Registered Bioimaging of Nanomaterials for Diagnostic and Therapeutic Monitoring
Summary
Bioimaging methods used to assess cell biodistribution of nanoparticles are applicable for therapeutic and diagnostic monitoring of nanoformulated compounds. The methods described herein are sensitive and specific when assessed by histological coregistration. The methodologies provide a translational pathway from rodent to human applications.
Abstract
Nanomedications can be carried by blood borne monocyte-macrophages into the reticuloendothelial system (RES; spleen, liver, lymph nodes) and to end organs. The latter include the lung, RES, and brain and are operative during human immunodeficiency virus type one (HIV-1) infection. Macrophage entry into tissues is notable in areas of active HIV-1 replication and sites of inflammation. In order to assess the potential of macrophages as nanocarriers, superparamagnetic iron-oxide and/or drug laden particles coated with surfactants were parenterally injected into HIV-1 encephalitic mice. This was done to quantitatively assess particle and drug biodistribution. Magnetic resonance imaging (MRI) test results were validated by histological coregistration and enhanced image processing. End organ disease as typified by altered brain histology were assessed by MRI. The demonstration of robust migration of nanoformulations into areas of focal encephalitis provides ‘”proof of concept” for the use of advanced bioimaging techniques to monitor macrophage migration. Importantly, histopathological aberrations in brain correlate with bioimaging parameters making the general utility of MRI in studies of cell distribution in disease feasible. We posit that using such methods can provide a real time index of disease burden and therapeutic efficacy with translational potential to humans.
Protocol
1. Introduction
The selective delivery of drugs and therapeutic macromolecules (peptides, proteins and nucleic acids) to cellular and tissue sites of active disease and ongoing microbial infections will improve pharmaceutical responses during disease1-3. One particular cellular site is the macrophage that is both highly mobile and immune engaging and is a consistent principal target for the human immunodeficiency virus (HIV).4 Importantly, macrophage engaged inflammation also underlies a broad range of disorders that include degenerative, inflammatory, infectious and cancerous diseases; and the cell’s mobility to disease sites underlies progression of tissue injuries5-9. Importantly, the use of blood borne macrophages as drug, macromolecule, and signal carriers has gained recent attention for its translational potential. However, a significant obstruction in realizing therapeutic potentials is the blood brain barrier (BBB) amongst other tissue barriers that are impermeable to a spectrum of macromolecules and proteins. These, barriers, nonetheless, do permit cell passage. All together it is projected that in the natural course of disease peripheral macrophages that bypass barriers can carry formulated drugs, markers, and peptides to sites of infection or inflammation. Nonetheless, such technologies remain only in development. It is through our works that cell-mediated delivery can be developed for diagnostic and therapeutic applications and such applications are supported by laboratory and animal models of human disease10-12.
2. Nanomaterial Preparations
Preparation of nanomaterials for drug delivery and biodistribution studies is the topic of a parallel manuscript in this issue (reference parallel manuscript). All procedures for crystalline nanoparticle manufacture are carried out in a laminar flow hood. All surfaces are disinfected prior to use with 70% alcohol. This includes working surface, the exterior of gloves and any spills. All are covered with solution of replicate 70% alcohol immediately with wipes. Gloves are discarded after use and are not worn when entering any other laboratory area. Excipient, drug, sterile water with/containing any/all reagents for manufacture of drug-laden particles are only brought into work areas when needed for procedures. Sterile wrapped pipettes are used only and discarded after use into a biohazard waste container. The wet willing apparatus is disinfected with alcohol prior to and following use. Work area is cleaned immediately before and after with 70% alcohol. Nanoparticle solution is tested for pyrogen in accordance with FDA guidelines to assess the absence of bacterial endotoxin in drug particle solutions used for animals. Briefly,
- Candidate nanoformulations for in-vivo use are replicated by replacing the drug core or droplet with an identical sized particle or milled piece of superparamagnetic iron oxide (SPIO) before coating with the appropriate surfactant.
- This is followed by measures of size, charge, shape, and cytotoxicity to determine whether the SPIO model system has the same properties as the candidate nanoformulated drug.
- Finally, cell loading assays are performed by incubation with the candidate SPIO model nanoformultation in order to determine the relaxivity within the cells using phantoms composed of labeled cells suspended in agar gel. Phantoms are prepared in triplicate and are prepared at a series of concentrations in order to quantify the relaxivity due to the SPIO uptake in cells. This provides an index of sensitivity and determines whether the nanoformulations may affect the oxidation state, and hence the visibility of the SPIO in magnetic resonance imaging (MRI) scans.
