High-Quality Brain and Bone Marrow Nuclei Preparation for Single Nuclei Multiome Assays
Summary
The success of single-cell/single-nuclei transcriptomics and multi-omics largely depends on the quality of cells/nuclei. Therefore, isolating cells/nuclei from tissue and their purification must be highly standardized. This protocol describes the preparation of nuclei from the brain and bone marrow for downstream single-nuclei multiome assay.
Abstract
Single-cell analysis has become the approach of choice for unraveling the complexity of biological processes that require assessing the variability of individual cellular responses to treatment or infection with single-cell resolution.
Many techniques for single-cell molecular profiling have been developed over the past 10 years, and several dedicated technologies have been commercialized. The 10X Genomics droplet-based single-cell profiling is a widespread technology that offers ready-to-use reagents for transcriptomic and multi-omic single-cell profiling. The technology includes workflows for single-cell and single-nuclei RNA sequencing (scRNA-Seq and snRNA-Seq, respectively), scATAC-Seq, single-cell immune profiling (BCR/TCR sequencing), and multiome. The latter combines transcriptional (scRNA-Seq) and epigenetic information (scATAC-Seq) coming from the same cell.
The quality (viability, integrity, purity) of single-cell or single-nuclei suspensions isolated from tissues and analyzed by any of these approaches is critical for generating high-quality data. Therefore, the sample preparation protocols should be adapted to the particularities of each biological tissue and ensure the generation of high-quality cell and nuclei suspensions.
This article describes two protocols for preparing brain and bone marrow samples for the downstream multiome 10X Genomics pipeline. The protocols are performed stepwise and cover tissue dissociation, cell sorting, nuclei isolation, and quality control of prepared nuclei suspension that is used as starting material for cell partitioning and barcoding, library preparation, and sequencing. These standardized protocols produce high-quality nuclei libraries and robust and reliable data.
Introduction
For many years, single-cell techniques have been the gold standard for the analysis of biological processes. They were initially restricted to single-cell phenotyping through microscopy, flow cytometry, and similar assays. A breakthrough in single-cell analysis came with the development of approaches for single-cell molecular profiling, in particular single-cell RNA sequencing (scRNA-Seq) that enabled characterizing the entire transcriptome of individual cells. Highly powerful, scRNA-Seq generates information about the transcriptional status of a cell in a specific condition and time point. However, it does not provide visibility on the gene regulation that drives transcription, or on the molecular modifications that occur over time. To overcome this limitation, many efforts have been invested in the development of single-cell multi-omics assays that enable the analysis of multiple factors and processes from the same cell1,2,3,4. The first successful measurement of two modalities within single cells came through linking multiplex surface protein expression patterns with the full transcriptome of individual cells in the CITE-Seq approach5. More recent evolutions combine gene expression with chromatin accessibility (Assay for Transposase-Accessible Chromatin using sequencing, ATAC-Seq), thereby simultaneously capturing transcriptomic and epigenomic modalities in the same cells (e.g., sci-CAR)6. The first commercial solutions that allowed associating transcriptomics with cell phenotype or with epigenetic changes of the same cell came from 10X Genomics.
Experiments for single-cell molecular profiling contain the following steps: (1) tissue dissociation or preparation of single-cell suspensions; (2) cell purification and/or nuclei isolation; (3) partitioning and barcoding; (4) library construction and quality control; (5) next-generation sequencing; (6) data analysis. While steps (3)-(6) may significantly vary depending on the technology employed, the initial steps are generally common to all of them. The quality of the prepared cell/nuclei suspension will determine the overall outcome of the experiment. Depending on the type of tissue, obtaining high-quality single-cell/nuclei suspensions may be challenging. The particularities of some tissues, such as the heart, the muscle, the brain, the lung, the intestine, and others, require methods for tissue disruption and nuclei isolation adapted to each type of sample in order to guarantee the production of high-quality nuclei for molecular analysis7,8,9,10. The tissue disruption methods and dissociation protocols can be mechanical, enzymatic (e.g., a mix of collagenases and DNase), or a combination of the two, and can be performed manually or by instruments (e.g., Qiagen DSC-400, gentleMACS).
Single-cell techniques have become a tool of choice for biomedical research. In neurobiology, the cell diversity in the brain and the complexity of their functions require high-resolution and high-throughput analysis for visualization of rare cell populations and for assessing their heterogeneity11,12,13,14. Linking cellular identity and gene regulatory mechanisms of individual cells provides insights into brain development and physiology. Another example is studies of immune response in the context of infectious, autoimmune, or cancer diseases, which strongly rely on single-cell analyses. The heterogeneity of immune cell subsets and the complexity of their activity and interactions with other cell types require single-cell resolution in deciphering the mechanisms underlying immune response. The immune cells originate from the bone marrow, where hematopoietic progenitors are composed of gradually differentiating cells that acquire and lose cell surface markers throughout a stepwise process prior to exiting the bone marrow to home in the periphery. Single-cell analysis allows for minute characterization of cellular developmental stages. It can be achieved through single-cell phenotyping, conventionally performed by multi-parameter flow cytometry. However, single-cell transcriptomic signatures have been shown to reveal a more precise identification of progenitor cell subtypes since these cells are distributed in clusters that fall into one another and can therefore be misidentified when using a coarse cell surface marker approach15. An increasing number of studies uncover the epigenetic modifications that hematopoietic stem and progenitor cells (HSPCs) can acquire from exposure to various agents, leading to a significant impact on the long-term responsiveness of the immune system16,17,18,19. The novel multi-omics technologies enable studying these processes with single-cell resolution.
Many protocols for cell and nuclei isolation have been described for brain11,20,21,22 and bone marrow samples23,24. To minimize bias due to experimental variability, it is necessary to validate optimized single-nuclei preparation protocols for joint single-cell transcriptomic and epigenomic sequencing, thereby ensuring the reproducibility of single-cell multiomic assays.
Here, two robust protocols for nuclei preparation from (1) fresh-frozen brain tissue and (2) fresh bone marrow HSPCs for downstream single-cell Multiome analysis are described (Figure 1).
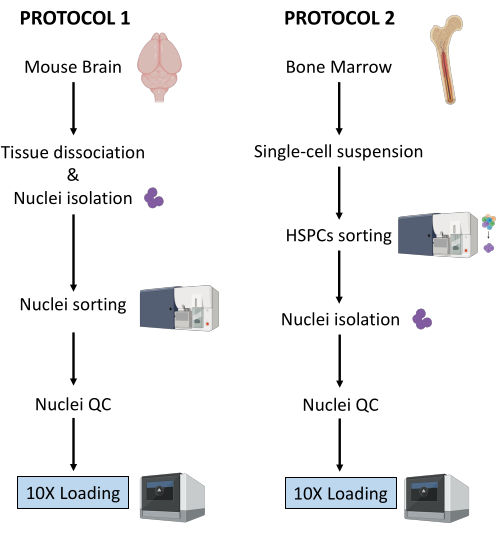
Figure 1: Schematic representation of protocols for nuclei isolation from fresh-frozen brain and bone marrow tissues. Please click here to view a larger version of this figure.
Protocol
Experimental procedures were conducted under the strict regulatory compliance of protocols approved by the Committee for Ethics in Animal Experimentation (CETEA). For the brain nuclei isolation, 3-month-old C57BL/6 mice were used. For the bone marrow isolation, 8-week-old female C57BL/6J mice weighing 18 g were used.
1. Purification of nuclei from mouse brain
NOTE: Wear latex or nitrile gloves at all times during the procedure. It is strongly advised to have two people performing the experiment, to have steps 1 to 3 (i.e., preparation of the single nuclei suspension) performed by one person, and step 4 (i.e., preparation of the sorter) performed in parallel by another person. Since the protocol is highly time-sensitive, it is critical to minimize sample processing time by having the sorter ready as soon as the single nuclei suspension is prepared.
