Microarray Polymer Profiling (MAPP) for High-Throughput Glycan Analysis
Summary
Microarray polymer profiling (MAPP) is a high-throughput technique for compositional analysis of glycans in biological samples.
Abstract
Microarray polymer profiling (MAPP) is a robust and reproducible approach to systematically determine the composition and relative abundance of glycans and glycoconjugates within a variety of biological samples, including plant and algal tissues, food materials, and human, animal, and microbial samples. Microarray technology underpins the efficacy of this method by providing a miniaturized, high-throughput screening platform, allowing thousands of interactions between glycans and highly specific glycan-directed molecular probes to be characterized concomitantly, using only small amounts of analytes. Constituent glycans are chemically and enzymatically fractionated, before being sequentially extracted from the sample and directly immobilized onto nitrocellulose membranes. The glycan composition is determined by the attachment of specific glycan-recognizing molecular probes to the extorted and printed molecules. MAPP is complementary to conventional glycan analysis techniques, such as monosaccharide and linkage analysis and mass spectrometry. However, glycan-recognizing molecular probes provide insight into the structural configurations of glycans, which can aid in elucidating biological interactions and functional roles.
Introduction
Glycans are ubiquitous in all domains of life and exhibit unparalleled diversity in structure and function compared to other macromolecules1. However, due to their complexity, variability in biosynthesis and glycosidic linkages, and the paucity of appropriate methods for dissecting glycan structures, our understanding of this diversity in structures and functions is relatively limited2.
Many glycan analysis techniques are destructive and necessitate the breakdown of glycans into their constituent monosaccharides, which can obscure relevant three-dimensional and biological contexts3. Conversely, monoclonal antibodies (mAbs), carbohydrate binding modules (CBMs), lectins, viral agglutinins, and microbial adhesins, known collectively as glycan-recognizing molecular probes (GRMPs)4, recognize and bind to specific epitopes and can be used as tools to detect and discriminate between glycans within complex multi-glycan matrices5,6.
Here, we present microarray polymer profiling (MAPP), a rapid, versatile, and nondestructive method for glycan analysis that is applicable to a broad spectrum of biological samples. The method aims to provide a robust and high-throughput technology for analyzing glycans from diverse biological and industrial/commercial systems. MAPP unites the recognition specificity of glycan-directed molecular probes with reproducible, high-performance microarray screening technology to allow thousands of molecular interactions to be profiled in parallel. The output of this approach is diagnostic insight into the composition and relative abundance of glycans within a sample or tissue of interest.
MAPP can be used as an independent, stand-alone method, or in conjunction with other biochemical techniques, such as immunofluorescence microscopy7,8,9 and monosaccharide or linkage analysis10,11. The technique can also be used to map epitope specificities of novel GRMPs, using arrays printed with pure and structurally well-defined oligosaccharide standards12. A major advantage of MAPP over other methods, such as enzyme-linked immunosorbent assay (ELISA), is its compatibility with small sample volumes13,14. Moreover, MAPP offers significantly higher-throughput analysis15 and provides an effective form of sample preservation, as printed samples are dry and stable when immobilized onto nitrocellulose16.
The binding of GRMPs is generally dependent on the presence of a number of contiguous sugar residues that collectively form a binding site (epitope) that is unique to a particular polysaccharide class (xylan, mannan, xyloglucan, etc.)17. In contrast, the individual sugar residues (xylose, mannose, glucose) that are quantified using most biochemical techniques, for example monosaccharide composition or methylation analysis, can be components of multiple polysaccharide classes and thus difficult to assign18.
MAPP has been developed in response to a technology gap, namely the ability to rapidly analyze multiple glycans from a variety of sources using small amounts of material. MAPP capitalizes on the extensive repertoire of GRMPs that have been developed and characterized over the last three decades12,19,20,21,22,23,24,25,26,27,28,29,30,31,32. The development of MAPP has been an iterative process, with the technique being steadily refined and optimized. There is now a substantial body of literature describing the application of MAPP to various natural and industrial systems where glycans play central roles 5,6,9,10,21,33,34,35,36,37,38,39. Here, we describe the current state of the art for MAPP.
Protocol
The main experimental stages of the MAPP method are summarized in Figure 1.
1. Preparation of samples
NOTE: Here, the method is applied to plant tissues for illustrative purposes. The plants selected were Coffea arabica, Allium sativum var. ophioscorodon, and several Thai mango varieties (Aokrong, Kam, Rad, Chokanan, Mamkamdang, Talabnak, Mahachanok, and Nga). The plants were selected for their commercial importance. Their processing for human consumption generates currently underutilized agro-industrial waste, which may provide a source of value-added products, including pure glycans. Thus, MAPP was applied to characterize the glycan composition of the waste plant biomass for bioprospecting purposes.
- Separate the plant material into different tissues (e.g., root, stem, and leaves).
- Dry the plant tissues in a hot air oven at 40 °C for 12-24 h (decrease/increase the time in accordance with the sample). Alternatively, snap freeze the samples in liquid nitrogen and subsequently lyophilize for ~4 days.
- Homogenise the samples to a fine powder using a pestle and mortar or a mechanical tissue lyser (see Table of Materials) with a ball bearing in each tube.
NOTE: For fresh plant tissue, drying or lyophilizing prior to homogenization is recommended. Generally, homogenization with a pestle and mortar is sufficient for most dry samples. For particularly resilient samples, such as grains, legumes, and processed food items like pasta, snap freezing in liquid nitrogen can increase the speed and efficiency of sample homogenization. We have found that mechanical homogenization is compatible with nearly all sample types, is considerably faster and less labor-intensive, and effectively minimizes the risk of sample cross-contamination from repeated use of the same equipment (i.e., pestle and mortar).
2. Preparation of alcohol insoluble residue (AIR)
- Add 1.5 mL of 70% (v/v) ethanol to 50-100 mg of air-dried and homogenized sample material.
