Transforming Static Barrier Tissue Models into Dynamic Microphysiological Systems
Summary
This protocol describes a reconfigurable membrane-based cell culture platform that integrates the open-well format with fluid flow capabilities. This platform is compatible with standard protocols and allows for reversible transitions between open-well and microfluidic culture modes, accommodating the needs of both engineering and bioscience laboratories.
Abstract
Microphysiological systems are miniaturized cell culture platforms used to mimic the structure and function of human tissues in a laboratory setting. However, these platforms have not gained widespread adoption in bioscience laboratories where open-well, membrane-based approaches serve as the gold standard for mimicking tissue barriers, despite lacking fluid flow capabilities. This issue can be primarily attributed to the incompatibility of existing microphysiological systems with standard protocols and tools developed for open-well systems.
Here, we present a protocol for creating a reconfigurable membrane-based platform with an open-well structure, flow enhancement capability, and compatibility with conventional protocols. This system utilizes a magnetic assembly approach that enables reversible switching between open-well and microfluidic modes. With this approach, users have the flexibility to begin an experiment in the open-well format using standard protocols and add or remove flow capabilities as needed. To demonstrate the practical usage of this system and its compatibility with standard techniques, an endothelial cell monolayer was established in an open-well format. The system was reconfigured to introduce fluid flow and then switched to the open-well format to conduct immunostaining and RNA extraction. Due to its compatibility with conventional open-well protocols and flow enhancement capability, this reconfigurable design is expected to be adopted by both engineering and bioscience laboratories.
Introduction
Vascular barriers serve as a critical interface that separates the blood compartment from the surrounding tissue. They play a critical role in preserving homeostasis by attracting immune cells, controlling molecular permeability, and shielding against the intrusion of pathogens into the tissue1,2. In vitro culture models have been developed to mimic the in vivo microenvironment, enabling systematic investigations into the factors and conditions that impact barrier properties in both healthy and diseased states3,4.
The most widely used approach for such culture models is the Transwell-like "open-well" configuration5, where a porous, track-etched culture membrane separates media-filled compartments (Figure 1A). In this format, cells can be seeded on either side of the membrane, and a wide range of experimental protocols has been developed. However, these systems are limited in their ability to provide the fluid flows essential for supporting barrier maturation and mimicking immune cell circulation seen in vivo5,6. Consequently, they cannot be used for studies requiring dynamic flows that introduce drug doses, mechanical stimulation, or fluid-induced shear stresses6,7,8.
To overcome the limitations of open-well systems, microfluidic platforms that combine porous culture membranes with individually addressable fluidic channels have been developed9. These platforms offer precise control over fluid routing, perfusion, and the introduction of chemical compounds, controlled shear stimulation, and dynamic cell addition capabilities7,10,11,12,13. Despite the advanced capabilities provided by microfluidic platforms, they have not seen widespread adoption in bioscience laboratories due to complex microfluidic protocols and their incompatibility with established experimental workflows4,10,14.
To bridge the gap between these technologies, we present a protocol that employs a magnetically reconfigurable, module-based system. This system can be easily switched between open-well and microfluidic modes based on the specific needs of the experiment. The platform features an open-well device, known as m-µSiM (modular microphysiological system enabled by a silicon membrane), with a 100 nm thick culture membrane (nanomembrane). This nanomembrane possesses high porosity (15%) and glass-like transparency, as illustrated in Figure 1B. It physically separates the top compartment from a bottom channel, allowing for molecular transport across physiological length scales15. Unlike conventional track-etched membranes, which have known challenges in imaging live cells with bright-field imaging, the nanomembrane's favorable optical and physical properties enable clear visualization of cells on either side of the membrane surface15,16,17.
The present protocol outlines the fabrication of specialized seeding and flow modules and explains the magnetic reconfiguration of the platform. It demonstrates how the platform can be employed to establish endothelial barriers under both static and dynamic conditions. This demonstration reveals that endothelial cells align along the flow direction, with an upregulation of shear-sensitive gene targets under shear stimulation.
Protocol
This design can be used in various modes based on experimental requirements and the preferences of the end user. Prior to each experiment, consult the decision flow chart presented in Figure 2 to determine the necessary steps and modules for the protocol. For example, if the user intends to maintain the open-well format throughout an experiment to directly compare it with the Transwell-type system, the patterning stencil is not required for cell seeding. The core module is commercially available (see Table of Materials), and the ultrathin nanomembrane can be selected from a library of materials with different porosity and pore sizes to suit experimental needs.
1. Fabrication of the patterning stencil
NOTE: The patterning stencil serves to position cells exclusively on the porous region of the membrane chip, preventing cells from settling onto the surrounding silicon layer where they could potentially experience damage after the flow module is added16 (refer to Figure 3). Damage to the monolayer can adversely affect barrier integrity and compromise experimental outcomes. The stencil is unnecessary in an open, static culture, as there is no risk of damage.
- Purchase, machine, or 3D print a mold using the design provided in Supplementary Coding File 1 (Figure 4A).