- Preparation of reagents and materials
- Carefully sterilize the dissection tools by autoclave (at 121 °C for 20 min) and wash them with ethanol 70% just before use. Prepare one Petri dish per sample, filled with 2-3 mL of ice-cold 1x Dulbecco's phosphate-buffered saline (DPBS).
- Cool the microcentrifuge to 4 °C, fill a bucket with ice, and put the glass dounce homogenizer on ice.
- Prepare nuclei lysis buffer by adding digitonin for a final concentration of 0.01%, 10 mL per sample.
- Prepare the staining buffer by adding RNase inhibitors to the cell staining buffer for a final concentration of 0.2 U/µL, 20 mL per sample.
- Prepare DPBS 0.04% BSA by adding RNase inhibitors for a final concentration of 0.2 U/µL, 2 mL per sample.
- Prepare 1 mL of diluted nuclei buffer according to the Multiome protocol25.
- Keep all the reagents and samples on ice.
- Tissue dissection
- Sacrifice mice using protocols approved by the institution. In this protocol, the mice were decapitated after a ketamine/xylazine overdose.
- Cut the mouse head with scissors and remove the brain from the skull as described in Meyerhoff et al.26. Immediately transfer the brain into a Petri dish prepared with the ice-cold 1x DPBS under a light-emitting diode (LED)-illuminated stereo microscope.
- Cut the brain tissue with a scalpel to separate brain areas of interest (e.g., Entorhinal cortex, hippocampus, prefrontal cortex) and transfer each region into a separate Petri dish containing ice-cold 1x DPBS. Keep on ice.
- With a scalpel, mince the tissue into <0.5 cm pieces to facilitate the homogenization in the following step.
- With a P1000 micropipette, transfer the minced tissue and the 1x DPBS from the Petri dish to a 1.5 mL tube. Make sure to use tubes made of protein-low-binding plastic. Allow the tissue pieces to separate by gravity. Carefully remove the excess of 1x DPBS using a P1000 micropipette.
NOTE: After this step, it is possible to snap-freeze the minced tissue by transferring the protein low-binding tubes to dry ice and then storing at -80 °C until proceeding with nuclei isolation.
- Nuclei isolation
- Fill the glass dounce with 2 mL of ice-cold nuclei lysis buffer with 0.01% digitonin. Add the tissue pieces in the dounce.
NOTE: if working with fresh-frozen tissue, add the minced frozen tissue directly to the nuclei lysis buffer 0.01% digitonin; do not let the tissue thaw before. - Homogenize using a glass dounce tissue homogenizer 25 times with pestle A and then 25 times with pestle B. Transfer the homogenate into a 15 mL tube.
- Add an additional 2 mL of ice-cold nuclei lysis buffer with 0.01% digitonin and incubate on ice for 5 min. Centrifuge nuclei at 500 x g for 5 min at 4 °C.
- Remove the supernatant with a micropipette and add 4 mL of ice-cold nuclei lysis buffer with 0.01% digitonin. Incubate on ice for 5 min and filter through a 40 µm cell strainer.
- Centrifuge nuclei at 500 x g for 5 min at 4 °C and remove the supernatant with a micropipette.
- Add 4 mL of staining buffer to wash the nuclei and centrifuge at 500 x g for 5 min at 4 °C. Remove the supernatant with a micropipette and resuspend the pellet in 4 mL of staining buffer.
- Filter through a 40 µm cell strainer and centrifuge at 500 x g for 5 min at 4 °C. Resuspend in 1 mL of PBS with 0.04% BSA.
- Count nuclei to ensure consistency of tissue/nuclei preparation across different samples. It is expected to obtain similar nuclei counts from the same brain regions:
- Add 10 µL of 0.4% trypan blue to an empty 0.5 mL tube. Add 10 µL of the nuclei and mix 5x by pipetting.
- Count nuclei using an automated cell counter following the supplier's recommendations. Keep the nuclei on ice.
- Prepare nuclei for sorting.
NOTE: The extracted nuclei incorporate 7-AAD, and this staining is used for their purification by fluorescence-activated cell sorter (FACS).- Transfer 100 µL of nuclei to a FACS tube for the unstained control. Add 10 µL of 7-AAD to the remaining nuclei, and keep 5 min at 4 °C.
- Sort a minimum of 0.5 x 106 nuclei by FACS to eliminate doublets and debris.
- Fill the glass dounce with 2 mL of ice-cold nuclei lysis buffer with 0.01% digitonin. Add the tissue pieces in the dounce.
- Nuclei sorting using a FACS
NOTE: While nuclei sorting can be performed on a wide variety of cell sorters, the procedure for using the BD FACSAria Fusion or BD FACSAria III instruments is described here. It is strongly recommended that the calibration and the setup of the cell sorter be performed under the supervision, or by an experienced user of the instrument. To reduce the sample processing time, it is critical to have the sorter ready as soon as the single nuclei suspension is prepared.- Calibration of FACS instrument
- Turn on the cell sorter and the computer. Once the software is connected to the instrument, launch the fluidic start-up procedure. Select Cytometer > Fluidic start-up in the main menu and follow the four steps. Click on Done after completing each.
- Insert the 70 µm nozzle, turn on the stream, and leave the stream to stabilize for 15 min. Adjust amplitude to get drop formation and click on Sweet Spot.
- Put the neutral density (N.D) filter 1.0 and open the cytometer setup and tracking (CST) interface.
- Daily quality control: Dilute CST beads in FACS medium (see supplier's recommendations) and perform CST control. Once completed, replace the N.D 1.0 with the N.D 2.0.
- Dilute Accudrops in FACS medium (see supplier's recommendations) and perform drop delay as described in steps 6 to 10.
- In the experiment template, select the Accudrop Drop Delay experiment and open the Sorting Layout for the tube.
- Inside the lower camera window, click on Voltage and then on Optical Filter to enable applying charge on the deflection plates and using a specific optical filter in front of the camera. Ensure that the quadrant on the right side indicates 100. If necessary, adjust the red laser screw to optimize the laser impact.
- Adjust the flow rate to reach the speed of 1,000 to 3,000 events per second.
- Click on Sort and Cancel. Ensure that the left quadrant equals 100, and the right quadrant is 0. If the left quadrant is below 95, perform Auto Delay.
- Click on Voltage, then Test Sort. Control the quality of the side streams deposit in the collection tubes. If needed, adjust the position of the side streams by moving the sliders.
- Set up of FACS instrument for nuclei sorting.
- Start the acquisition of the unstained nuclei. These are used to define the forward and side scatters, and the detector voltage for the 7-ADD parameter. Set the parameters so that the 7-AAD signal of the unstained sample falls inside the first decade of the log scale on the dot plot.
- Start acquiring the tube of 7-AAD-stained nuclei and define the nuclei populations by using a gating strategy based on (1) FSC-A/SCC-A and then FSC-H/SSC-H for size and granularity, (2) FSC-H/FSC-A for doublets discrimination, and (3) SSC-A/7-AAD for 7-AAD positive nuclei (see Figure 2A).
- Make sure that the stream and the deflection are stable.
- In the side stream camera, turn on the test sort, voltage ON, and confirm the accurate drop sorting in a 1.5 mL tube fitted on the left side.
- In the Sorting Layout window, select the population of interest, as defined in step 2 (above). In Target Events, select the threshold in Continuous to obtain a minimum of 0.5 x 106 nuclei per sample. Under Precision, select 4-Way Purity.
- Once ready, click Sort and OK to start with nuclei sorting.
- Calibration of FACS instrument
- Quality control and counting of purified nuclei
NOTE: This step is to be done only during the pilot experiment for optimization of sample preparation steps, with the goal of testing the purity of the sorted nuclei that are to be loaded on the 10X chromium chip. Once the protocol is fully optimized, it is not advised to perform this quality control step in the follow-up experiments to avoid unnecessary waste of collected nuclei that may be available in low numbers.- Purity control by flow cytometry
- Transfer 10 µL of the sorted nuclei into a new FACS tube containing 90 µL of DPBS with 2% heat-inactivated Fetal Bovine Serum (HI-FBS).