- Vortex thoroughly to mix and then centrifuge at 10,000 x g for 10 min at room temperature. Discard the resulting supernatant using a pipette and retain the pellet.
- To the remaining pellet, add 1.5 mL of methanol and chloroform (1:1 [v/v]). Vortex, centrifuge, and discard the supernatant, as per step 2.2.
- To the remaining pellet, add 1.5 mL of 100% acetone. Vortex, centrifuge, and discard the supernatant, as per step 2.2.
- Place the resultant pellet either overnight in a fume hood to allow the residual acetone to evaporate or in a vacuum centrifuge until dry.
NOTE: AIR material can be stored at ambient temperature until required.
3. Glycan extraction
NOTE: If possible, perform all extraction steps in a tissue lyser with a ball bearing in each tube to aid resuspension. If a tissue lyser is unavailable, extractions can be performed instead with continuous stirring or shaking. It may be necessary to extend the extraction time if this is not possible.
- To 10 mg of AIR material, add 30 µL/mg of 50 mM cyclohexanediaminetetraacetic acid (CDTA; see Table of Materials), pH 7.5.
- Shake at 27 Hz for 2 min, followed by 10 Hz for 2 h.
- Centrifuge at 10,000 x g for 10 min at 4 °C. Retain the resultant supernatant, add it to a sterile microcentrifuge tube, and store it at 4 °C on a rotary shaker.
- To the residual pellet, add 30 µL/mg 4 M NaOH + 0.1 % (w/v) NaBH4.
CAUTION: NaBH4 is toxic if swallowed. Use personal protective equipment (PPE). Handle under a fume hood. Avoid dust formation. Avoid breathing dust. Do not allow the product to come into contact with water. - Repeat steps 3.2 to 3.3. Wash the residual pellet two or three times with dH2O to remove the residual NaOH.
- Add 30 µL/mg cellulase (preferably GH5 endo-1,4-β-glucanase, in appropriate enzyme buffer-as recommended by the manufacturer; see Table of Materials) to the pellet and incubate at the enzyme optimal temperature for 16 h.
- Centrifuge the samples at 10,000 x g for 10 min at 4 °C . Retain the supernatant, transfer to clean microcentrifuge tubes, and store at 4 °C on a rotary shaker. Once extracted, the samples must be printed as soon as possible.
- Centrifuge all the stored extracts again at 10,000 x g for 10 min at 4 °C.
NOTE: The samples must be free of particulates and debris. Pass through a 0.2 µm spin filter prior to microarray printing if necessary. Particularly viscous samples are incompatible with microarray analysis as the microarrayer capillaries and the print head can become easily clogged. As a general rule, users should be able to pipette all the samples intended for printing with a standard low-volume pipette.
4. Preparation of standards
- Prepare 1 mg/mL solutions of defined glycan standards (Table 1) in sterile dH2O. If using pachyman as a standard, dissolve in 4 M NaOH instead and neutralize with glacial acetic acid following solubilization.
- Store the prepared standards overnight at 4 °C on a rotary shaker to allow complete solubilization.
- Centrifuge all the standards for 10 min at 10,000 x g at 4 °C to pellet any debris. The resultant supernatant is used for subsequent printing.
5. Microarray printing
- Prepare a 1:20 (v/v) dilution of black Indian ink/drawing ink in glycerol system buffer (GSB; combine 47% glycerol, 52.9% dH2O, 0.06% Triton X-100, and 0.04% biocide [0.15%-0.17% cupric nitrate and 1.4%-2.0% magnesium nitrate in water] and filter sterilize) (see Table of Materials) and centrifuge for 10 min at 15,000 x g (room temperature).
NOTE: Ink solution is necessary to create a top and bottom border around the printed samples, so that the printed microarrays can be visually detected on the membrane. However, the ink solution is likely to contain sediment. All solutions must be free of particulates for printing, so avoid disturbing the sediment when pipetting. Discard and prepare fresh when this is no longer possible. The solution cannot easily be filtered to remove particulates. - Add 40 µL of ink solution and GSB to the first section of the first 384-well plate (Figure 2).
- Add 25 µL of GSB to all dilution 1 (D1) wells. Add 40 µL of GSB to all dilution 2, 3, and 4 (D2-D4) wells.
- Dilute the extracted glycan samples and defined glycan substrates 1:1 (v/v) with GSB by adding 25 µL of extracted glycan sample to the D1 wells in order.
- Serially dilute each sample four times by taking 10 µL of the sample from the D1 well and adding to the D2 well. Aspirate gently with a pipette to mix.
- Repeat the process by taking 10 µL of sample from the D2 well and adding it to the D3 well.
- Repeat for the D4 well. After mixing, discard 10 µL from the D4 well so that each well contains a final volume of 40 µL.
- Add 40 µL of ink solution and GSB to the final block of the final plate.
- Cover the plates with an adhesive plate cover and centrifuge for 10 min at 3,000 x g (room temperature). Ensure no bubbles remain following centrifugation and repeat if necessary.
- Using a non-contact piezoelectric microarray printing robot, print the samples onto the nitrocellulose membrane (see Table of Materials) following the steps below.
NOTE: Below are actions recommended for optimum print quality; however, the specific parameters required will ultimately depend on the instrument used. We advise that users contact the instrument manufacturer to discuss the appropriate customizations and print settings required for their device.- Prior to printing, empty the waste buffer reservoir and fill the clean buffer reservoir with clean GSB, as necessary. Switch on the instrument and allow it to stabilize for >10 min if it has an integrated humidity and temperature control system. Switch on the microarrayer and initialize the system.
NOTE: Running a test to determine the internal pressure of the instrument is recommended. If the pressure is too low, then it may be necessary to perform a high-pressure purge. Again, contact the instrument manufacturer to discuss the specific operation of the specific instrument. - Purge the print head and capillaries several times with GSB to remove debris and potential contaminants. Perform a test print run by either loading a plate of GSB alone or setting the instrument up to print from the clean buffer reservoir directly, bypassing sample aspiration from a loaded plate.