NOTE: The stencil walls are tapered to increase media capacity and minimize bubble trapping compared to high aspect ratio straight-walled features. - Attach a 130 µm pressure-sensitive adhesive (PSA) film to a 3.2 mm acrylic sheet using a cold roller (see Table of Materials).
- Laser cut the acrylic sheet according to the design provided in Supplementary Coding File 2 to create an array of cavities (Figure 4B).
- Peel off the protective layer from the PSA film and attach the acrylic sheet to the mold.
NOTE: Ensure that the laser-cut sheet is aligned with the mold so that the mold features are centered within the cavity (Figure 4C). The accuracy of cell positioning on the membrane depends on this step. - Thoroughly mix the PDMS prepolymer (using a 10:1 base to catalyst ratio) (see Table of Materials) and degas it in a vacuum chamber until no visible bubbles remain (5-15 min).
- Slowly pour the PDMS into the mold cavities, leveling it with a flat edge.
NOTE: To prevent bubble entrapment, pour the PDMS into each cavity slowly. If bubbles are present after pouring, place the mold in the vacuum chamber and degas it again. - Cure the PDMS on a hotplate at 70 °C for 1 h, and then allow it to cool to room temperature.
- Extract the stencils from the mold cavities using flat-tipped tweezers.
2. Fabrication of the flow module
NOTE: The flow module shares a similar footprint with the clover-shaped well of the core module and includes a molded microchannel (width = 1.5 mm, height = 0.2 mm, length = 5 mm). The clover shape aids in aligning the channel over the porous culture region (Figure 5).
- Use standard soft lithography techniques18 to create a silicon mold with features defined by the design provided in Supplementary Coding File 3 (Figure 4D).
- Attach a 130 µm PSA film to a 3.2 mm thick acrylic sheet.
- Laser cut the acrylic sheet based on the design given in Supplementary Coding File 4 to create an array of clover-shaped cavities (Figure 4E).
NOTE: The height of the cavity determines the height of the flow module, which must be slightly taller (by 0-0.1 mm) than the height of the core module well to ensure proper sealing. - Remove the protective layer from the PSA film and then attach the laser-cut sheet to the silicon mold by aligning the triangular alignment marks on the silicon mold with those on the laser-cut sheet.
NOTE: Similar to step 1.4, ensure that the laser-cut acrylic sheet is aligned with the silicon mold so that the mold features are centered within each cavity (Figure 4F). This step determines the position of the microchannel relative to the footprint. - Slowly pour the degassed PDMS onto the mold cavities and level it with a flat edge.
NOTE: If bubbles are present after pouring, degas the mold until the bubbles are removed. - Place the mold on a hotplate and bake the PDMS at 70 °C for 1 h, then allow the mold to cool to room temperature.
- Carefully remove the flow modules from the mold cavities using flat-tipped tweezers.
- Orient the flow module with the microchannel features facing upward, and position the core inlet and outlet ports at the end of the channel using a biopsy punch (see Table of Materials).
3. Fabrication of lower and upper acrylic housings
NOTE: The core module fits into the lower housing. The attraction between embedded magnets in the housings compresses and seals the flow module to the core module (Figure 6).
- Laser-cut a 2 mm acrylic sheet using the design provided in Supplementary Coding File 5 to create the upper housing with holes for magnet insertion.
- Attach a 130 µm PSA film to a 3.5 mm acrylic sheet.
- Laser-cut the acrylic sheet based on the design given in Supplementary Coding File 6 to create the lower housing with holes for magnet insertion.
NOTE: The thickness of the lower housing is a critical parameter and must match the overall thickness of the core module to prevent leakage during flow. - Remove the PSA protective layer and attach the lower housing to a glass coverslip.
- Press-fit 4.75 mm nickel-plated neodymium magnets (see Table of Materials) into the laser-cut holes of the housings.
NOTE: Ensure that magnets in the lower and upper housings are placed with opposite polarity to ensure magnetic attraction (Figure 6).
4. Fabrication of the flow circuit
NOTE: The closed-loop flow circuit contains two sample collection vials as reservoirs (Figure 7). The inlet reservoir has a polyvinylidene difluoride (PVDF) filter to allow the cell media to equilibrate with the CO2 concentration in the incubator.
- Use a 1 mm biopsy punch to create two holes in the cap of a sample collection vial.
- Use biopsy punches to create two holes (1 mm and 4 mm) in the cap of a separate sample collection vial, and create a 1 mm hole in the lower part of this vial.
- Insert 20 G dispensing tips into each of the 1 mm holes and seal them with hot glue.
- Place a 0.22 µm PVDF filter (see Table of Materials) in the 4 mm hole and seal it with hot glue.
- Laser-cut a 1.5 mm acrylic sheet using the design provided in Supplementary Coding File 7 to create a sample holding stage.
- Position the reservoirs on the acrylic stage.
- Connect the dispensing tips using micro flow tubes (see Table of Materials).
- Link the tubes to the inlet and outlet of the flow module using 21 G dispensing tips.
- Utilize a peristaltic pump (see Table of Materials) for flow circulation.