- Acquire and record post-sort data to verify the sorting purity and viability. Ensure at least 98% of the nuclei appear in the gate of interest, as defined in 4.2 (see Figure 2B).
- Counting the purified nuclei
- Centrifuge sorted nuclei for 5 min at 500 x g and at 4 °C and carefully completely remove the supernatant using a micropipette. Resuspend in 100 µL of diluted nuclei buffer.
- Add 10 µL of 0.4% trypan blue to an empty 0.5 mL tube. Add 10 µL of the sorted nuclei and mix 5x by pipetting.
- Count nuclei using an automated cell counter following the supplier's recommendations. Adjust the nuclei concentration to 3.5 x 106/mL, i.e., 16,000 nuclei per 5 µL.
- Quality control of purified nuclei by microscopy
NOTE: This step is to be done only during the pilot experiment for optimization of sample preparation steps to test the quality of the nuclei that are to be loaded on the 10X chromium chip. Once the protocol is fully optimized, it is not advised to perform this quality control step in the follow-up experiments to avoid unnecessary waste of collected nuclei that may be available in low numbers.- Ensure that microscope slides and cover slips are clean and dust-free. If needed, wash and rinse the coverslips with absolute ethanol and dry them with lint-free wipes.
- Distribute 25 µL of poly-l-lysine in the slide wells that will be used and incubate for 10 min at room temperature (RT), protected from dust.
- Remove the excess poly-l-lysine and add 10 µL of the purified nuclei suspension. Incubate for 5 min at RT, protected from dust.
- Add a drop of mounting medium to each well, avoiding bubbles.
- Place a coverslip on top of the seeded wells. Cover with paper wipes and press the coverslip firmly to remove the excess mounting medium. Be careful not to move the coverslip, and do not clean the excess mounting medium.
- Take several images with an inverted microscope with brightfield light and a minimum magnification of 40x.
- Purity control by flow cytometry
- Perform multiome assay.
- Proceed immediately to Chromium Next GEM Single Cell Multiome ATAC + Gene Expression User Guide (CG000338 – Rev F)25.
2. Purification of nuclei from mouse bone marrow hematopoietic stem and progenitor cells (HSPCs)
NOTE This protocol describes the purification of nuclei from three subsets of the bone marrow HSPCs: lineage–c-Kit+Sca-1+ hematopoietic stem cells (HSC), lineage–c-Kit+Sca-1–CD34+FcγR– common myeloid progenitors (CMP), and lineage– c-Kit+Sca-1–CD34+FcγR+ granulocyte-monocyte progenitors (GMP). Wear latex or nitrile gloves at all times during the procedure. This protocol is an adaptation of the 10X Genomics Demonstrated Protocol – Nuclei Isolation for Single Cell Multiome ATAC + GEX sequencing (CG000365 – Rev C)27. Modifications have been introduced in the original protocol to maximize nuclei recovery. It is strongly advised to have two people performing the experiment, to have steps 1. to 3. (i.e., preparation of the single-cell solution) performed by one person, and step 4 (i.e., preparation of the sorter) performed in parallel by another person. Since the protocol is highly time-sensitive, it is critical to minimize sample processing time by having the sorter ready as soon as the single-cell suspension is prepared.
- Preparation of reagents and materials
- Fill a bucket with ice.
- Prepare the FACS buffer: DPBS with 2% HI-FBS solution (approximately 500 mL for 6 samples) and filter through a 0.2 µm filter.
- Prepare the collection medium: DPBS with 10% HI-FBS solution (500 µL per sample), and filter through a 0.2 µm filter.
- Isolation of bone marrow cells
- Sacrifice mice using protocols approved by the institution. In this experiment, the mice were sacrificed by cervical dislocation after a ketamine/xylazine overdose.
- Spray the mice's abdomen and hind legs with 70% ethanol.
- Use sterile forceps and scissors to make a small incision in the middle of the lower abdomen and open the peritoneum from the base of the hind legs to the diaphragm (Supplementary Figure 1).
- Make an additional cut for each hind leg perpendicular to the opened peritoneum, then grab either side of one of these additional cuts and pull it apart to peel off the skin from both hind legs past the ankle joint to expose the muscles of both hind legs (Supplementary Figure 1A).
- Line the scissors along the spine at the hip joint of one hind leg to cut out the leg without cutting through the femur (Supplementary Figure 1B, C). Repeat the same for the other leg.
- To isolate the femur, cut most of the muscle tissue out, then hold the femur and tibia in each hand with fingertips at the joint (Supplementary Figure 1D, E). Gently fold the leg against the natural bend to dislocate the tibia from the femur (Supplementary Figure 1E) and then carefully cut the connective tissue with scissors to separate the femur and tibia.
- Use the scissors with light twisting motions to dislocate the bit of backbone from the top end of the femur (Supplementary Figure 1E).
- Clean the isolated femur with tissue paper to remove the remaining muscle and connective tissue.
- Keep cold on ice in a 12-well plate well filled with 2 mL of DMEM (1x) + GlutaMAX-I.
- Once all the femurs are collected, ensure that muscle and fibrous tissues are completely removed from the bone. Do not cut the bone open to (a) keep the marrow inside sterile and (b) avoid losing cells in the well. Use the following steps for flushing the cells from two femurs of one mouse, adapted from Haag and Murthy28.
- Prepare one 1.5 mL and one 0.5 mL tube. Add 150 µL of the FACS buffer to the 1.5 mL tube, then poke a hole at the bottom of the 0.5 mL tube using an 18 G needle and fit the 0.5 mL tube into the 1.5 mL tube.
- Open the distal part of each femur using mouse surgical scissors (Supplementary Figure 1F): Lock in the distal epiphysis between the blades and apply gentle pressure while flipping the scissors to smoothly detach the distal epiphysis without cutting open the bone harshly. If successful, 4 protrusions should be visible at the now exposed physis end (Supplementary Figure 1G).
- Fit the two femurs with the open-end facing downwards into the prepared 0.5 mL tube placed inside a 1.5 mL tube containing FACS buffer (Supplementary Figure 1H).
- Place a 70 µm cell strainer onto a 50 mL tube and pre-wet the strainer with 2 mL of the FACS buffer.
- To flush the bone marrow, centrifuge the tubes (caps open) at 12,000 x g until the centrifuge reaches the 12,000 x g value, then immediately stop the centrifuge.
- Verify that the bone marrow cells are pelleted in the 1.5 mL tube and that the femurs are white (before cell flushing, they are red) (Supplementary Figure 1I). Discard the 0.5 mL tubes with the 2 femurs.
- Discard the 150 µL supernatant using a pipette.
- Resuspend the pellet with a micropipette in 1 mL of Ammonium-Chloride-Potassium (ACK) lysing buffer for 1-2 min at RT to lyse red blood cells. Avoid longer incubation times as they may result in lowered viability of nucleated cells.
- Transfer into the 50 mL tube through the pre-wetted 70 µm cell strainer.
- Add 10 mL of FACS buffer to dilute the ACK lysing buffer and hereby stop the lysis.
- Centrifuge at 400 x g for 5 min at 4 °C. Resuspend in 10 mL FACS buffer by first resuspending in 1 mL then topping up with 9 mL.
- Prepare the cells for counting as described in 1.3.8.
- Count the cells using an automated cell counter following the supplier's recommendations. It is expected to collect approximately 40 million cells from 2 femurs.
- Staining of bone marrow HSPC
- Centrifuge the cells at 400 x g for 5 min at 4 °C and resuspend the pellet with a micropipette in FACS buffer to a final concentration of 1 x 107 cells/mL.
- With a P1000 micropipette, transfer the suspension into a FACS tube, filtering through a 35 µm cell strainer cap.