NOTE: It is not necessary to perform the test print using a nitrocellulose membrane; clean microscope slides are sufficient and offer the advantage that the spot size, shape, and quality can be visually assessed prior to sample printing. - When printing extracted glycan samples, program the system to flush with clean GSB between each sample.
NOTE: Typically, the sample volume per printed spot is 100 pL to 10 nL, and the spot size ranges from 20 µm to 100 µm, depending on the sample volume selected. Printing 100 microarrays from one source 384-well plate takes approximately 40 min, including system flushing. The resultant microarrays will be approximately 1 cm2 in size. Additional plates will increase the length of the array by approximately 1 cm per plate. A schematic of the printed microarray design is shown in Figure 3. Once printed, the microarrays are ready to use immediately and can be stored for several years. Should the print job need to be repeated for any reason, due to the potential evaporation of samples during printing, it is recommended that a new sample plate be loaded and the original plate discarded. - Once a week, thoroughly clean the print head and capillaries. Do this by loading a 384-well containing a 1:20 dilution of concentrated NaOH in GSB and perform a 40 min to 1 h print run onto microscope slides. Users should ensure this cleaning solution is compatible with their instrument before proceeding.
- Prior to printing, empty the waste buffer reservoir and fill the clean buffer reservoir with clean GSB, as necessary. Switch on the instrument and allow it to stabilize for >10 min if it has an integrated humidity and temperature control system. Switch on the microarrayer and initialize the system.
- Save the unique grid file (.gal file) produced for the printed microarray ready for downstream analysis.
6. Microarray probing
- Cut out individual, identical printed microarrays from the nitrocellulose membrane and place them into an appropriately sized vessel for probing (e.g., a 12- or 24-well microtiter plate) (see Table of Materials). The array should lie flat on the base of the vessel. One microarray is required per probe and represents one technical replicate.
- To reduce non-specific binding, incubate the microarrays for 1 h in MP-TBST blocking buffer (1x Tris-buffered saline, pH 7.5, + 0.1% [v/v] TWEEN 20 [TBST] and supplemented with 5% [w/v] skimmed-milk powder; see Table of Materials) on a rotating/rocking shaker. Ensure that the volume is sufficient to submerge the entire array.
- Following incubation, remove the MP-TBST and replace it with a fresh volume of MP-TBST.
- Incubate the arrays with monoclonal antibodies (mAbs) or His-tagged CBMs, or other GRMPs (e.g., lectins), diluted 1:10-1:1,000 (as specified by the manufacturer; see Table of Materials) in MP-TBST for 2 h on a rotating/rocking shaker.
- Following incubation, remove the molecular probe solution and cover the arrays in clean TBST, ensuring the arrays are fully submerged. To remove residual probe solution, immediately remove the TBST and replace with a fresh volume. Place the arrays on a rotating/rocking shaker for 5 min. After 5 min, remove the TBST, replace with a fresh volume, and place on a rotating/rocking shaker for 5 min.
NOTE: This process should be repeated three times, not including the initial addition and immediate removal of TBST. - Incubate the arrays with alkaline-phosphatase conjugated secondary antibodies (anti-mouse, anti-rat, anti-rabbit, anti-His, as appropriate; see Table of Materials) diluted 1:1,000 in MP-TBST for 2 h on a rocking/rotating shaker.
NOTE: Horseradish peroxidase-conjugated secondary antibodies are also suitable, used in conjunction with tetramethyl benzidine (TMB)/hydrogen peroxide substrate for color development. - Following incubation, repeat the wash procedure, as per step 6.5, to remove non-specifically bound secondary antibodies.
- Cover the arrays in nitro-blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl-phosphate (BCIP) color development solution (see Table of Materials) for the chromogenic detection of antibody binding. Leave until purple precipitate spots develop at the antigen binding sites (typically 5-30 min, however the arrays should be monitored closely to avoid oversaturation, as the reaction can occur rapidly).
CAUTION: BCIP is harmful when in contact with the skin and may cause respiratory irritation. Use PPE. Avoid dust formation. Avoid breathing dust. - To terminate the reaction, submerge the arrays in clean tap water and wash extensively.
- Place the arrays between blotting paper overnight at ambient temperature to dry.
- Discard the arrays with obvious defects and repeat the probing protocol in such instances (Figure 4).
7. Analysis and quantification
- Scan the developed arrays at a 2,400 dots per inch (dpi) resolution using a desktop scanner. Convert the images to TIFF files and then to negatives.
- Using microarray analysis software (see Table of Materials), overlay the unique .gal grid file onto each microarray image to calculate the color intensity of the spot produced at each antigen binding site and subtract the local background.
- Export the grid data as a .txt file. These can then be manually imported into an Excel sheet for analysis.
- Generate a mean spot signal intensity value for each sample by averaging the spot intensity firstly across each sample dilution and then across any biological replicates included.
- Assign a value of 100 to the highest mean spot signal intensity and normalize the remaining data accordingly.
NOTE: Normalized mean spot signal intensities can then be presented as a heatmap of relative glycan epitope abundance using the conditional formatting function in Excel33, or using the geom_tile function in R ggplot2 package40,41.
Representative Results
MAPP was applied to determine the glycan composition of agricultural biomass waste, comprising mango peels from several northern Thai varieties, Coffea arabica cherry pulp and coffee bean processing waste, and root, stem, and leaf tissue from Thai black garlic, Allium sativum var. ophioscorodon. Several plant-derived polysaccharides are used in the food industry as functional ingredients42,43. Thus, the aim of this experiment was to deduce whether these abundant and currently underutilized agro-industrial waste materials may provide a source of value-added pure polysaccharides.
AIR material is routinely employed to prepare samples intended for glycan analysis44. There are several advantages to using AIR; treatment with solvents effectively removes endogenous CAZymes, metabolites, small saccharides, lipids, and pigments, resulting in samples enriched with polysaccharides and structural proteins34. Furthermore, producing AIR is a fast and effective way to increase sample longevity, as it is thermostable and can be stored for several years.