NOTE: Syringe or pneumatic pumps can also be used to introduce flow if desired. The flow rate should be determined based on experimental needs. A comprehensive table, derived from an experimentally validated COMSOL model, is provided in Supplementary Table 1 to assist users in selecting the necessary flow rate to achieve the desired shear stress at the cell monolayer16.
5. Cell seeding
NOTE: Similar to conventional membrane inserts, different cell types can be cultured on the nanomembrane. A secondary cell type can also be co-cultured on the other side of the membrane in the bottom channel15.
- Coat the membrane with 5 µg/cm2 of fibronectin (see Table of Materials) at room temperature for 1 h to improve cell attachment.
NOTE: The chosen cell type may influence the required coating and cell density. The procedure described here is for Human umbilical vein endothelial cells (HUVECs, see Table of Materials). - Aspirate the coating solution using a P200 pipette and place a patterning stencil in the core module well.
- Add cell media to the well and bottom channel of the device.
- Seed HUVECs at a density of 40,000 cells/cm2 into the well.
NOTE: HUVECs at this density typically form a confluent monolayer after 24 h of incubation16,19. - Place the device in a Petri dish. Add a 15 mL conical tube cap with DI water to the Petri dish to maintain local humidity.
- Transfer the Petri dish to the incubator.
NOTE: Cell media in the top well and bottom channel of the device should be changed every 24 h. To change the media in the bottom channel, gently inject fresh media into one of the ports to displace the old media from the other port.
6. Reconfiguration to microfluidic mode
- Sterilize the upper housing, lower housing, flow module, reservoirs, tubes, and dispensing tips by placing them in a UV sterilization chamber for 30 min prior to assembly. Circulate 70% ethanol and then sterile PBS in the flow circuit.
- Fill the reservoirs and tubes with Endothelial Cell Growth media (see Table of Materials).
- Place the lower housing on the sample stage. Insert the open-well core module into the lower housing.
- Aspirate cell media from the well of the module. Place the flow module in the well.
- Place the upper housing to seal the flow module in the core module well. Connect the inlet and outlet dispensing tips to the flow module ports.
- Start the peristaltic pump to introduce fluid flow into the system.
7. Conducting downstream analysis in open-well format after flow introduction
NOTE: The culture time here depends on the experimental goals. Users can conduct downstream analysis (e.g., Immunocytochemistry, RNA extraction) either in the open-well or microfluidic formats based on their preference. For instance, if an open-well format is preferred, the system should be reconfigured to conduct assays based on standard protocols16,19.
- Stop the pump when the desired time for shear flow exposure is reached. Remove the inlet and outlet dispensing tips.
- Remove the upper housing. Gently remove the flow module from the well using tweezers.
- Add 100 µL of cell media to the well. Conduct desired assays based on standard protocols.
Representative Results
The open-well core module is initially positioned within a specific cavity created by a lower housing and a coverslip, as illustrated in Figure 6A. Subsequently, the flow module, which includes a microchannel and access ports, is inserted into the well of the core module. The flow module is securely sealed against the silicon support layer of the membrane due to the magnetic attraction force between magnets embedded in the lower and upper housings, as depicted in Figure 6B. To evaluate the effectiveness of this magnetic latching mechanism, a burst pressure test was conducted, demonstrating that the system can withstand dead-ended pressures of up to 38.8 ± 2.4 kPa. This pressure tolerance significantly exceeds the typical operating pressures encountered in cell culture applications. Furthermore, the system remains free of leaks when subjected to flow rates of up to 4000 µL/min, which is equivalent to a shear stress of 74 dynes/cm2 at the culture region16.
When developing a platform that can switch between open-well and microfluidic modes, careful consideration must be given to the cell seeding approach, which is not typically a concern for conventional static open-well platforms16. Damage to the monolayer around the channel boundary could introduce complications in experimental results20. To address this issue, a removable stencil was designed that fits within the open well of the core module and provides a specific window for cells to settle preferentially on the membrane surface (Figure 3). Once the cell monolayer is patterned and reaches confluency, the user has the flexibility to continue the experiment in the open-well format or reconfigure the platform into microfluidic mode to expose the cell monolayer to physiological shear stress (Figure 3). The magnetic latching mechanism provides the ability to easily switch between the open-well and microfluidic formats as required. For instance, the device can be reverted to the open-well format after a flow stimulation, offering users the flexibility to conduct a variety of assays (such as immunostaining, RNA extraction, and molecular permeability measurements) using standard experimental protocols15,16.
In the physiological setting of the human body, the vascular barrier is exposed to flow-induced shear stress, which serves as a key biophysical cue that affects the structure and function of the barrier5,21,22. Thus, the addition of fluid flow in microphysiological systems is a key requirement. To demonstrate the versatility of the platform, an HUVEC monolayer was established in an open-well format using standard protocols. After 24 h of static culture, the platform was reconfigured into microfluidic mode to expose the cell monolayer to 10.7 dynes/cm2 shear stress for 24 h. The results indicated that cells cultured under flow aligned along the flow direction while cells cultured without flow remained randomly oriented (Figure 8A,B). After shear stimulation, the platform was reconfigured to the open-well format to extract RNA using standard protocols. The results indicated that the exposure of cells to shear stress resulted in the upregulation of Kruppel-like factor 2 (KLF2) and endothelial nitric oxide synthase (eNOS), which serve critical roles such as anti-thrombotic and atheroprotective functions in healthy blood vessels23,24(Figure 8C).
