Immunoaffinity Based Extraction of Ubiquitinylated Peptides: A Technique to Selectively Extract Ubiquitin Tagged Peptides from Purified Peptide Fractions
Abstract
Source: Bezstarosti, K. et al. Detection of Protein Ubiquitination Sites by Peptide Enrichment and Mass Spectrometry. J. Vis. Exp. (2020)
This video describes a method to extract and purify ubiquitinylated peptides containing remnant di-glycine peptides from a complex peptide mixture. The presented method may help in identifying original ubiquitination sites in the protein.
Protocol
1. Extraction of Ubiquitinylated Peptides
- Use high pH reverse-phase (RP) C18 chromatography with polymeric stationary phase material (300 Å, 50 µM; see Table of Materials) loaded into an empty column cartridge to fractionate the tryptic peptides. The stationary phase bed size must be adjusted to the amount of protein digest to be fractionated. Prepare an empty 6 mL column cartridge (see Table of Materials) filled with 0.5 g of stationary phase material for ~10 mg of protein digest. The protein digest to stationary phase ratio should be approximately 1:50 (w/w).
- Load the peptides onto the prepared column and wash the column with approximately 10 column volumes of 0.1% TFA, followed by approximately 10 column volumes of H2O.
- Elute the peptides into three fractions with 10 column volumes of 10 mM ammonium formate solution (pH = 10) with 7%, 13.5%, and 50% acetonitrile (AcN), respectively. Lyophilize all fractions to completeness.
- Use ubiquitin remnant motif (K-ε-GG) antibodies conjugated to protein A agarose beads for the immunoenrichment of diGly peptides. Because the exact amount of antibody per batch of beads is proprietary information and not disclosed by the manufacturer, it is recommended to use the same definition for a batch of beads as the manufacturer does in order to avoid confusion. Wash one batch of these beads 2x with PBS and split the bead slurry into six equal fractions. See Figure 1 for a detailed experimental scheme.
- Dissolve the three peptide fractions collected in step 1.3 in 1.4 mL of a buffer composed of 50 mM MOPS, 10 mM sodium phosphate, and 50 mM NaCl (pH = 7.2), and spin down the debris.
- Add the supernatants of the fractions to the diGly antibody beads and incubate for 2 h at 4 °C on a rotator unit. Spin down the beads and transfer the supernatant to a fresh batch of antibody beads and incubate again for 2 h at 4 °C.
- Store the supernatants for subsequent global proteome (GP) analysis.
- Transfer the beads from every fraction into a 200 µL pipette tip equipped with a GF/F filter plug to retain the beads. Put the pipette tip with the beads into a 1.5 mL microcentrifuge tube equipped with a centrifuge tip adapter. Wash the beads 3x with 200 µL of ice-cold IAP buffer and subsequently 3x with 200 µL of ice-cold Milli-Q H2O. Spin down the column at 200 x g for 2 min before every wash step but be careful not to let the column run dry. Elute the peptides using 2 cycles of 50 µL of 0.15% TFA.
- Desalt the peptides using a C18 stage tip (essentially a 200 µL pipette tip with two C18 disks) and dry them to completeness using vacuum centrifugation.
Representative Results
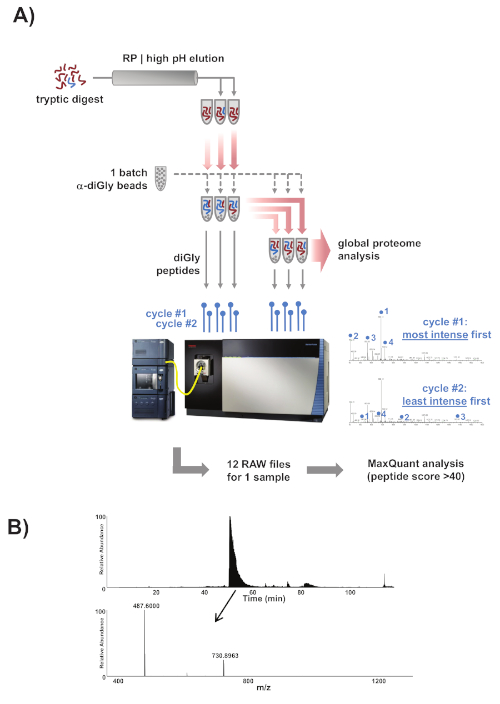
Figure 1: Experimental overview. (A) Overview of the experimental approach. Samples were prepared, trypsinized, and fractionated into three fractions using reverse-phase chromatography with high pH elution. One batch of commercial α-diGly peptide antibody beads was split into six equal fractions and the three peptide fractions were then loaded on three of the bead fractions. The diGly peptides were immunopurified, eluted, and collected, and the flowthrough was subsequently transferred to the three remaining fresh beads fractions. The collected diGly peptides were analyzed by mass spectrometry on a Lumos Orbitrap mass spectrometer according to a two-tier scheme combining one cycle in which the most intense peaks were first selected for peptide fragmentation and the next cycle in which the least intense peaks were selected first. The complete set of nLC-MS/MS runs were then analyzed using MaxQuant. (B) One of the fractions should contain ubiquitin's own K48 modified tryptic diGly peptide LIFAGK(GG)QLEDGR (m/z 730.39). This is by far the most abundant peptide in the immunoprecipitated fraction and was characterized by the intense and broad peak in the LC chromatogram between 50–55 min on a 120 min gradient. If this benchmark peak is absent from the chromatogram the IP was most likely unsuccessful.
Divulgazioni
The authors have nothing to disclose.
Materials
| Lysyl Endopeptidase(LysC) | Wako Pure Chemicals | 129-02541 | |
| NanoLC oven | MPI design, MS Wil GmbH | ||
| N-Lauroylsarcosine sodium salt | Sigma-Aldrich | L-5125 | |
| Orbitrap Fusion Lumos mass spectrometer |
ThermoFisher | ||
| PLRP-S (300 Å, 50 µm) polymeric reversed phase particles |
Agilent Technologies | PL1412-2K01 | |
| PTMScan Ubiquitin Remnant Motif (K-ε-GG) Kit |
Cell Signaling Technologies | 5562 | |
| Sep-Pak tC18 6 cc Vac Cartridge | Waters | WAT036790 | Remove the tC18 material from the cartridge before filling the cartridge with PLRP-S |
| Sodium deoxycholate | Sigma-Aldrich | 30970 | |
| Tris-base | Sigma-Aldrich | T6066 | |
| Tris-HCl | Sigma-Aldrich | T5941 | |
| Trypsin, TPCK Treated | ThermoFisher | 20233 |