Three mixed fractions of constituent glycans were sequentially extracted from plant AIR material using CDTA, NaOH, and cellulase. CDTA chelates Ca2+ ions, which allow the removal of Ca2+ crosslinked de-esterified pectins from plant cell walls45. Alkaline conditions allow predominantly hemicelluloses, such as mannan, xylan, and β-glucan, to be released due to the disruption of hydrogen bonding and saponification of ester linkages between cellulose microfibrils and hemicellulose, and lignin and hemicellulose, respectively46. A recombinant endo-1,4-β-glucanase from Bacillus spp. was used to degrade amorphous regions of the structural cellulose microfibrils, releasing residual glycans bound to cellulose within the cell walls47. Although this method effectively separates glycans into these three broad groups, it should be noted that the samples are not pure; by the very nature of the extraction method, hemicellulose, if present in the sample, will inevitably be extracted and subsequently detected to varying degrees in the CDTA and cellulase fractions. Likewise, some pectin will be detected in the NaOH extraction if present in the sample.
A non-contact, piezoelectric microarray printing robot was used to immobilize extracted glycan fractions onto nitrocellulose via non-covalent attachment11, forming 300 identical microarrays. Defined glycan standards (Table 1) were also included in the printed microarrays as positive controls (Figure 5). The MAPP binding profile obtained for the selected glycan standards corresponds to previously reported epitope specificities. For example, LM21 exhibited strong binding to multiple mannan polysaccharides (galactomannan and glucomannan), while LM22 exhibited only weak binding to galactomannan25. Similarly, LM19 preferentially bound to de-esterified homogalacturonan48 and LM15 bound to tamarind seed xyloglucan23.
The relative abundance of 16 epitopes, diagnostic of non-cellulosic plant cell wall polysaccharides, were detected by the attachment of glycan-directed monoclonal antibodies (Table 2) to printed extracts (Figure 6). The majority of extracted glycans were detected within the alkaline NaOH fraction. Strong binding signals were recorded for mAbs LM10 and LM11, representing xylan/arabinoxylan, within the peels of all the mango varieties tested. Within the garlic samples, LM10 and LM11 bound preferentially to root tissue extract (Garlic R) and exhibited only weak binding to the leaf tissue extract (Garlic L). LM19, representing partially methyl-esterified or un-esterified homogalacturonan, bound strongly to some mango variety extracts (Aokrong and Talabnak), but bound only weakly, or its binding was undetectable, in other varieties (Chokanan, Mamkamdang, Mahachanok, and Nga). In addition, LM19 bound only to the coffee pulp fractions and did not bind to the coffee bean processing waste material, previously thought to be composed of semi-purified coffee pectin (unpublished data).
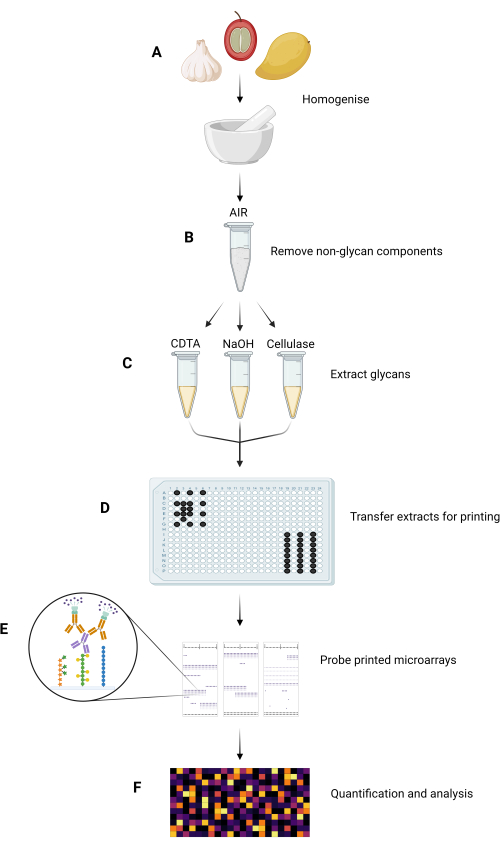
Figure 1: Major experimental steps in the MAPP method. (A) Samples are homogenized to form fine powders. (B) The homogenized samples are processed to isolate their AIRs. (C) The constituent glycans are sequentially extracted using a tailored extraction regime. (D) The extracted glycan fractions, ink, and GSB are transferred into 384-well plates, according to the plate layout, for printing onto nitrocellulose. (E) The printed microarrays are probed with selected GRMPs. (F) GRMP binding to the printed glycan fractions is quantified and analyzed before data are presented as a heatmap. Please click here to view a larger version of this figure.
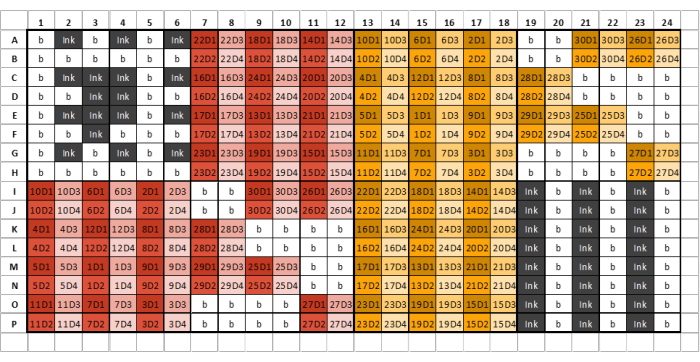
Figure 2: Example of a 384-well plate layout for sample, ink, and GSB loading with four dilutions per extracted glycan sample/standard. Different colors denote samples arising from different extraction reagents, while different shades represent serial dilutions. The first number in the code represents the sample number, while the end number represents the dilution number (D1 denotes dilution one, D2 denotes dilution two, and so on). For example, a well labeled '12D3' represents glycan sample 12, dilution three. Well plates should be divided into eight identical sections comprising six columns and eight rows. The first section of the first plate should contain only ink and buffer and resemble the example plate layout. Extracted glycan samples can then be loaded into subsequent plate sections according to the plate layout. Different extraction reagents should not be loaded into the same plate section. If there are insufficient samples to fill an entire section, fill all remaining wells in that section with buffer; do not leave any wells empty. If multiple plates are required, the next section after all samples have been loaded should contain three alternating columns of ink and GSB-this may not be section eight, depending on the number of samples being printed. Please click here to view a larger version of this figure.