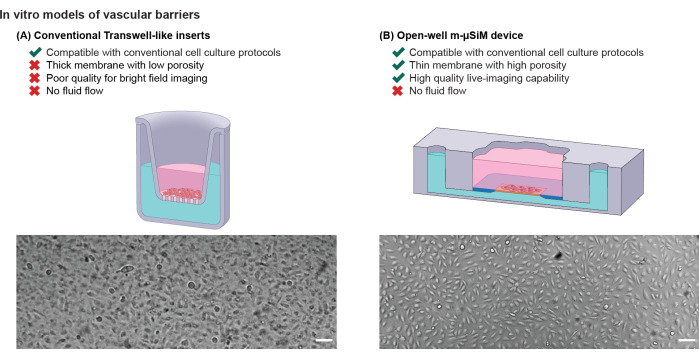
Figure 1: Comparison of in vitro vascular barrier models. Schematic illustration of (A) conventional Transwell-like inserts and (B) the open-well m-µSiM. Bright-field images of a confluent HUVEC monolayer highlight the difference in bright-field imaging quality between a track-etched membrane and an ultrathin nanomembrane. Scale bars = 100 µm. Adapted from Mansouri et al.16. Please click here to view a larger version of this figure.
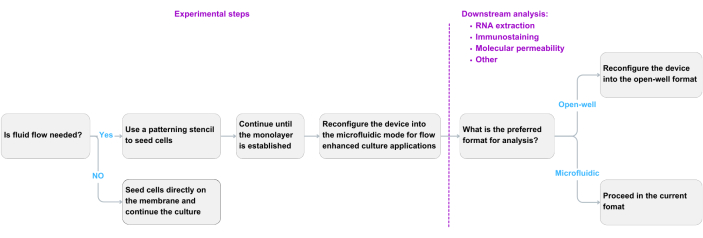
Figure 2: Decision-making flow chart. A flow chart based on experimental needs and downstream analysis preferences. Please click here to view a larger version of this figure.
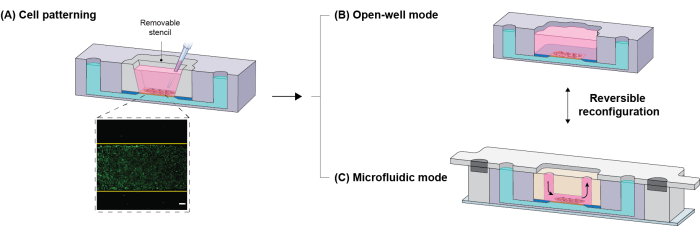
Figure 3: Experimental workflow of the platform. (A) To directly position cells on the porous membrane, a removable patterning stencil is inserted in the well of the core module (the inset shows patterned cells, yellow lines exhibit microchannel boundaries). (B) The stencil can be kept or removed in the device for static cell culture. (C) To reconfigure the platform into microfluidic mode, the stencil is replaced with the flow module. Because of the magnetic sealing mechanism, the configuration is reversible; housings and the flow module can be removed to switch into open-well mode. Scale bar = 200 µm. Adapted from Mansouri et al.16. Please click here to view a larger version of this figure.
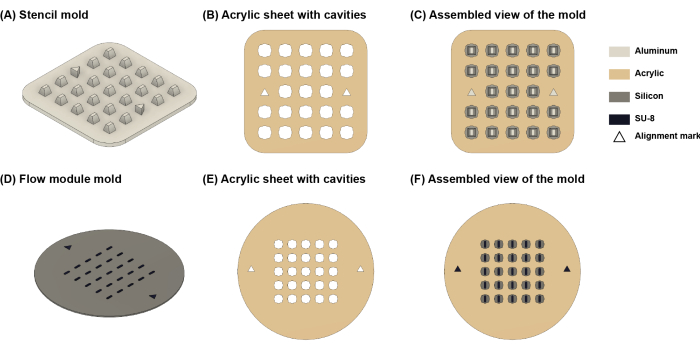
Figure 4: Schematic illustration of the molds. (A) The stencil mold. (B) Laser-cut acrylic sheet. (C) assembled view of the stencil mold. (D) Flow module mold. (E) Laser-cut acrylic sheet. (F) Assembled view of the flow module mold. Triangle-shaped features are alignment marks to facilitate attaching acrylic sheets to the molds. Please click here to view a larger version of this figure.