- Prepare single stain test tube samples for each antibody listed in Table 1 to set up compensations of fluorochromes on the cell sorter:
- Prepare one FACS tube per antibody and fill the tubes with 200 µL of PBS.
- Add 15 µL of fluorochrome-compensation beads in each FACS tube of fluorochrome-conjugated antibody. In the FACS tubes for unstained and for Live/Dead single stained cells, add 500,000 cells instead of beads.
- Add 1 µL of each fluorochrome-conjugated antibody (see Table 1) into its corresponding FACS tube. Add 0.5 µL of Live/Dead stain into Live/Dead single stain FACS tube.
- Keep on ice for 15 min protected from light.
- Prepare Mixes 1 and 2 as indicated in Table 2.
NOTE: The antibody volumes indicated in Table 2 are valid for the antibodies referenced in the Table of Materials. They need to be optimized for any new antibody reference or a different lot of the same antibody reference. - Add 300 µL of Mix 1 into the sample tube, resuspend, and keep for 15 min on ice protected from light.
- Add 300 µL of Mix 2 into the sample tube, resuspend, and keep for 20 min on ice protected from light.
- Add 3 mL of the FACS buffer to the single-stained tubes and the Mix-stained sample tubes. Spin down at 400 x g for 5 min at 4°C.
- Carefully discard the supernatant using a micropipette and resuspend the pellet in 500 µL of the FACS buffer.
- Prepare a 1.5 mL tube prefilled with 500 µL of collection medium.
NOTE: Mix 1 is prepared in DPBS since it contains the Live/Dead stain significantly affected by HI-FBS. Once the cells are stained by Live/Dead, Mix 2 is added, which contains the fluorochrome-conjugated antibodies resuspended in FACS buffer containing HI-FBS. The only exception is the anti-CD16/32 antibody that is included in Antibody Mix 1 to serve as Fc Receptor blocker that prevents the non-specific binding of the other antibodies added in the following step.
- Cell sorting using a FACS
NOTE: While cell sorting can be performed on a wide variety of cell sorters, here, the procedure for using the BD FACSAria Fusion or BD FACSAria III instruments is described. It is strongly recommended that the calibration and the setup of the cell sorter be performed under supervision or by an experienced user of the instrument.- Calibration of FACS instrument: Refer to Protocol 1 step 4.1.
- Set up of FACS instrument for cell sorting:
- Start the acquisition of the unstained cells. These are used to define the forward and side scatters and the detector voltage for each fluorophore. Set the parameters so that the fluorescent signal of each fluorophore falls inside the first decade of the log scale on the dot plot.
- Acquire single color controls to set up compensations manually (median of positive and negative populations should be aligned) or use the automatic calculation software (slope measurements). Ensure that the compensation controls match the experimental fluorochromes and detector settings. Record 10,000 events for cells and 5,000 events for beads.
- Use the sample tube (i.e., multi-stained cells) to define cell populations of interest by using the gating strategy shown in Figure 3A. Follow steps 4 – 6 (below).
- For identifying the three bone marrow HSPCs of interest (HSC, CMP, and GMP), start the gating by using the size (FSC-A) and granularity (SSC-A) to gate on leukocytes, then FSC-H/FSC-A to discriminate doublets.
- Based on SSC-A/dead cell marker, gate live cells. Use Lineage/c-Kit to select cells that are lineage-negative and expressing intermediate to high levels of c-Kit. Through c-Kit/Sca-1, gate on lineage–c-Kit+ Sca-1+ (LKS+) HSCs, one of the three populations of interest.
- Amongst the myeloid progenitors (lineage-c-Kit+Sca-1-), use FcγR/CD34 to exclude CD34–FcγR– megakaryocyte and erythroid progenitors (MEP), while including CD34+ FcγR– CMP, as well as CD34+FcγR+ GMP in the cell populations to be sorted.
- Make sure that the stream and deflection are stable.
- In the side stream camera, turn on the test sort, voltage ON, and confirm the accurate drop sorting in a 1.5 mL tube fitted on the left side.
- In the Sorting Layout window, select the population(s) of interest (i.e., "LKS+" and "CD34+ myeloid progenitors" shown in this example). Under Device, select 2 Tube. Under Precision, select Purity. In Target Events, select Continuous to sort between 160,000 and 200,000 LKS+ and CD34+ myeloid progenitors.
- Add 500 µL of FACS buffer to the cell suspension and transfer the 1 mL of the sample by filtering into a new 35 µm-cell strainer-capped FACS tube to ensure all cells are in a single suspension just before acquisition. This eliminates cell clumps that could clog the instrument.
- Once ready, click Sort and OK to start sorting. Adjust the Flow Rate to maintain the speed below 10,000 events per second.
NOTE: The expected ratio of LKS+ to CD34+ myeloid progenitors is 1:3 for an adult (8-12 weeks old) C57BL/6J female mouse at a steady state. The targeted sorted cell numbers are usually reached within 30 min of sorting.
- Quality control and counting of sorted cells
NOTE: This step is to be done only during the pilot experiment for optimization of sample preparation steps, with the goal of testing the purity of the sorted cells that are to be used for nuclei isolation. Once the protocol is fully optimized, it is not advised to perform this quality control step in the follow-up experiments to avoid unnecessary waste of starting material that may be available in low numbers for nuclei isolation.- Purity control by flow cytometry
- Transfer 10 µL of the sorted cells into a new FACS tube containing 90 µL of FACS buffer.
- Acquire and record post-sort data to verify the sorting purity and viability. Ensure that at least 95% of the cells appear in the gate of interest, as defined in 3 – 6 and illustrated in Figure 3B.
- Purity control by flow cytometry
- Nuclei isolation from sorted bone marrow HSPCs
- Use the "Low Cell Input Nuclei Isolation" protocol of the Appendix from the 10X Genomics Demonstrated Protocol – Nuclei Isolation for Single Cell Multiome ATAC + GEX sequencing (CG000365 – Rev C)27, with the following modifications made to optimize nuclei recovery:
- Lysis time: Run a pilot experiment for this protocol to identify the best lysis time for nuclei isolation. Ensure to achieve a complete cell lysis while maintaining intact nuclei.
NOTE: Step f of the abovementioned 10X Genomics protocol27 instructs to "incubate [in Lysis Buffer] for 3-5 min on ice". During the pilot experiment, test at least for 3 min, 4 min, and 5 min and assess the recovered nuclei quantity by counting and quality by flow cytometry and microscopy imaging to choose the optimal lysis duration (see the description of these quality control checks below). To spare reagents, replace the diluted nuclei buffer with PBS 0.04% BSA in the pilot experiment. For bone marrow HSPCs, 3 min was identified as the optimal lysis duration. - Cell centrifugations: For all cell suspension centrifugations, centrifuge at 300 x g for 7 min (instead of the 5 min in CG000365 – Rev C)27 at 4 °C.
- Nuclei centrifugations: Perform all nuclei suspension centrifugations at 500 x g for 5 min as per CG000365 – Rev C27.
- Nuclei collection: At step b, after resuspending in 50 µL of PBS 0.04% BSA and transferring to a 0.2 mL tube, add 50 µL of PBS 0.04% BSA to the original tube and pipette-mix to collect any leftover cells. Transfer to the 0.2 mL tube to reach a total of 100 µL volume.
- Henceforth, the total volume will be 100 µL instead of the 50 µL of the protocol. Adjust the downstream steps accordingly (e.g., for step d, remove 90 µL instead of 45 µL; for step e, add 90 µL of lysis buffer instead of 45 µL).
- For step m, resuspend the nuclei pellet in 12 µL of diluted nuclei buffer instead of 7 µL.
- Count the isolated nuclei. Into an empty 0.5 mL tube, add 10 µL of 0.4% trypan blue and 8 µL of PBS 0.04% BSA.
- Add 2 µL of nuclei to the tube and count nuclei as described in 1.3.8. Use an automated cell counter following the supplier's recommendations.
