High-Resolution Melting PCR to Determine Sequence Variations in Mutant DNA Sequences
Abstract
Source: Kisserli, A., et al. High-resolution Melting PCR for Complement Receptor 1 Length Polymorphism Genotyping: An Innovative Tool for Alzheimer's Disease Gene Susceptibility Assessment. J. Vis. Exp. (2017).
In this video, we describe a high-resolution melting PCR technique to determine the length polymorphism of DNA fragments of varying sequence lengths, based on melting temperature analysis.
Protocol
1. HRM-PCR Protocol
- Thaw the DNA samples. Dilute the DNA samples in 1.5 mL tubes with water to adjust them to a concentration of 10 ng/µL
NOTE: The total volume of diluted DNA should be between 2 µL and 10 µL. - Thaw the primer solutions. Dilute the primer solutions in 1.5-mL tubes with water to adjust them to the same concentration of 6 µM.
NOTE: The primer sequences and reaction conditions are provided in Table 1. - Thaw the HRM-PCR kit solutions and mix carefully by vortexing to ensure the recovery of all contents. Briefly spin the three vials containing the enzymatic mixture with DNA binding dye, MgCl2, and water in a microcentrifuge before opening them. Store them at room temperature.
- In a 1.5 mL tube at room temperature, prepare the PCR mix for one 20 µL reaction by adding the following components in the order listed below:
- 10 µL of enzymatic mixture with DNA binding dye;
- 2 µL of 25 mM MgCl2;
- 1 µL of primer 1, 6 µM (final concentration: 300 nM);
- 1 µL of primer 2, 6 µM (final concentration: 300 nM); and
- 5 µL of water.
NOTE: To prepare the PCR mix for more than one reaction, multiply the volumes above by the number of reactions to be run, plus one additional reaction.
- Mix carefully by vortexing.
- Pipette 19 µL of PCR mix, prepared above, into each well of a white multiwell plate.
- Add 1 µL of concentration-adjusted DNA template.
NOTE: For control reactions, always run a negative control with the samples. To prepare a negative control, replace the template DNA with water. - Seal the white multiwell plate with sealing foil.
- Place the white multiwell plate in the centrifuge and balance it with a suitable counterweight (i.e., another multiwell plate). Centrifuge for 1 min at 1,500 x g in a standard swing-bucket centrifuge containing a rotor for multiwell plates with suitable adaptors.
- Load the white multiwell plate into the HRM-PCR instrument.
- Start the HRM-PCR program with the following PCR conditions:
Denaturation: 95 °C for 10 min; 1 cycle.
Amplification: 95 °C for 10 s, 62 °C for 15s, and 72 °C for 20s; 47 cycles.
Melting curve: 95 °C; ramp rate: 0.02 °C/s; 25 acquisitions per °C; 1 cycle.
Cooling: 40 °C for 30 s; ramp rate 2.2 °C/s; 1 cycle.
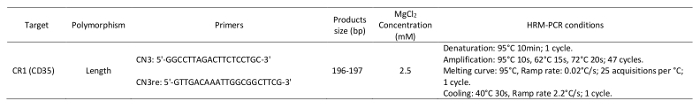
Table 1: Primers and parameters used in the high-resolution melting analysis.
2. HRM Analysis to Determine the CR1 Length Polymorphism
NOTE: The methodology described (Figure 1) is specific to our software (See the Table of Materials), although other software packages may be used.
- Open a gene scanning software to perform the CR1 length polymorphism scanning analysis.
- Open the experiment containing the amplification program and the melting curve program.
- Click Sample editor in the Module bar and then select the Scanning workflow.
- Define the properties of the samples (i.e., name; unknown or negative control).
- Click Analysis in the Module bar.
- In the Create New Analysis list, select Gene Scanning.
- Click the Normalization tab to normalize the melting curves.
- Click the Temperature shift tab to reset the temperature axis (x-axis) of the melting curves.