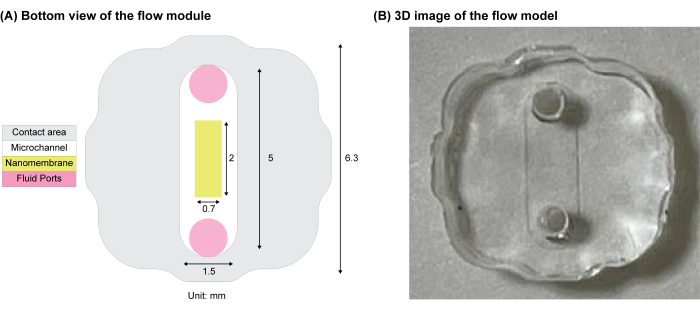
Figure 5: Schematic of the clover-shape flow module. (A) The contact interface between the flow module and the membrane chip. The inlet and outlet ports for fluid flow are shown in pink. (B) 3D image of the PDMS flow module. Adapted from Mansouri et al.16. Please click here to view a larger version of this figure.
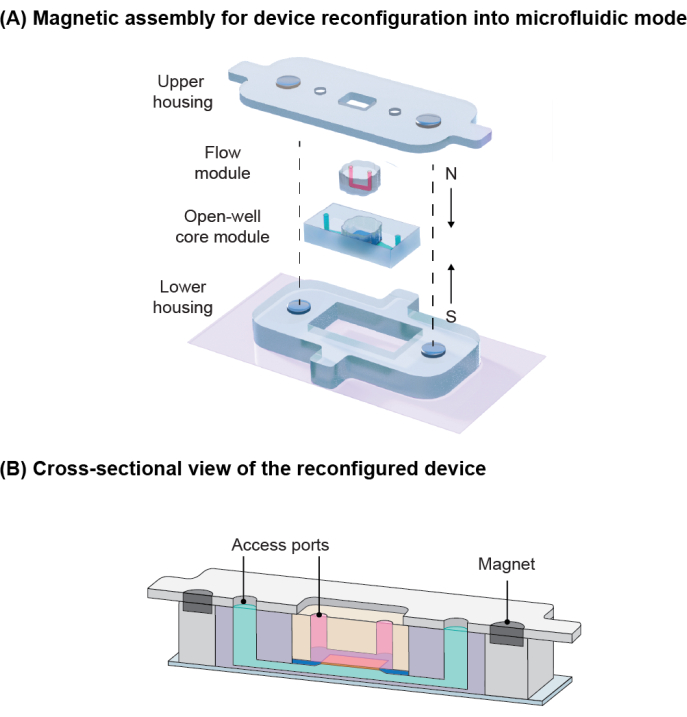
Figure 6: Magnetic assembly for device reconfiguration. (A) Schematic demonstration of components for device reconfiguration into microfluidic mode. Embedded magnets with opposite poles induce attraction for the sealing. (B) Cross-sectional view of the reconfigured device showing the vascular channel in pink and the tissue compartment in green. Adapted from Mansouri et al.16. Please click here to view a larger version of this figure.
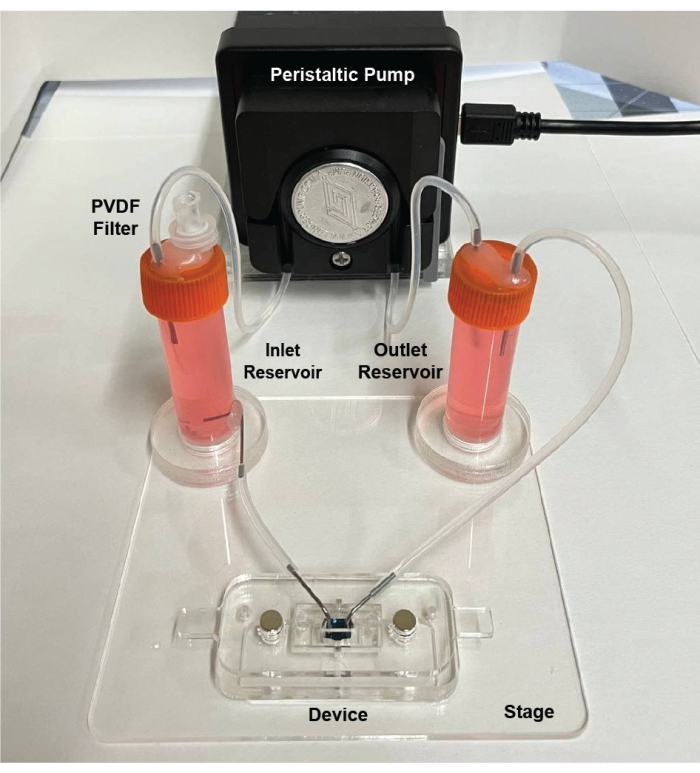
Figure 7: Assembled view of the flow circuit. The circuit consists of a peristaltic pump, two reservoirs for supplying cell media and damping fluctuations, tubing, and an acrylic stage to hold the components in place. Adapted from Mansouri et al.16. Please click here to view a larger version of this figure.