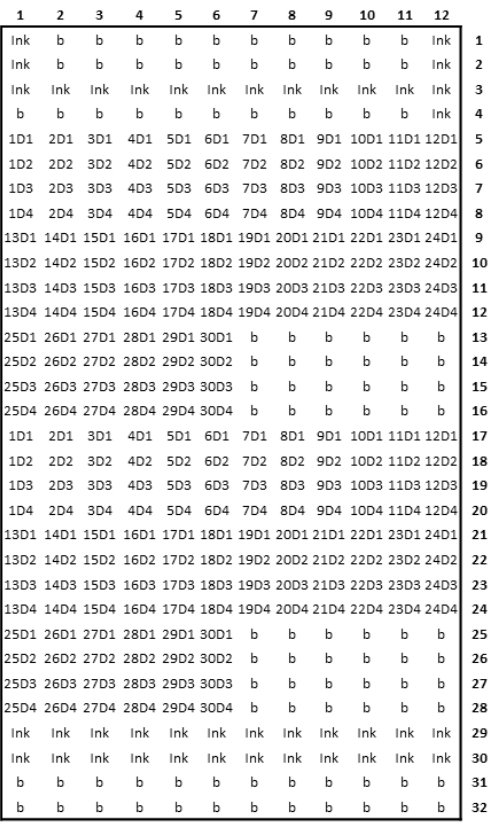
Figure 3: Schematic representation of printed microarray design. Please click here to view a larger version of this figure.
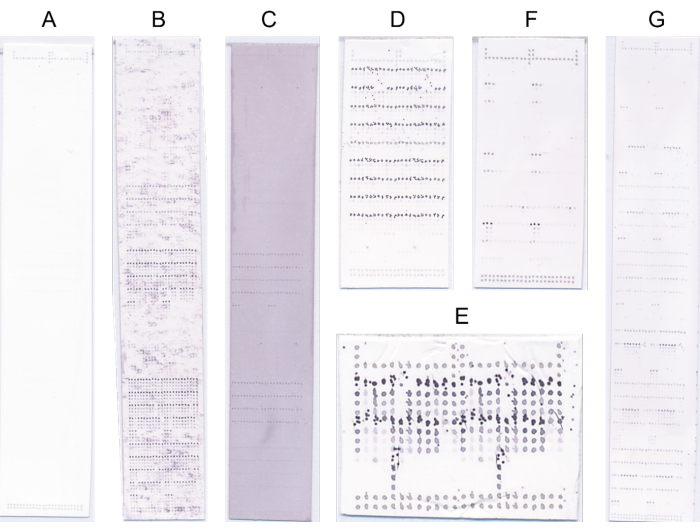
Figure 4: Representative microarrays. (A) No binding. (B) The binding signal is obscured by high background signal. (C) Generalized blue/purple staining due to oversaturation with NBT/BCIP. (D) Defective probing due to high substrate concentration. (E) Defective printing due to the unclean print head. (F) Strong binding to few samples. (G) Strong binding to many samples. Please click here to view a larger version of this figure.
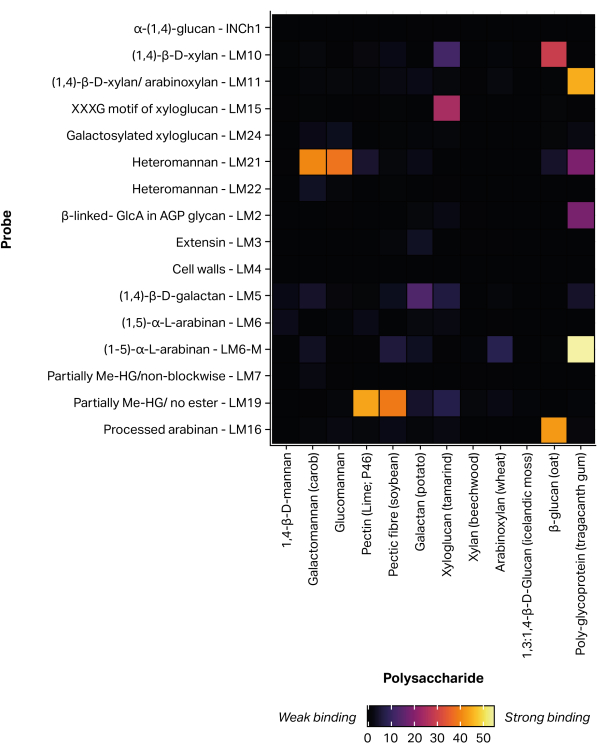
Figure 5: Monoclonal antibody binding to defined glycan standards, included to validate the printing and probing process. Please click here to view a larger version of this figure.
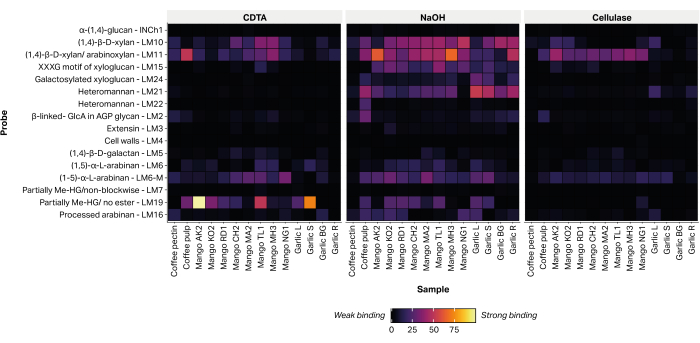
Figure 6: MAPP of glycans extracted from agricultural biomass waste. The samples include coffee pulp waste (coffee pulp and coffee pectin), mango peels from several Thai varieties (AO, Aokrong; KO, Kam; RD, Rad; CH, Chokanan; MA, Mamkamdang; TL, Talabnak; MH, Mahachanok; NG, Nga) and black garlic leaves (Garlic L), stem (Garlic S), bulb (Garlic BG), and roots (Garlic R), using CDTA, NaOH, and cellulase (Bacillus spp. cellulase 5A). Please click here to view a larger version of this figure.