- Lysis time: Run a pilot experiment for this protocol to identify the best lysis time for nuclei isolation. Ensure to achieve a complete cell lysis while maintaining intact nuclei.
- Use the "Low Cell Input Nuclei Isolation" protocol of the Appendix from the 10X Genomics Demonstrated Protocol – Nuclei Isolation for Single Cell Multiome ATAC + GEX sequencing (CG000365 – Rev C)27, with the following modifications made to optimize nuclei recovery:
- Purity control by flow cytometry
NOTE: This step is to be done only during the pilot experiment for optimization of sample preparation steps to test the purity of the nuclei that are to be loaded on the 10X Chromium chip. Once the protocol is fully optimized, it is not advised to perform this quality control step in the follow-up experiments to avoid unnecessary waste of collected nuclei that may be available in low numbers.- After completing nuclei isolation, transfer 6 µL of nuclei resuspension in a new FACS tube prefilled with 150 µL of FACS buffer. Add 3 µL of 7-AAD and incubate for 5 min on ice.
- Acquire and record post-sort data to verify the sorting purity and viability. Ensure at least 95% of the nuclei appear in the gate of interest, as defined in Protocol 1 step 4.2 (see Figure 4).
- Quality control of purified nuclei by microscopy:
NOTE: This step is to be done only during the pilot experiment for optimization of sample preparation steps to test the quality of the nuclei that are to be loaded on the 10X Chromium chip. Once the protocol is fully optimized, it is not advised to perform this quality control step in the follow-up experiments to avoid unnecessary waste of collected nuclei that may be available in low numbers.- Proceed as described in step 1.5.3.
- Perform multiome assay
- Proceed immediately to Chromium Next GEM Single Cell Multiome ATAC + Gene Expression User Guide (CG000338 – Rev F)25.
Representative Results
The two protocols described above detail the isolation of nuclei starting from two different types of tissue. The differences and similarities between the two protocols are schematically represented in Figure 1.
Purification of nuclei from mouse brain
In the protocol described here, we propose a gentle method for nuclei preparation from brain samples. It starts with a mechanical dissociation of the brain tissue in a lysis buffer, followed by washing and strainer filtering steps that remove the remaining tissue from the suspension. The subsequent removal of debris, non-lysed cells, and small particles is achieved by FACS to guarantee that only purified nuclei are loaded for downstream Multiome protocol. Figure 2 shows the profile of nuclei before and after sorting. After the filtering and before nuclei sorting, the sample contains a high quantity of debris, with more than 99% of the "singlets" positive for nuclear stain (7-AAD), indicating optimal cell lysis (Figure 2A). Nuclei are sorted based on the 7-AAD positive gate. A fraction of sorted material is acquired to verify the purity of the prepared nuclei. Figure 2B shows the profile of the brain nuclei after sorting. The nuclei sorting allowed an increase in the nuclei purity from the initial 36% (Figure 2A) to almost 100% (Figure 2B).
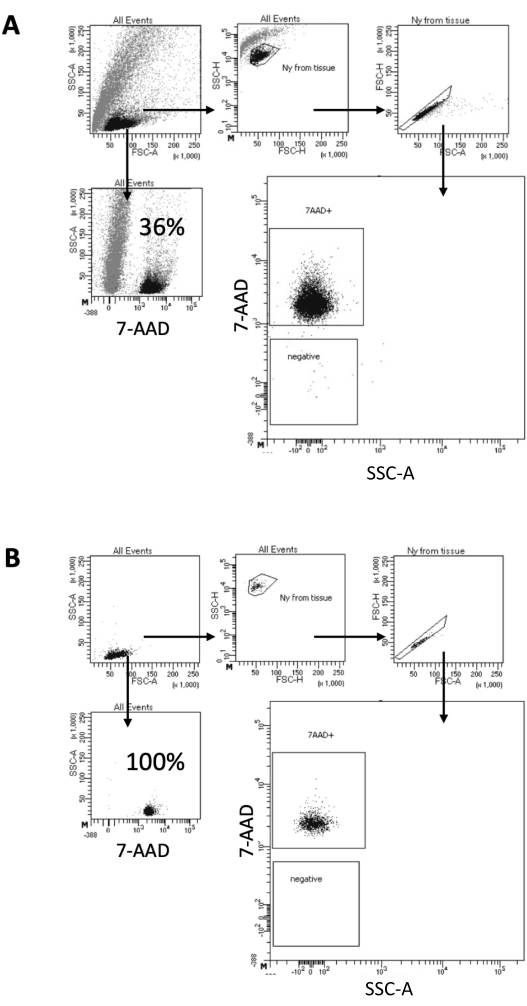
Figure 2: Gating strategy for nuclei sorting and purity test after sorting. Nuclei were stained with 7-AAD and acquired by the cell sorter. (A) Nuclei are first gated based on their size and granularity (FSC-A and SSC-A, respectively). Single particles are then selected based on their FSC-A/FSC-H properties and 7-AAD staining. (B) After cell sorting, a fraction of the nuclei from the collection tube is tested for purity using the same gating strategy as in A. Please click here to view a larger version of this figure.
Purification of mouse bone marrow hematopoietic stem cell progenitors (HSPCs)
Upon isolation from the bone marrow, up to 2 x 105 HSPCs are sorted by FACS, according to the gating strategy shown in Figure 3A. The sorting efficacy and sample purity are assessed (Figure 3B).
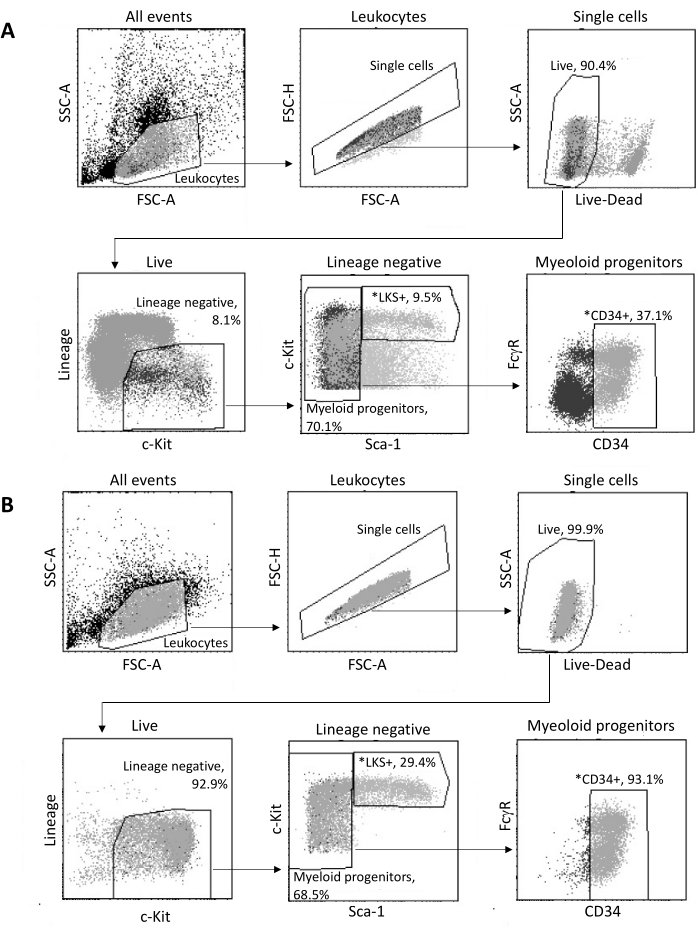
Figure 3: Gating strategy for sorting of bone marrow HSPCs. (A) A representative FACS gating strategy for sorting viable LKS+ hematopoietic stem cells and CD34+ myeloid progenitors for nuclei isolation. (B) Representative FACS plots used for verification of sorted cell population purity. Shown are the proportions of different cell subsets with respect to the parent population. *The two sorted populations. Please click here to view a larger version of this figure.