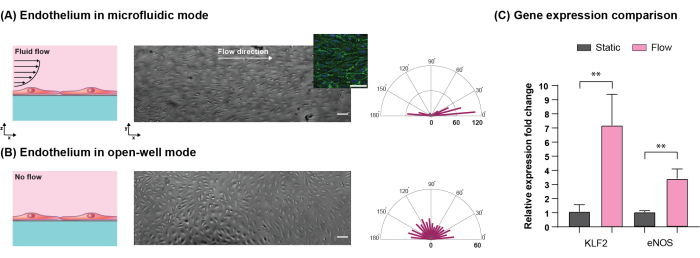
Figure 8: Comparison of HUVECs cultured in open-well and microfluidic modes. Cells were seeded and cultured in open-well for 24 h to establish a confluent monolayer. During the subsequent 24 h period, one set of devices was reconfigured into microfluidic mode. (A) Cells cultured under flow (10.7 dynes.cm-2 shear stress) aligned along the flow direction (the inset shows actin and nuclei of cells in green and blue, respectively). (B) Cells cultured without flow in open-well format showed no alignment. The length of bars in radar plots shows the number of cells in the corresponding direction. (C) Cells cultured under flow showed higher upregulation of KLF2 and eNOS genes compared to the no-flow condition (**p < 0.01, n = 3, mean ± SD). Scale bars = 100 µm. Adapted from Mansouri et al.16. Please click here to view a larger version of this figure.
Supplementary Table 1: Shear stress on the nanomembrane surface at different flow rates. This table provides information about shear stress values on the nanomembrane surface at various flow rates. Please click here to download this File.
Supplementary Coding File 1: CAD model of the stencil mold. Please click here to download this File.
Supplementary Coding File 2: CAD model of laser cut cavities for the stencil mold. Please click here to download this File.
Supplementary Coding File 3: CAD model of the flow module. Please click here to download this File.
Supplementary Coding File 4: CAD model of laser cut cavities for the flow module mold. Please click here to download this File.
Supplementary Coding File 5: CAD model of the upper housing. Please click here to download this File.
Supplementary Coding File 6: CAD model of the lower housing. Please click here to download this File.
Supplementary Coding File 7: CAD model of the acrylic stage. Please click here to download this File.
Discussion
The aim of this protocol is to develop a practical method for incorporating flow capabilities into an open-well platform featuring an ultrathin nanomembrane. In this design, a magnetic latching approach is utilized, allowing switching between open-well and fluidic modes during experiments and combining the advantages of both approaches. Unlike conventional permanently bonded platforms, magnetic latching allows the platform to be disassembled at convenient points during the experimental workflow16,25,26,27. In this platform, cells are seeded, and the barrier is established in the familiar open-well format, and then the system is reconfigured to a fluidic mode by simply adding a flow module and magnetically sealing it. This reconfiguration offers two key benefits: first, it enables the simulation of physiological conditions by exposing the cell monolayer to fluid-induced shear stress; second, it facilitates the introduction of secondary components like immune cells under flow conditions16. The magnetic latching feature offers a tool-free assembly method, enabling on-demand switching between open-well and microfluidic formats. Consequently, downstream analyses, such as immunocytochemistry and RNA extraction, can be conducted in the familiar open-well format using standard techniques or using microfluidic techniques as desired16.
Magnetic sealing is a critical parameter of the design that enables the reconfigurability of the platform, and thus, several considerations are required for its accurate function: (1) The height of the lower housing must match the height of the core module (tolerance: ±0.1 mm) to enclose the module properly. (2) The height of the flow module must be slightly taller than the well of the core module (0-0.1 mm). (3) This design is adaptable to a wide range of materials and doesn't mandate that the flow module be constructed from elastomeric materials. The crucial element is to incorporate a soft, gasket-like material at the interface between the flow module and the membrane chip to establish a reliable seal. (4) The diameter of the laser-cut cavities in the housings for press-fitting magnets must be optimized based on the instrument used28. Magnets might detach and lead to flow leakage if the cavity diameter is too big, or housings might crack during magnet insertion if cavities are too small. By following the aforementioned considerations, the magnetic assembly approach introduced here can be applied to reconfigure other open-well systems into microfluidic mode.
To have the reconfiguration capability, it is necessary to use a patterning stencil that allows precise positioning of cells onto the membrane surface. The stencil ensures that no cells are outside the porous culture region (i.e., on the silicon support region), which can be damaged upon the insertion of the flow module. This aspect becomes particularly crucial in co-culture models because cells inadvertently seeded in unintended locations cannot actively participate in the development of the barrier tissue. Nonetheless, these misplaced cells are usually retrieved during the lysis process and can potentially introduce bias in gene expression studies that investigate interactions between cells across the membrane20. Open-well cell seeding using the stencil enhances seeding efficiency by mitigating cell losses along the flow path, which are inherent in conventional microfluidic platforms4,16. This platform is also capable of incorporating multiple cell types and ECM materials15,16,17. For instance, a cell-laden hydrogel can be injected into the lower channel of the device to mimic the tissue compartment while an endothelium is established on top of the membrane.
Although the magnetic modules and components can be reused, maintaining effective sealing over several uses can be an issue due to the magnets loosening in the housing and decreasing the sealing force. This problem can be mitigated by making new laminated modules for each experiment or mass-producing components using injection molding or 3D printing techniques once designs have been established. In the method described in this protocol, the flow module is manually placed in the core module, and the multiplexing and automation capability of the system is limited. To improve experimental throughput, the flow module and upper housing can be integrated into one functional module containing internal routing channels and standardized tubing connections that simultaneously perfuse several modules in parallel.