Table 1: Defined commercial polysaccharide standards used in MAPP analysis as positive controls. Please click here to download this Table.
Table 2: Glycan-directed monoclonal antibodies selected for the interrogation of extracted plant glycan microarrays. Please click here to download this Table.
Discussion
The MAPP technique described here is now a well-established method for glycan analysis. The basic principles were first described in 200711, but the technique has undergone continuous development in order to capitalize on the latest innovations in microarray technology, molecular probe development, and advances in our understanding of glycan biochemistry. In general, glycans, especially polysaccharides, are more challenging to analyze than proteins and nucleotides due to their structural complexity and heterogeneity45, as well as the fact that they cannot readily be sequenced or synthesized1. In many cases, no single technique can decipher glycan complexity conclusively; thus, MAPP is often used with other methods. This is one of the reasons why AIR preparation is usually chosen as a starting point for MAPP, since AIR is compatible with most other glycan analysis methods34, facilitating the subsequent comparison of datasets.
Due to homogenization of the sample prior to AIR preparation, some spatial information is invariably lost. However, as polysaccharides are sequentially released from samples, the presence of epitopes in the fractions obtained provides information about the molecular architecture and composition of that sample17. Thus, selecting an appropriate extraction regime is critical to the success of the method. Multiple parameters determine the suitability of the extraction method: cellular structure, time, temperature, pH, pressure, ionic strength of the solvent, and fineness of the solid particulate sample49. It is recommended that a range of increasingly more aggressive solvents is used to maximize the likelihood of successfully extracting constituent glycans and building a representative compositional picture of the sample. For most samples, CDTA, NaOH, and cellulase are sufficient to remove plant-derived storage and cell wall polysaccharides33,50,51,52. For some tissue samples, a hybrid extraction regime that also includes CaCl2, HCl, and Na2CO3 has been shown to be successful53, while marine microalgal samples may require the addition of ethylenediaminetetraacetic acid (EDTA)10.
Microarrays should include a range of pure, defined glycan standards to be used as positive controls5. Included standards should be modified according to the nature of the sample. Once printed, appropriate GRMPs need to be selected. The generation of hybridoma mAbs to polysaccharide structures is challenging54; glycan-binding antibodies are difficult to raise and can have low affinity55. Fortunately, gene sequence information for CBMs can be obtained with relative ease for recombinant expression4 and engineering their binding specificities56,57. Whilst an impressive catalog of GRMPs has been developed, with most now available from commercial sources, relative to the diversity of glycan structures existing in nature, only a small proportion have been produced and successfully characterised58. This can limit the ability to detect and discriminate between certain structures. It is advisable to perform an initial probing experiment using one or two probes representative of each major glycan structure anticipated to be present, for which the binding specificity is well characterized. In subsequent probing experiments, the probe list can be expanded to cover a broader range of glycans and delve deeper into fine structures.
Although mundane, ensuring that microarrays are washed thoroughly after each incubation step is fundamental to the success of the probing procedure. The ineffective removal of non-specifically bound probes is likely to obscure the result by causing a high background signal following color development. In this case, it is necessary to repeat the probing procedure, starting with a new microarray. Furthermore, arrays should be touched sparingly and only by holding the edges with forceps; the nitrocellulose membrane is brittle and easily damaged. The color development solution collects in cracks and creases, causing oversaturation, which impedes array analysis.
MAPP is quick, adaptable, and convenient. This method is compatible with animal, microbial, or plant glycans derived from any biological or industrial system, so long as they can be extracted and immobilized onto nitrocellulose, and for which one has appropriate molecular probes. The data generated provide detailed, semi-quantitative, compositional insight, which cannot readily be obtained via other glycan analysis methods.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge ArrayJet for their expert advice regarding microarray robotics. SS and JS would like to acknowledge the support from Fundamental Fund 2022 (FF65/004), Chiang Mai University.