The "Low Cell Input Nuclei Isolation" protocol allows nuclei isolation from samples with a maximum of 105 cells. It includes a low number of centrifugation steps, thereby minimizing cell/nuclei loss. We have adjusted the volume of lysis and wash buffers proportionally to the cell input and increased the centrifugation time for maximal nuclei recovery. We have performed a pilot experiment to assess the quantity of recovered nuclei by counting and their quality by flow cytometry and microscopy imaging. Figure 4 shows the HSPCs sample after cell lysis. This protocol generated high-quality nuclei, as observed in Figure 5A, without any debris that could impact the downstream multiome protocol.
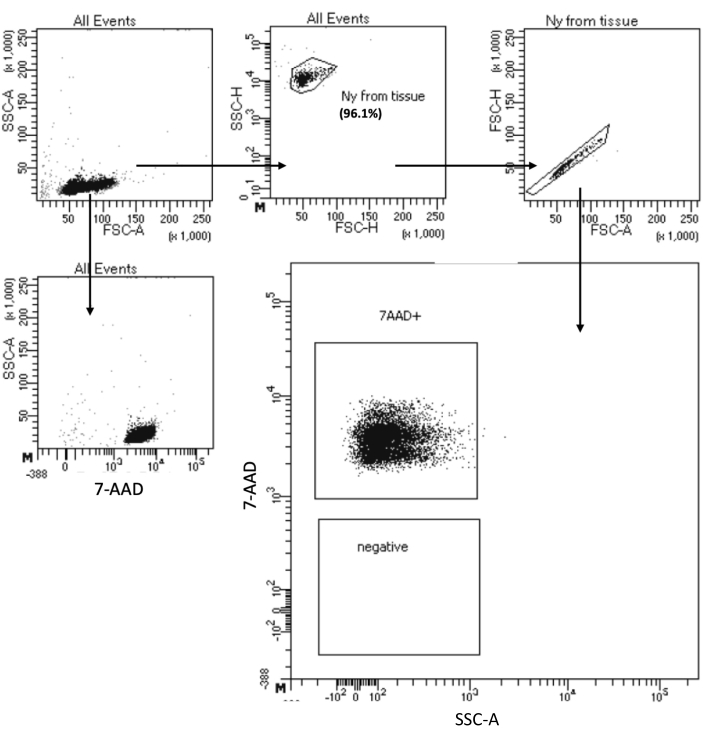
Figure 4: Purity test sorting of isolated bone marrow HSPC nuclei. Nuclei were stained with 7-AAD and acquired by the cell sorter. Nuclei were first gated based on their size and granularity (FSC-A and SSC-A, respectively) to assess the purity of the sample. The proportion of nuclei is indicated with respect to the parent population. Please click here to view a larger version of this figure.
| Tube number | Tube name | Stained entity | Amount of stained entity | Antibody/dye (µL) | Collection Buffer (µL) |
| 1 | Unstained | Cells | 5,00,000 | N/A | 200 |
| 2 | LIVE/DEAD Fixable Aqua Dead Cell Stain | Cells | 5,00,000 | 0.5 | |
| 3 | APC/Cyanine 7 anti-mouse CD16/32 (FcγR) | OneComp eBeads | 15 µL | 1 | |
| 4 | Pacific Blue anti-mouse Lineage Cocktail | OneComp eBeads | 15 µL | 1 | |
| 5 | PE anti-mouse Ly-6A/E (Sca-1) | OneComp eBeads | 15 µL | 1 | |
| 6 | APC anti-mouse CD117 (c-Kit) | OneComp eBeads | 15 µL | 1 | |
| 7 | FITC anti-mouse CD34 | OneComp eBeads | 15 µL | 1 |
Table 1: Single stain controls for compensation settings on the flow cytometer. Indicated are the required single stain controls, the number of cells or beads to be stained, and the antibody quantities.
| Master Mix | Reagent | Final dilution | Antibody/dye (µL) | Buffer type | Buffer (µL) |
| Mix 1 | APC/Cyanine 7 anti-mouse CD16/32 (FcγR) | 1/500 | 1.2 | DPBS | 300 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain | 1/250 | 2.4 | |||
| Mix 2 | Pacific Blue anti-mouse Lineage Cocktail | 1/20 | 30 | FACS buffer | 300 |
| PE anti-mouse Ly-6A/E (Sca-1) | 1/200 | 3 | |||
| APC anti-mouse CD117 (c-Kit) | 1/200 | 3 | |||
| FITC anti-mouse CD34 | 1/50 | 12 | |||
| Total staining volume | 600 |
Table 2: Composition of the staining mix for bone marrow HSPCs. Shown are volumes of reagents required for the staining of one sample containing 40 million cells. For staining a higher number of samples, multiply the indicated volume by the required number of samples and add half of an extra sample volume to ensure a sufficient volume of the master mix.
Supplementary Figure 1: Bone marrow cell isolation protocol. (A) Open the peritoneum. White dotted lines indicate the line to cut along. (B) After peeling the skin off from the hind leg, line the scissors along the spine at the hip joint to cut out the leg without cutting through the femur. (C) The appearance of the leg separated from the body before the removal of muscle. (D) The appearance of the leg after removal of muscle. (E) The procedure for separating the femur at the knee joint, then at the hip joint, taking care not to cut open the femur. White curved arrows show the motion required. White dotted arrows indicate the area to separate gently using scissors to pinch off. (F) Procedure to open the distal part of the femur (i.e., the part previously attached to the tibia at the knee joint) by securely grabbing the cartilage and distal epiphysis with scissors and flipping it back to expose the bone marrow. (G) Four protrusions, indicated by black arrows, should be visible at the exposed physis end. (H) The appearance of a femur with the open-end facing downwards into the prepared 0.5 mL tube placed inside a 1.5 mL tube containing 150 µL of FACS buffer. (I) The appearance of the pelleted bone marrow cells and the now white femur after quick centrifugation at 12,000 x g. Please click here to download this File.
Discussion
Preparation of high-quality cell or nuclei suspension is of crucial importance for the success of single-cell or single-nuclei RNA-Seq and single-cell multi-omic analyses29,30,31. Here, we have described protocols for sample preparation and nuclei isolation for multiome assays from two types of tissue: brain and bone marrow.
The brain protocol described in this paper allows the recovery of high-quality nuclei from fresh-frozen brain tissue. It includes the following steps: frozen tissue disruption, isolation of nuclei, purification of nuclei, and quality control of the prepared material. The brain tissue is composed of many different cell types, and the procedure of tissue dissociation and nuclei isolation should preserve the proportions of cell populations as present in the initial tissue. Here, the lysis buffer composition and incubation time were optimized to enable complete and gentle lysis of all cell populations that compose the tissue.
The bone marrow HSPCs protocol is somewhat different since it requires one additional step at the beginning of the experiment to isolate the cell population of interest from a heterogeneous cellular suspension. After the collection of fresh tissue, red blood cells are lysed, and the sample is enriched for the cell subset of interest. The targeted cells are lysed, the nuclei are isolated, and the quality of the prepared material is controlled.
10X Genomics provides several protocols validated for nuclei isolation in numerous different tissues32,33. The company also commercializes a nuclei isolation kit with a straightforward pipeline for isolating nuclei from validated tissues34. However, these protocols need additional optimization to tailor the particularities of certain samples. An example is the samples that require working with low cell input. For these samples, the most challenging steps are centrifugations that need to be sufficiently stringent to clean the sample and gentle enough to avoid cell/nuclei loss. With the protocol described here, we have adapted the 10X Genomics Demonstrated Protocol – Nuclei Isolation for Single Cell Multiome ATAC + GEX Sequencing (CG000365 – Rev C)27 to find a fine balance between these two requirements. As demonstrated in the example of the preparation of nuclei from sorted HSPCs, we have improved the nuclei recovery with no impact on the quality of the sample.