Components of the open-well core module are mass-produced and commercially available. Other components of the system, such as the flow module, housings, and patterning stencil, can also be fabricated on a large scale. Furthermore, the reconfigurability of the platform allows the addition of sensors and actuators (e.g., transendothelial electrical resistance module, electroporation module) for real-time stimulation and measurements. Overall, this platform combines conventional and microfluidic approaches to help support widespread use outside of engineering laboratories.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This research was funded in part by the National Institute of Health under award numbers R43GM137651, R61HL154249, R16GM146687, and NSF grant CBET 2150798. The authors thank the RIT Machine Shop for aluminum mold fabrication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| 0.5 x 0.86 Micro Flow tubes | Langer Instruments | WX10-14 & DG Series | |
| 1 mm Disposable Biopsy Punches, Integra Miltex | VWR | 95039-090 | |
| 1x PBS 7.4 pH | ThermoFisher Scientific | 10010023 | |
| 20 GAUGE IT SERIES DISPENSING TIP | Jensen Global | JG20-1.5X | |
| 21 GAUGE NT PREMIUM SERIES ANGLED DISPENSING TIP | Jensen Global | JG21-1.0HPX-90 | |
| 3M 467 MP Pressure senstitive adhesive (PSA) | DigiKey | 3M9726-ND | |
| 3M 468 MP Pressure senstitive adhesive (PSA) | DigiKey | 3M9720-ND | |
| AlexaFluor 488 conjugated phalloidin | ThermoFisher Scientific | A12379 | |
| Applied Biosystems TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | 4444556 | |
| Bovine Serum Albumin (BSA), Fraction V, 98%, Reagent grade, Alfa Aesar, Size = 10 g | VWR | AAJ64100-09 | |
| Clear Scratch- and UV-Resistant Cast Acrylic Sheet | McMaster-Carr | 8560K171 | 12" x 12" x 1/16" |
| Clear Scratch- and UV-Resistant Cast Acrylic Sheet | McMaster-Carr | 8589K31 | 12" x 12" x 3/32" |
| Clear Scratch- and UV-Resistant Cast Acrylic Sheet | McMaster-Carr | 8560K191 | 12" x 12" x 7.64" |
| Corning Fibronectin, Human, 1 mg | Corning | 47743-728 | |
| Cover Glasses, Globe Scientific, L x W = 24 x 60 mm | VWR | 10118-677 | |
| DOW SYLGARD 184 SILICONE ENCAPSULANT CLEAR 0.5 KG KIT | Ellsworth Adhesives | 4019862 | |
| EGM-2 Endothelial Cell Growth Medium-2 BulletKit | Lonza | CC-3162 | |
| Fixture A1&A2 | SiMPore Inc. | NA | |
| Fixture B1&B2 | SiMPore Inc. | NA | |
| High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor | Thermo Fisher Scientific | 4374966 | |
| Human umbilical vein endothelial cells (HUVEC) | ThermoFisher Scientific | C0035C | |
| LIVE/DEAD Cell Imaging Kit (488/570) | Thermo Fisher Scientific | R37601 | |
| Molecular Probes Hoechst 33342, Trihydrochloride, Trihydrate | Thermo Fisher Scientific | H3570 | |
| Nickel-plated magnets (4.75 mm diameter, 0.34 kg pull force) | K&J Magnetics | D31 | 3/16" dia. x 1/16" thick |
| Paraformaldehyde, 4% w/v aq. soln., methanol free, Alfa Aesar | Fisher Scientific | aa47392-9M | |
| Peristaltic Pump | Langer Instruments | BQ50-1J-A | |
| Photoresist SU-8 developer solution | Fisher Scientific | NC9901158 | |
| PVDF syringe filters | PerkinElmer | 2542913 | |
| Silicon wafer | University wafer,USA | 1196 | |
| SU-8 3050 | Fisher Scientific | NC0702369 | |
| Target gene: eNOS (Hs01574659_m1) | ThermoFisher Scientific | 4331182 | |
| Target gene: GAPDH (Hs02786624_g1) | ThermoFisher Scientific | 4331182 | |
| Target gene: KLF2 (Hs00360439_g1) | ThermoFisher Scientific | 4331182 | |
| Thermo Scientific Pierce 20x PBS Tween 20 | Thermo Fisher Scientific | 28352 | |
| Transport Tube Sample White caps, 5 mL, Sterile | VWR | 100500-422 | |
| TRI-reagent | ThermoFisher Scientific | AM9738 | |
| Ultrathin Nanoporous Membrane Chip | SiMPore Inc. | NPSN100-1L | The design is compatible with all of SiMPore membranes |
| uSiM component 1 | SiMPore Inc. | NA | |
| uSiM component 2 | SiMPore Inc. | NA |
Riferimenti
- Claesson-Welsh, L., Dejana, E., McDonald, D. M. Permeability of the Endothelial Barrier: Identifying and Reconciling Controversies. Trends in Molecular Medicine. 27 (4), 314-331 (2021).