NOTE: The lower graph shows melting curves that are both normalized and temperature-shifted. - Click the Calculate button to analyze the results and determine the grouping.
- Click the Difference plot tab in the charts area to view the Normalized and Shifted Melting Curves and the Normalized and Temperature Shifted Difference Plot.
Representative Results
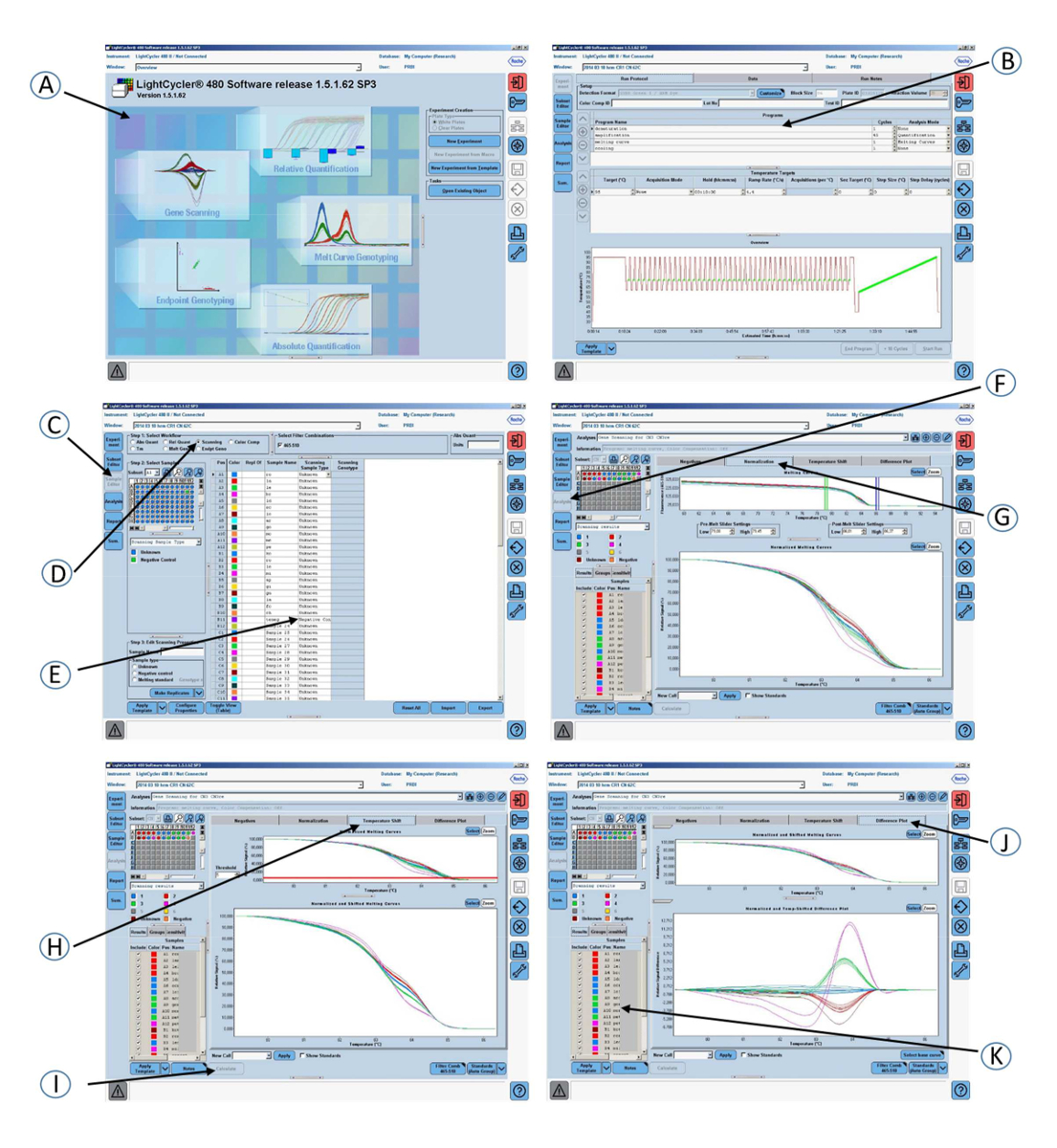
Figure 1: Screenshots of the graphical interface of the software used in step 2 of the protocol. (A) Open the gene scanning software. (B) Amplification program and melting curve program. (C) Click on Sample editor in the Module bar. (D) Select Scanning. (E) Define the properties of the samples. (F) Click on Analysis in the Module bar. (G) Click on the Normalization tab to normalize the melting curves. (H) Click on the Temperature shift tab to show the melting curves that are both normalized and temperature-shifted. (I) Click on the Calculate button to analyze the results and determine the grouping. (J) Click on the Difference plot tab to view the Normalized and Shifted Melting Curves and the Normalized and Temperature Shifted Difference Plot. (K) Colored grouping of the samples according to the CR1 length genotypes.