3. Methods and Procedures: Animal Preparation
- Injections/Catheters. Depending on time of interest, the injections may require use of a catheter to inject the animal within the MRI. Catheters are prepared using a non-magnetic needle and a tubing extension with minimum diameter to minimize dead space in the injection line. The catheter should be prefilled with either the solution containing the nanomaterial to be injected or saline, depending on the dead space and the total acceptable volume of the injection. If possible, the injection may be followed with a physiological saline flush. If acute times are not of extreme importance, a pre-scan can be performed, and the injection may be done outside of the magnet at a predetermined time before the follow up scans for biodistribution measures. Catheters are typically inserted into the tail vein for I.V. injections.
- Anesthesia and monitoring. Prior to scanning, the animal is placed into a chamber to induce anesthesia. This chamber is prefilled with the 1.5% isoflurane in 70% nitrous oxide and 30% oxygen in order to speed the onset of anesthesia in the animal and minimize the amount of time required to ensure that the animal will not wake up upon removal from the chamber. Once the animal is fully anesthetized, the animal is removed from the chamber and placed into the stereotactic holder equipped to monitor the breath rate and temperature of the animal while continuing to supply isoflurane during set-up and scanning.
- Animal Holders and Adjustment Considerations: Set-up includes eye lubricant to protect against corneal ulcers. The animal is lightly wrapped with gauze and the gauze is taped in place to minimize heat loss during scanning and to provide a positive pressure against the breathing monitor. Animal holders are equipped with adjustable tooth bars, allowing for vertical and horizontal alignment of the head. This is particularly important for high field MRI, as angulation of the head in the caudal-rostral direction will cause additional difficulties with magnetic field inhomogeneity due to magnetic susceptibility. Magnetic field inhomogeneity is deleterious to high quality T2* MRI as well as 1H magnetic resonance spectroscopy (1H MRS) and magnetic resonance diffusion tensor imaging (DTI). In addition to proper positioning of the head angle in the caudal-rostral direction, rotations of the head should be avoided to the degree that is feasible. Allowing for rotation of the animal holder in the magnet will provide compensation for minor rotations, which may occur from animal to animal. This may be further minimized by careful placement of the head and attention to the angulation prior to inserting the animal into the MRI system.
- Calibration and Shimming: Once the animal is in the holder and the surface coil is properly placed on the head, the initial position of the animal is determined by a real-time one-dimensional readout in the caudal-rostral direction. Signal is restricted to the area around the surface coil used for reception, limiting the need for interpretation of the observed wave forms. Once the initial position is determined, a 3-plane localizer image is taken to determine the precise position of the animal in the scanner and to allow for movement to the precise location required for the scan(s) of interest. This is followed by adjustment of the homogeneity of the magnetic field or “shimming” the magnet. This is done by mapping the field distribution and calculation of a precisely determined spatial correction based on measured responses of a series of electromagnets or “shim coils” within the system designed to adjust the field homogeneity. Shimming is accomplished using a multi-gradient echo sequence and mapping software developed by Dr. Hetherington13. Regions of homogeneity are matched to the region examined by each individual imaging method. Once shimming is complete, we can acquire the scan(s) of interest from the animal.
4. Data Acquisition
- High Resolution T2* weighted MRI. Biodistribution of SPIO containing nanoparticles can be determined by detecting regions of signal loss in high resolution 3D T2* weighted MRI. The region of the brain is determined from the localizer scans and prescribed on the localizer scans or additional scans as needed. A T2* weighted MRI scan with 150 micron isotropic resolution is then acquired. A high resolution 3D gradient recalled echo MRI scan of the mouse head is acquired using a 25 mm birdcage volume coil with acquisition parameters of echo time= 5 ms, repetition time = 50 ms, 30% echo, flip angle = 35 degrees, averages = 2, field of view = 20 x 20 x 20 mm with a resolution of 128 x 128 x 128 (voxel size = 150 x 150 x 150 μm3), total acquisition time = 30 min.
- Diffusion Tensor Imaging (DTI): Diffusion tensor images are quantitative measures of the direction and magnitude of diffusion of water within the cells of the tissue. As a result, the phase of the signal is extremely susceptible to motion, as the scans are sensitized or “weighted” for microscopic water motion. As a result, single shot acquisitions are desired to prevent phase shifts between acquisitions from causing signal smearing; and respiratory gating is required to prevent gross motion during signal acquisition. Therefore, a respiratory gated spin-echo diffusion weighted echo planar imaging (EPI) MR sequence is employed. Again, shimming the region of the scans is very important, as off-resonance effects during the evolution of the signal causes misregistration of the signal frequency, and hence position, in the plane of the image. EPI acquisition parameters included 14 slices, 200 KHz bandwidth, 96 x 96 in plane acquisition zero-filled to 256 x 256, and a 0.5 mm slice thickness. The diffusion encoding used was a balanced, rotationally-invariant and alternating polarity icosahedral scheme (12 directions) 14,15. The encoding scheme was designed to reduce background-diffusion gradient couplings 16. Diffusion weighting b-factor = 800 s mm-2, δ=4 ms, Δ=15 ms, Gdmax= 40 G/cm, 200 μs rise time, 7 averages for b=0 acquisition, 3 averages for each b=800 encoding direction, for a total acquisition time of 20-40 min, depending upon respiration rate.