Materials
| 1,3:1,4-β-D-Glucan, Lichenan (icelandic moss) | Megazyme | P-LICHN | |
| 1,4-β-D-Mannan | Megazyme | P-MANCB | |
| 384-well microtiter plate | Greiner Bio-One | M1686 | |
| 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) | Melford | B74100-1.0 | |
| Acetone | Sigma | 270725 | |
| Alkaline Phosphatase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 115-055-003 | |
| Alkaline Phosphatase AffiniPure Goat Anti-Rat IgG (H+L) | Jackson ImmunoResearch | 112-055-003 | |
| Alkaline Phosphatase AffiniPure Rabbit Anti-His Tag | Jackson ImmunoResearch | 300-055-240 | |
| Arabinoxylan (wheat) | Megazyme | P-WAXYL | |
| Array-Pro Analyzer Software | Media Cybernetics | Version 6.3 | |
| Bacillus sp. Cellulase 5A (BCel5A) | NZYTech | CZ0564 | |
| BAM antibodies | SeaProbes | Various | |
| Black drawing ink (indian ink) | Winsor & Newton | GWD030 | |
| Carbohydrate binding modules | NZYTech | Various | |
| CCRC antibodies | CarboSource | Various | |
| CDTA | Sigma | 319945 | |
| Chloroform | Sigma | PHR1552 | |
| Ethanol | Sigma | 1.11727 | |
| Galactan (potato) | Megazyme | P-GALPOT | |
| Galactomannan (carob) | Megazyme | P-GALML | |
| Glycerol solution | Sigma | 49781-5L | |
| Gum tragacanth (legumes) | Sigma-Aldrich | G1128 | |
| INCh antibodies | INRA | Various | |
| LM and JIM antibodies | PlantProbes | Various | |
| Marathon Argus Microarray Printer | ArrayJet | ||
| Methanol | Sigma | 34860 | |
| Monoclonal antibodies | Biosupplies Australia | Various | |
| NaBH4 | Sigma | 452882 | |
| NaOH | Sigma | S5881 | |
| Nitro-blue tetrazolium (NBT) | Melford | N66000-1.0 | |
| Nitrocellulose membrane | Thermo Fisher Scientific | 88018 | |
| Pectin (degree of methyl esterification 46%) | Danisco | NA | |
| ProClin 200 | Sigma | 48171-U | |
| Rhamnogalacturonan (soybean pectic fibre) | Megazyme | P-RHAGN | |
| Rotating mixer | Fisher Scientific | 88-861-050 | |
| Rotating/rocking Shaker | Cole-Parmer | ||
| Skimmed milk powder | Marvel | ||
| Spin filter | Costar Spin-X | 8160 | |
| Stainless steel beads | Qiagen | 69989 | |
| TissueLyser II | Qiagen | 85300 | |
| Tris | Sigma | 93362 | |
| Triton X-100 | Sigma | T8787-250ML | |
| Tween 20 | Sigma | P9416-100ML | |
| Xylan (beechwood) | Megazyme | P-XYLNBE | |
| Xyloglucan (tamarind) | Megazyme | P-XYGLN | |
| β-Glucan (oat) | Megazyme | P-BGOM |
References
- Amicucci, M. J., et al. A nonenzymatic method for cleaving polysaccharides to yield oligosaccharides for structural analysis. Nature Communications. 11 (1), 3963 (2020).
- Gagneux, P., Panin, V., Hennet, T., Aebi, M., Varki, A. Evolution of glycan diversity. Essentials of Glycobiology [Internet]. 4th edition. , (2022).
- Willats, W. G. T., McCartney, L., Knox, J. P. Pectin cell biology: complexity in context. Advances in Pectin and Pectinase Research. , 147-157 (2003).
- Cummings, R. D., et al. Glycan-recognizing probes as tools. Essentials of Glycobiology [Internet]. 4th edition. , (2022).
- Bakshani, C. R., et al. Analysis of glycans in a Burnt-on/Baked-on (BoBo) model food soil using Microarray Polymer Profiling (MAPP) and immunofluorescence microscopy. Food Chemistry. 410, 135379 (2023).
- Salmeán, A. A., Willats, W. G. T., Ribeiro, S., Andersen, T. J., Ellegaard, M. Over 100-year preservation and temporal fluctuations of cell wall polysaccharides in marine sediments. Frontiers in Plant Science. 13, 785902 (2022).
- Cid, M., et al. Recognition of the helical structure of β-1,4-galactan by a new family of carbohydrate-binding modules. Journal of Biological Chemistry. 285 (46), 35999-36009 (2010).
- Runavot, J. -. L., et al. Non-cellulosic polysaccharides from cotton fibre are differently impacted by textile processing. PLoS One. 9 (12), e115150 (2014).
- Ahl, L. I., et al. Analyses of aloe polysaccharides using carbohydrate microarray profiling. Journal of AOAC International. 101 (6), 1720-1728 (2018).
- Vidal-Melgosa, S., et al. Diatom fucan polysaccharide precipitates carbon during algal blooms. Nature Communications. 12 (1), 1150 (2021).
- Moller, I., et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. The Plant Journal. 50 (6), 1118-1128 (2007).
- Ruprecht, C., et al. A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiology. 175 (3), 1094-1104 (2017).
- Cummings, R. D., et al. Principles of glycan recognition. Essentials of Glycobiology [Internet]. 4th edition. , (2022).
- Gao, C., et al. Glycan microarrays as chemical tools for identifying glycan recognition by immune proteins. Frontiers in Chemistry. 7, 833 (2019).
- Vidal-Melgosa, S., et al. A new versatile microarray-based method for high throughput screening of carbohydrate-active enzymes. Journal of Biological Chemistry. 290 (14), 9020-9036 (2015).
- Willats, W. G. T., Rasmussen, S. E., Kristensen, T., Mikkelsen, J. D., Knox, J. P. Sugar-coated microarrays: A novel slide surface for the high-throughput analysis of glycans. Proteomics. 2 (12), 1666-1671 (2002).
- Sørensen, I., et al. The charophycean green algae provide insights into the early origins of plant cell walls. The Plant Journal. 68 (2), 201-211 (2011).
- Liu, D., Tang, W., Yin, J. -. Y., Nie, S. -. P., Xie, M. -. Y. Monosaccharide composition analysis of polysaccharides from natural sources: Hydrolysis condition and detection method development. Food Hydrocolloids. 116, 106641 (2021).
- Pattathil, S., et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiology. 153 (2), 514-525 (2010).
- Verhertbruggen, Y., et al. Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. The Plant Journal. 59 (3), 413-425 (2009).
- Rydahl, M. G., et al. Development of novel monoclonal antibodies against starch and ulvan – implications for antibody production against polysaccharides with limited immunogenicity. Scientific Reports. 7 (1), 9326 (2017).
- McCartney, L., Marcus, S. E., Knox, J. P. Monoclonal antibodies to plant cell wall xylans and arabinoxylans. The Journal of Histochemistry and Cytochemistry. 53 (4), 543-546 (2005).
- Marcus, S. E., et al. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biology. 8, 60 (2008).
- Pedersen, H. L., et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. The Journal of Biological Chemistry. 287 (47), 39429-39438 (2012).
- Marcus, S. E., et al. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. The Plant Journal. 64 (2), 191-203 (2010).