An additional challenge is the step of lysis of purified cells for nuclei isolation. Harsher lysis conditions and longer incubation times can damage nuclei and thereby impact the quality of sequencing data. Figure 5 shows representative nuclei imaging from bone marrow samples upon different incubation times with lysis buffer and illustrates how different the state of nuclei could be depending on the cell lysis. In the example of the HSPCs, we have identified 3 min lysis as the condition that results in the highest proportion of healthy-looking, intact nuclei and the lowest proportion of damaged nuclei. The lysis incubation times should be optimized for each new type of sample.
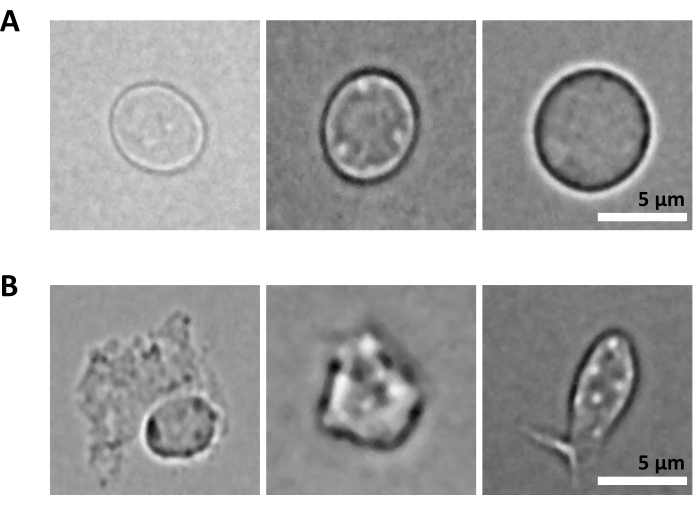
Figure 5: Nuclei quality control by microscopy. Shown are representative brightfield images of isolated nuclei from mouse bone marrow with (A) intact and (B) damaged nuclei. Scale bar 5 µm. Images were taken with an inverted microscope using a 40x ELWD NA 0.60 objective and 1.5x digital zoom. Please click here to view a larger version of this figure.
Both protocols detailed in this work rely on purifying targeted cells or nuclei by high-throughput FACS instruments. This step is of crucial importance for single-cell/nuclei preparation protocols where rare subsets of cells are to be isolated from heterogeneous suspensions. In these, like in the example shown here for HSPCs sorting, a high-dimensional flow cytometry panel may be required to enable "gating" on the cell population of interest. The sorting is extremely fast and accurate, leading to over 95% purity of the sorted cell subsets. This approach exposes the cellular suspension to a pressure of up to 70 psi and may therefore, be limiting for sorting of fragile cells (e.g., dendritic cells, neutrophils) since it may cause the rupture of their cell membrane. In these cases, alternative solutions should be selected for cellular purification, including magnetic sorting, application of new generation instruments (e.g., CellenOne, Cellenion; MACSQuant Tyto, Miltenyi)35,36, or droplet-based systems (e.g., ODIN, Sensific)37. Nevertheless, the slow sorting speed of these technologies, with cell sorting that lasts for hours instead of minutes, is a strong limiting factor for the application of these approaches in the preparation of viable cells for Multiome and other single-cell applications based on analysis of large cell numbers.
For the purification of nuclei isolated from the tissue, FACS is the method of choice due to its throughput and the purity of the isolated material. Nuclei are not sensitive to pressure, and filtered tissue isolates can easily be purified through the cell sorter. If the laboratory is not equipped with a FACS instrument, other alternatives exist, somewhat less efficient but sufficiently good. Examples include ultracentrifugation or the use of small equipment such as MARS (Applied Cell) that separates particles based on their difference in size, using acoustic waves; CURIOX laminar washer that uses hydrophobic properties of cell/nuclei suspensions; or LEVITAS bio that relies on physical properties of cells (levitation) to separate them from the debris.
Here, we describe protocols to obtain a high number of nuclei and the best purity for the downstream Multiome protocol. FACS sorting and repeated centrifugation steps result in a substantial loss of the initial material. For this reason, in the protocol for nuclei preparation from the brain that we describe here requires sufficiently abundant starting material to result in the collection of at least 500,000 nuclei after the FACS sorting. Alternative protocols should be applied if this criterion cannot be matched. When working with rare cell populations or small tissue sections, the available amount of initial material can be a limiting factor. To address this issue, it is possible to improve nuclei recovery by (a) reducing the volume of lysis, (b) reducing the washing volume, (c) using a single wash with extended centrifugation time to try to improve recovery as indicated in 10X Genomics protocols for low cell input nuclei isolation. For multiomic analysis of low-content material, it is worth considering plate-based applications such as scNMT, SNARE-seq, and Paired-seq38 that require much fewer input samples.
In summary, we have described two robust protocols for the preparation of nuclei from the brain and the bone marrow HSPCs for downstream Multiome analysis. These protocols are applicable in any scientific project that requires high-quality single nuclei suspensions from these two types of tissue, irrespective of the scientific question posed. Our group has been applying the brain nuclei isolation protocol in studies of brain development upon inactivation of various targeted genes and in studies of immune response in the context of neurological diseases. We are using the bone marrow nuclei isolation protocol for deciphering the participation of various hematopoietic subpopulations in the establishment of the immune system.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Ana Jeemin Choi was supported by a stipend from the Pasteur – Paris University (PPU) International Ph.D. Program.
Materials
| 18 G x 1 ½ (1.2 mm x 38 mm) Agani needles | Terumo | AN*1838S1 | |
| 15 mL tubes | Falcon | 352097 | |
| 5 mL round bottom FACS tube with cell strainer cap 35 µm | falcon | 352235 | |
| 50 mL tubes | Falcon | 352070 | |
| 7-AAD | BD pharmagen | 559925 | |
| ACK Lysing Buffer | Gibco | A10492-01 | |
| APC anti-mouse CD117 (c-Kit) | BioLegend | 105812 | Clone: 2B8 |
| APC/Cyanine 7 anti-mouse CD16/32 (FcγR) | BioLegend | 101328 | Clone: 93 |
| BD FACSAria III | BD Biosciences | non-applicable | |
| BD FACSDiva Software v8.0.1 | BD Biosciences | non-applicable | |
| Bovine Serum Albumin stock solution 10% | Miltenyi Biotec | 130-091-376 | |
| Cell staining buffer | Biolegend | 420201 | |
| CFI Suprplan Fluor ELWD 40XC ON 0.6 | Nikon | non-applicable | |
| CMOS camera Prime 95B 25 mm | Photometrix | non-applicable | |
| Countess II FL Automated Cell Counter | Invitrogen | AMQAF1000 | |
| Countess cell counting chamber slide | Invitrogen | C10283 | |
| Coverglass 24 mm x 24 mm 0.13-0.17 mm | Brand | BR470819 | |
| Digitonine 5% | Invitrogen | BN2006 | |
| Disposable Scalpels | Swann-Morton | 0508 | |
| DMEM (1x) + GlutaMAX-I | Gibco | 31966-021 | |
| DPBS (10x) | Gibco | 14200-067 | |
| DTT | Sigma aldrich | 646563 | |
| Epifluorescence inverted microscope Nikon Ti2 -E | Nikon | non-applicable | |
| Eppendorf Safe-Lock Tubes 0.5 mL | Eppendorf | 30123603 | |
| Ethanol 70% | VWR | 83801.290 | |
| FITC anti-mouse CD34 | Invitrogen | 11-0341-85 | Clone: RAM34 |
| Forceps for dissection | FST | 11152-10 | |
| Heat-inactivated Fetal Bovine Serum (FBS) | Gibco | 11533387 | |
| Dounce Homogeniser 2 mL | Bellco glass | 1984-10002 | Pestle “A” Large Clearance: .0030-.0050″ and Pestle “B” Small Clearance: .0005-.0025″ |
| LIVE/DEAD fixable aqua dead cell stain kit | Invitrogen | L34957 | |
| Magnesium chloride solution 1 M | Sigma aldrich | M1028 | |
| Microcentrifuge | Eppendorf | 5424R | |
| Mounting medium Fluoromount-G | invitrogen | 00-4958-02 | |
| Nonidet P40 substitute | Sigma aldrich | 74385 | |
| Nuclease free water | ThermoFischer | AM9932 | |
| Nuclei buffer 20x | 10X Genomics | 2000153/2000207 | |
| Nuclei isolation kit EZ prep | Sigma Aldrich | NUC-101 | |
| OneComp eBeads compensation beads | Invitrogen | 01-1111-41 | |
| Pacific Blue anti-mouse lineage cocktail (including anti-mouse CD3, Ly-6G/Ly-6C, CD11b, CD45R/B220, TER-119) |
BioLegend | 133310 | Clones (in the same order as the antibodies listed): 17A2, RB6-8C5, M1/70, RA3-6B2, Ter-119 |
| PCR Tube Strips 0.2 mL | Eppendorf | 951010022 | |
| PE anti-mouse Ly-6A/E (Sca-1) | BioLegend | 122507 | Clone: E13-161.7 |
| Petri dish 100 mm x 20 mm OPTILUX | Falcon | 353003 | |
| Ply-L-lysine 0.01% sterile-filtered suitable for cell culture | Sigma | P4707 | |
| Printed microscope slides 8 well 6 mm numbered | Epredia | ER-301B-CE24 | |
| Protein LoBind Tubes 1.5 mL | Eppendorf | 30108116 | |
| Recombinant Rnase inhibitor 5000 U | Takara | 2313A | |
| Scissors for dissection | FST | 14090-09 | |
| Sodium chloride solution 5 M | Sigma aldrich | 59222C | |
| Syringe filters, PES, 0.2 µm | Fisher Scientific | 15206869 | |
| Transparent nail polish | any | non-applicable | |
| Trizma Hydrochloride solution pH 7.4 | Sigma aldrich | T2194 | |
| Trypan Blue 0.4% | gibco | 15250061 | |
| Tween 20 | Biorad | 1662404 | |
| UltraPure Distilated Water Dnase/Rnase Free | Invitrogen | 10977-035 |
References
- Clark, S. J., et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nature Communications. 9 (1), 781 (2018).
