 (8), 687-692 (2016).
- Zhu, Y., Dong, Z., Wejinya, U. C., Jin, S., Ye, K. Determination of mechanical properties of soft tissue scaffolds by atomic force microscopy nanoindentation. Journal of Biomechanics. 44 (13), 2356-2361 (2011).
- Norman, M. D. A., Ferreira, S. A., Jowett, G. M., Bozec, L., Gentleman, E. Measuring the elastic modulus of soft culture surfaces and three-dimensional hydrogels using atomic force microscopy. Nature Protocols. 16 (5), 2418-2449 (2021).
- Thomas, G., Burnham, N. A., Camesano, T. A., Wen, Q. Measuring the mechanical properties of living cells using atomic force microscopy. Journal of Visualized Experiments. (76), e50497 (2013).
- McKee, C. T., Last, J. A., Russell, P., Murphy, C. J. Indentation versus tensile measurements of Young’s modulus for soft biological tissues. Tissue Engineering. Part B, Reviews. 17 (3), 155-164 (2011).
- Scholten, D., Trebicka, J., Liedtke, C., Weiskirchen, R. The carbon tetrachloride model in mice. Laboratory Animals. 49, 4-11 (2015).
- Nevzorova, Y. A., Boyer-Diaz, Z., Cubero, F. J., Gracia-Sancho, J. Animal models for liver disease – A practical approach for translational research. Journal of Hepatology. 73 (2), 423-440 (2020).
- Ribeiro, J. F., dos Anjos, E. H. M., Mello, M. L. S., de Campos Vidal, B. Skin collagen fiber molecular order: A pattern of distributional fiber orientation as assessed by optical anisotropy and image analysis. PLoS One. 8 (1), 54724 (2013).
- Ozkan, A., et al. The influence of chronic liver diseases on hepatic vasculature: A liver-on-a-chip review. Micromachines. 11 (5), 487 (2020).
- Guimarães, C. F., Gasperini, L., Marques, A. P., Reis, R. L. The stiffness of living tissues and its implications for tissue engineering. Nature Reviews Materials. 5, 351-370 (2020).
- Khajehahmadi, Z., et al. Liver stiffness correlates with serum osteopontin and TAZ expression in human liver cirrhosis. Annals of the New York Academy of Sciences. 1465 (1), 117-131 (2020).
- Tian, M., et al. The nanomechanical signature of liver cancer tissues and its molecular origin. Nanoscale. 7 (30), 12998-13010 (2015).
- Zhao, G., et al. Mechanical stiffness of liver tissues in relation to integrin β1 expression may influence the development of hepatic cirrhosis and hepatocellular carcinoma. Journal of Surgical Oncology. 102 (5), 482-489 (2010).
- Gang, Z., Qi, Q., Jing, C., Wang, C. Measuring microenvironment mechanical stress of rat liver during diethylnitrosamine induced hepatocarcinogenesis by atomic force microscope. Microscopy Research and Technique. 72 (9), 672-678 (2009).
- Sader, J. E., et al. Spring constant calibration of atomic force microscope cantilevers of arbitrary shape. The Review of Scientific Instruments. 83 (10), 103705 (2012).
- JPK Instruments. A practical guide to AFM force spectroscopy and data analysis. JPK Instruments Technical Note. JPK Instruments. , 1-8 (2016).
- van Eysden, C. A., Sader, J. E. Frequency response of cantilever beams immersed in viscous fluids. Journal of Applied Physics. 101, 044908 (2015).
- Kim, Y., Yang, Y. I., Choi, I., Yi, J. Dependence of approaching velocity on the force-distance curve in AFM analysis. Korean Journal of Chemical Engineering. 27, 324-327 (2010).
- Hermanowicz, P., Sarna, M., Burda, K., Gabryś, H. AtomicJ: An open source software for analysis of force curves. The Review of Scientific Instruments. 85 (6), 063703 (2014).
- Hermanowicz, P. . AtomicJ 2.3.1 User’s Manual. , (2021).
- Iwashita, M., et al. Comparative Analysis of Brain Stiffness Among Amniotes Using Glyoxal Fixation and Atomic Force Microscopy. Frontiers in Cell and Developmental Biology. 8, 574619 (2020).
- Crichton, M. L., et al. The viscoelastic, hyperelastic and scale dependent behaviour of freshly excised individual skin layers. Biomaterials. 32 (20), 4670-4681 (2011).
- Puricelli, L., Galluzzi, M., Schulte, C., Podestà, A., Milani, P. Nanomechanical and topographical imaging of living cells by atomic force microscopy with colloidal probes. The Review of Scientific Instruments. 86 (3), 033705 (2015).
- Hermanowicz, P. Determination of Young’s modulus of samples of arbitrary thickness from force distance curves: Numerical investigations and simple approximate formulae. International Journal of Mechanical Sciences. 193, 106138 (2021).
- Han, R., Chen, J. A modified Sneddon model for the contact between conical indenters and spherical samples. Journal of Materials Research. 36, 1762-1771 (2021).