- Smallwood, M., Martin, H., Knox, J. P. An epitope of rice threonine-and hydroxyproline-rich glycoprotein is common to cell wall and hydrophobic plasma-membrane glycoproteins. Planta. 196 (3), 510-522 (1995).
- Smallwood, M., Yates, E. A., Willats, W. G. T., Martin, H., Knox, J. P. Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta. 198 (3), 452-459 (1996).
- Wisniewski, J. P., Rathbun, E. A., Knox, J. P., Brewin, N. J. Involvement of diamine oxidase and peroxidase in insolubilization of the extracellular matrix: implications for pea nodule initiation by Rhizobium leguminosarum. Molecular Plant-Microbe Interactions. 13 (4), 413-420 (2000).
- Jones, L., Seymour, G. B., Knox, J. P. Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1[->]4)-β-D-galactan. Plant Physiology. 113 (4), 1405-1412 (1997).
- Willats, W. G., Marcus, S. E., Knox, J. P. Generation of a monoclonal antibody specific to (1→5)-α-L-arabinan. Carbohydrate Research. 308 (15), 149-152 (1998).
- Cornuault, V., et al. LM6-M: a high avidity rat monoclonal antibody to pectic α-1, 5-L-arabinan. BioRxiv. , 161604 (2017).
- Sutherland, P., Hallett, I., Jones, M. Probing cell wall structure and development by the use of antibodies: a personal perspective. New Zealand Journal of Forestry Science. 39, 197-205 (2009).
- Mikkelsen, M. D., et al. Ancient origin of fucosylated xyloglucan in charophycean green algae. Communications Biology. 4 (1), 754 (2021).
- Fangel, J. U., Jones, C. Y., Ulvskov, P., Harholt, J., Willats, W. G. T. Analytical implications of different methods for preparing plant cell wall material. Carbohydrate Polymers. 261, 117866 (2021).
- Moore, J. P., et al. Analysis of plant cell walls using high-throughput profiling techniques with multivariate methods. The Plant Cell Wall: Methods and Protocols. , 327-337 (2020).
- Gao, Y., Fangel, J. U., Willats, W. G. T., Moore, J. P. Tracking polysaccharides during white winemaking using glycan microarrays reveals glycoprotein-rich sediments. Food Research International. 123, 662-673 (2019).
- Solden, L. M., et al. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nature Microbiology. 3 (11), 1274-1284 (2018).
- Fangel, J. U., et al. Tracking polysaccharides through the brewing process. Carbohydrate Polymers. 196, 465-473 (2018).
- Mravec, J., et al. Pea border cell maturation and release involve complex cell wall structural dynamics. Plant Physiology. 174 (2), 1051-1066 (2017).
- Wickham, H. . ggplot2: elegant graphics for data analysis. , (2009).
- Gu, Z. Complex heatmap visualization. iMeta. 1 (3), 43 (2022).
- Nasrollahzadeh, M., Nezafat, Z., Shafiei, N., Soleimani, F. . Polysaccharides in Food Industry. , (2021).
- Shao, P., et al. Recent advances in improving stability of food emulsion by plant polysaccharides. Food Research International. 137, 109376 (2020).
- Sanz, M. L., Martínez-Castro, I. Recent developments in sample preparation for chromatographic analysis of carbohydrates. Journal of Chromatography. A. 1153 (1-2), 74-89 (2007).
- Bethke, G., Glazebrook, J. Cyclohexane diamine tetraacetic acid (CDTA) extraction of plant cell wall pectin. Bio-Protocol. 4 (24), e1357 (2014).
- Lu, Y., He, Q., Fan, G., Cheng, Q., Song, G. Extraction and modification of hemicellulose from lignocellulosic biomass: A review. Green Processing and Synthesis. 10 (1), 779-804 (2021).
- Jayasekara, S., Ratnayake, R. Microbial cellulases: an overview and applications. Cellulose. 22, 92 (2019).
- Verhertbruggen, Y., Marcus, S. E., Haeger, A., Ordaz-Ortiz, J. J., Knox, J. P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydrate Research. 344 (14), 1858-1862 (2009).
- Villares, A., Mateo-Vivaracho, L., Guillamón, E. Structural features and healthy properties of polysaccharides occurring in mushrooms. Agriculture. 2 (4), 452-471 (2012).
- Kračun, S. K., et al. Carbohydrate microarray technology applied to high-throughput mapping of plant cell wall glycans using comprehensive microarray polymer profiling (CoMPP). Methods in Molecular Biology. 1503, 147-165 (2017).
- Rajasundaram, D., et al. Understanding the relationship between cotton fiber properties and non-cellulosic cell wall polysaccharides. PLoS One. 9 (11), e112168 (2014).
- Michalak, L., et al. Microbiota-directed fibre activates both targeted and secondary metabolic shifts in the distal gut. Nature Communications. 11 (1), 5773 (2020).
- Salmeán, A. A., Hervé, C., Jørgensen, B., Willats, W. G., Mravec, J. Microarray glycan profiling reveals algal fucoidan epitopes in diverse marine metazoans. Frontiers in Marine Science. 4, 293 (2017).
- Knox, J. P. Revealing the structural and functional diversity of plant cell walls. Current Opinion in Plant Biology. 11 (3), 308-313 (2008).
- Manimala, J. C., Roach, T. A., Li, Z., Gildersleeve, J. C. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 17 (8), 17-23 (2007).
- Stephen, P., Tseng, K. -. L., Liu, Y. -. N., Lyu, P. -. C. Circular permutation of the starch-binding domain: inversion of ligand selectivity with increased affinity. Chemical Communications. 48 (20), 2612-2614 (2012).
- Gunnarsson, L. C., Dexlin, L., Karlsson, E. N., Holst, O., Ohlin, M. Evolution of a carbohydrate binding module into a protein-specific binder. Biomolecular Engineering. 23 (2-3), 111-117 (2006).
- Moller, I., et al. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconjugate Journal. 25 (1), 37-48 (2008).