Divulgazioni
The authors have nothing to disclose.
Materials
| Lab coat | protection | ||
| SensiCareIce powder-free Nitrile Exam gloves | Medline Industries, Inc, Mundelein, IL 60060, USA | 486802 | sample protection |
| Eppendorf Reference 2 pipette, 0.5-10µL | Eppendorf France SAS, F-78360 Montesson, France | 4920000024 | sample pipetting |
| Eppendorf Reference 2 pipette, 20-100µL | Eppendorf France SAS, F-78360 Montesson, France | 4920000059 | sample pipetting |
| Eppendorf Reference 2 pipette, 100-1000µL | Eppendorf France SAS, F-78360 Montesson, France | 4920000083 | sample pipetting |
| TipOne 10µL Graduated, filter tip | Starlab GmbH, D-22926 Ahrenburg, Germany | S1121-3810 | sample pipetting |
| TipOne 1-100µL bevelled, filter tip | Starlab GmbH, D-22926 Ahrenburg, Germany | S1120-1840 | sample pipetting |
| ART 1000E Barrier Tip | Thermo Fischer Scientific , F-67403 Illkirch, France | 2079E | sample pipetting |
| Eppendorf Safe-Lock Tubes, 1.5 mL, Eppendorf Quality | Eppendorf France SAS, F-78360 Montesson, France | 30120086 | mix |
| Vortex-Genie 2 | Scientific Industries, Inc, Bohemia, NY 111716, USA | SI-0236 | mix |
| Mikro 200 centrifuge | Hettich Zentrifugen, D-78532, Germany | 0002020-02-00 | centrifugation |
| Multipette E3 | Eppendorf France SAS, F-78360 Montesson, France | 4987000010 | distribution |
| Light Cycler 480 multiwell plate 96, white | Roche Diagnostics GmbH, D-68305 Mannheim, Germany | 4729692001 | reaction place |
| Light Cycler 480 sealing foil | Roche Diagnostics GmbH, D-68305 Mannheim, Germany | 4429757001 | coverage |
| LightCycler 480 Instrument II, 96-well | Roche Diagnostics GmbH, D-68305 Mannheim, Germany | 05015278001 | high resolution melting polymerase chain reaction |
| Heraeus Megafuge 11R centrifuge | Thermo Fischer Scientific , F-67403 Illkirch, France | 75004412 | centrifugation |
| LightCycler 480 High Resolution Melting Master | Roche Diagnostics GmbH, D-68305 Mannheim, Germany | 04909631001 | reaction reagents |
| CN3 primer: 5'ggccttagacttctcctgc 3' | Eurogentec Biologics Division, B4102 Seraing, Belgium | reaction reagent | |
| CN3re primer: 5'gttgacaaattggcggcttcg 3' | Eurogentec Biologics Division, B4102 Seraing, Belgium | reaction reagents | |
| light cycler 480 SW 1.5.1 software | Roche Diagnostics GmbH, D-68305 Mannheim, Germany | software used for HRM-PCR CR1 polymorphism data analysis |