- Vera, D., et al. Engineering tissue barrier models on hydrogel microfluidic platforms. ACS Applied Materials & Interfaces. 13 (12), 13920-13933 (2021).
- Wang, Y. I., Abaci, H. E., Shuler, M. L. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnology and Bioengineering. 114 (1), 184-194 (2017).
- Sakolish, C. M., Esch, M. B., Hickman, J. J., Shuler, M. L., Mahler, G. J. Modeling barrier tissues in vitro: methods, achievements, and challenges. eBioMedicine. 5, 30-39 (2016).
- Kaisar, M. A., et al. New experimental models of the blood-brain barrier for CNS drug discovery. Expert Opinion on Drug Discovery. 12 (1), 89-103 (2017).
- Tan, K., et al. A high-throughput microfluidic microphysiological system (PREDICT-96) to recapitulate hepatocyte function in dynamic, re-circulating flow conditions. Lab on a Chip. 19 (9), 1556-1566 (2019).
- Ayuso, J. M., Virumbrales-Muñoz, M., Lang, J. M., Beebe, D. J. A role for microfluidic systems in precision medicine. Nature Communications. 13 (1), 3086 (2022).
- Katt, M. E., Shusta, E. V. In vitro models of the blood-brain barrier: building in physiological complexity. Current Opinion in Chemical Engineering. 30, 42-52 (2020).
- Ingber, D. E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nature Reviews Genetics. 23 (8), 467-491 (2022).
- Duncombe, T. A., Tentori, A. M., Herr, A. E. Microfluidics: reframing biological enquiry. Nature reviews. Molecular cell biology. 16 (9), 554-567 (2015).
- Williams, M. J., et al. A low-cost, rapidly integrated debubbler (rid) module for microfluidic cell culture applications. Micromachines. 10 (6), 360 (2019).
- Ahmed, A., et al. Engineering fiber anisotropy within natural collagen hydrogels. American Journal of Physiology-Cell Physiology. 320 (6), C1112-C1124 (2021).
- Ahmed, A., et al. Microengineered 3D collagen gels with independently tunable fiber anisotropy and directionality. Advanced Materials Technologies. 6 (4), 2001186 (2021).
- Łach, A., Wnuk, A., Wójtowicz, A. K. Experimental models to study the functions of the blood-brain barrier. Bioingegneria. 10 (5), 519 (2023).
- McCloskey, M. C., et al. The Modular µSiM: A mass produced, rapidly assembled, and reconfigurable platform for the study of barrier tissue models in vitro. Advanced Healthcare Materials. 11 (18), 2200804 (2022).
- Mansouri, M., et al. The modular µsim reconfigured: integration of microfluidic capabilities to study in vitro barrier tissue models under flow. Advanced Healthcare Materials. 11 (21), 2200802 (2022).
- Hudecz, D., et al. Modelling a human blood-brain barrier co-culture using an ultrathin silicon nitride membrane-based microfluidic device. International Journal of Molecular Sciences. 24 (6), 5624 (2023).
- Joshi, I. M., et al. Microengineering 3D Collagen Matrices with Tumor-Mimetic Gradients in Fiber Alignment. bioRxiv. , (2023).
- Hsu, M. C., et al. A miniaturized 3D printed pressure regulator (µPR) for microfluidic cell culture applications. Scientific Reports. 12 (1), 10769 (2022).
- Rogers, M. T., et al. A high-throughput microfluidic bilayer co-culture platform to study endothelial-pericyte interactions. Scientific reports. 11 (1), 12225 (2021).
- Wettschureck, N., Strilic, B., Offermanns, S. Passing the vascular barrier: endothelial signaling processes controlling extravasation. Physiological Reviews. 99 (3), 1467-1525 (2019).
- Wang, Y. I., Shuler, M. L. UniChip enables long-term recirculating unidirectional perfusion with gravity-driven flow for microphysiological systems. Lab on a Chip. 18 (17), 2563-2574 (2018).
- Nayak, L., Lin, Z., Jain, M. K. 34;Go with the flow": how Krüppel-like factor 2 regulates the vasoprotective effects of shear stress. Antioxidants & Redox Signaling. 15 (5), 1449-1461 (2011).
- Satoh, T., et al. A pneumatic pressure-driven multi-throughput microfluidic circulation culture system. Lab on a chip. 16 (12), 2339-2348 (2016).
- Abhyankar, V. V., Wu, M., Koh, C. Y., Hatch, A. V. A Reversibly sealed, easy access, modular (seam) microfluidic architecture to establish in vitro tissue interfaces. PLOS ONE. 11 (5), e0156341 (2016).
- Ahmed, A., et al. Local extensional flows promote long-range fiber alignment in 3D collagen hydrogels. Biofabrication. 14 (3), 035019 (2022).
- Hasan, M. R., et al. One-step fabrication of flexible nanotextured PDMS as a substrate for selective cell capture. Biomedical Physics & Engineering Express. 4 (2), 025015 (2018).
- Ahmed, A., et al. Microengineering 3D collagen hydrogels with long-range fiber alignment. Journal of Visualized Experiments. 187, e64457 (2022).