- Lee, J., Hyeon, D. Y., Hwang, D. Single-cell multiomics: technologies and data analysis methods. Experimental & Molecular Medicine. 52 (9), 1428-1442 (2020).
- Cerrizuela, S., et al. High-throughput scNMT protocol for multiomics profiling of single cells from mouse brain and pancreatic organoids. STAR Protocols. 3 (3), 101555 (2022).
- Dimitriu, M. A., Lazar-Contes, I., Roszkowski, M., Mansuy, I. M. Single-cell multiomics techniques: From conception to applications. Frontiers in Cell and Developmental Biology. 10, 854317 (2022).
- Stoeckius, M., et al. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods. 14 (9), 865-868 (2017).
- Cao, J., et al. . Joint profiling of chromatin accessibility and gene expression in thousands of single cells. 361 (6409), 1380-1385 (2018).
- Narayanan, A., et al. Nuclei Isolation from Fresh Frozen Brain Tumors for Single-Nucleus RNA-seq and ATAC-seq.Journal of Visualized Experiments. JoVE. 162, 61542 (2020).
- Kim, M., et al. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nature Communications. 11 (1), 6375 (2020).
- Santos, M. D., et al. Extraction and sequencing of single nuclei from murine skeletal muscles. STAR Protocols. 2 (3), 100694 (2021).
- Safabakhsh, S., et al. Isolating nuclei from frozen human heart tissue for single-nucleus RNA sequencing. Current Protocols. 2 (7), (2022).
- Lau, S. -. F., Cao, H., Fu, A. K. Y., Ip, N. Y. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 117 (41), 25800-25809 (2020).
- Armand, E. J., Li, J., Xie, F., Luo, C., Mukamel, E. A. Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron. 109 (1), 11-26 (2021).
- Morabito, S., et al. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nature Genetics. 53 (8), 1143-1155 (2021).
- Chen, S., et al. Spatially resolved transcriptomics reveals genes associated with the vulnerability of middle temporal gyrus in Alzheimer’s disease. Acta Neuropathologica Communications. 10 (1), (2022).
- Paul, F., et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 163 (7), 1663-1677 (2015).
- Kaufmann, E., et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 172 (1-2), 176-190 (2018).
- Christ, A., et al. diet triggers NLRP3-dependent innate immune reprogramming. Cell. 172 (1-2), 162-175 (2018).
- Moorlag, S. J. C. F. M., et al. β-Glucan Induces protective trained immunity against mycobacterium tuberculosis infection: A key role for IL-1. Cell Reports. 31 (7), 107634 (2020).
- de Laval, B., et al. C/EBPβ-dependent epigenetic memory induces trained immunity in hematopoietic stem cells. Cell Stem Cell. 26 (5), 657-674 (2020).
- Renthal, W., et al. Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nature Neuroscience. 21 (12), 1670-1679 (2018).
- Yang, A. C., et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature. 603 (7903), 885-892 (2022).
- Lee, D. R., Zhang, Y., Rhodes, C. T., Petros, T. J. Generation of single-cell and single-nuclei suspensions from embryonic and adult mouse brains. STAR Protocols. 4 (1), 101944 (2022).
- Corces, M. R., et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature Genetics. 48 (10), 1193-1203 (2016).
- Ranzoni, A. M., et al. Integrative single-cell RNA-seq and ATAC-seq analysis of human developmental hematopoiesis. Cell Stem Cell. 28 (3), 472-487 (2021).
- . 10X Genomics Chromium Next GEM Single Cell Multiome ATAC + Gene Expression User Guide. Document Number CG000338 Rev F. At. , (2022).
- J, , et al. Microdissection of mouse brain into functionally and anatomically different regions. Journal of Visualized Experiments: JoVE. 168, 61941 (2021).
- . . 10X Genomics 10X Genomics Demonstrated Protocol – Nuclei Isolation for Single Cell Multiome ATAC + GEX sequencing (CG000365 – Rev C). At. , (2022).
- Haag, S., Murthy, A. Murine monocyte and macrophage culture. Bio-Protocol. 11 (6), (2021).
- Haque, A., Engel, J., Teichmann, S. A., Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Medicine. 9 (1), (2017).
- Jiang, P. Quality control of single-cell RNA-seq. Methods in Molecular Biology. , 1-9 (1935).
- Regan, C., Preall, J. Practical considerations for single-cell genomics. Current Protocols. 2 (8), (2022).
- . . 10X Genomics 10X Genomics Demonstrated Protocol – Nuclei Isolation for Single Cell ATAC Sequencing (CG000169 – Rev E). At. , (2022).
- . 10X Genomics 10X Genomics Demonstrated Protocol – Nuclei Isolation from Complex Tissues for Single Cell Multiome. ATAC + Gene Expression Sequencing. (CG000375 – Rev C). At. , (2022).
- . . 10X Genomics 10X Genomics – Chromium Nuclei Isolation Kit (CG000505 – Rev A). AT. , (2022).
- Shomroni, O., et al. A novel single-cell RNA-sequencing approach and its applicability connecting genotype to phenotype in ageing disease. Scientific Reports. 12 (1), 4091 (2022).
- Ocañas, S. R., Pham, K. D., Blankenship, H. E., Machalinski, A. H., Chucair-Elliott, A. J., Freeman, W. M. Minimizing the ex vivo confounds of cell-isolation techniques on transcriptomic and translatomic profiles of purified microglia. 9 (2), 0348-0321 (2022).
- Gérard, A., et al. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nature Biotechnology. 38 (6), 715-721 (2020).
- Vandereyken, K., Sifrim, A., Thienpont, B., Voet, T. Methods and applications for single-cell and spatial multi-omics. Nature Reviews. Genetics. 24, 494-515 (2023).