- Localized 1H Magnetic Resonance Spectroscopy (1H MRS): 1H MRS can be obtained from brain regions prescribed on images acquired during the same imaging session. Anatomical locations are found on images to prescribe the region of interest for acquiring spectra. Once the region is identified, shimming is performed on a region matching the volume of acquisition, checked using a localized water spectrum. Then, the power of the water suppression pulses are optimized, water frequency is measured to ensure on-resonance water signal, and a short test spectrum is acquired to provide quality control. If spectra are of insufficient quality, system settings, including radiofrequency (RF) power and shim settings, are checked. Finally, if quality is still insufficient, a second 3-plane localizer is run to ensure that the animal has not moved from the initial scans. In our experience, this provides a very high degree of reproducibility and accuracy for spectroscopic acquisitions. Finally, the spectra are acquired in short blocks with resetting of the system frequency between acquisitions to eliminate effects of magnetic field drift and to ensure reproducibility and quality of the final scans. At the end of the acquisition, a single pulse water spectrum at a predefined preamplifier gain is used as a quantitative signal amplitude reference.
- Histology and Blockface Imaging: After the final MRI scanning session in the time series of experiments, the mouse is perfused, the brain is removed and embedded into a block of OCT, compound which has been darkened using a drop of India ink. The block is placed into a cryostat for slicing and histological analysis. Blockface images are acquired using a digital camera (Canon EOS Digital Rebel 300D with a Canon Ultrasonic EFS 60mm f/2.8 Macro USM lens) mounted to the front of the cryostat with a custom mount and triggered by a remote switch. Digital images are acquired every 50 micrometers through the entire brain volume. Slices are numbered to allow registration within the volume after histological processing and staining. Individual blockface slices were aligned to reconstruct the 3D volume using the block outlines to account for jitter in the position of the cryostat head. The brain volume is then automatically segmented using seed based region growing algorithm in the Analyze software package (AnalyzeDirect, Lexena, KS).
5. Data Analyses
- SPIO detection using MRI: SPIO causes signal loss in T2* weighted MRI; and, as such, the MRI signal void is a sensitive, but not specific marker for SPIO presence in tissue. The sensitivity is dependent on the spatial resolution of the MRI scan and the size of the SPIO particle, with a single micron sized particle detected with 100 micron isotropic resolution. In these works, 150 micron isotropic resolution with 200 nanometer sized SPIO particles are used. To provide both sensitivity and specificity for the presence of SPIO in brain, mice were scanned prior to injection of the SPIO labeled cells to allow subtraction images to be used for positive identification of the cells in the brain at later time points. 3D MRI scans were subimaged using the constrained level set method developed in our laboratory as previously described17. Subimaged brain volumes were then coregistered, signal intensity normalized, and the volumes subtracted to detect regions within the brain volume with signal loss (presence of SPIO), which was not along the edges to eliminate positive false signal from any subpixel registration errors.
- Coregistration of Histology and MRI: Coregistration between histology and MRI was accomplished by using the blockface image as a common reference. This approach reduces the complexity of the major problem of correcting the asymmetrical shrinkage of tissue slices during preparation and staining to a two dimensional problem as described in our previous works18,19. Here, MRI and histology detection of SPIO containing macrophages in brain have shown excellent spatial correlation, with an expected overestimation of the volume by the loss in MRI signal and greater sensitivity to few cells demonstrated by histology12. Accurate co-localization of these two signals provides one measure of the accuracy of coregistration and warping of histology back to the original shapes of the slices.
- Region of Interest (ROI) Analyses of DTI: DTI scans are typically analyzed by selection of an anatomical ROI to determine the average diffusion properties of the tissue in a particular anatomical substructure.
Analyses of the diffusion-weighted data are performed using custom programs written in IDL (Interactive Data Language, ITT Visual Information Solutions, Boulder, CO) as previously described 15,20. Analyses produce maps of the tensor diffusivities (λ1, λ2, λ3), average diffusivity (Dav) where: Dav = (λ1+λ2+λ3)/3 and fractional anisotropy (FA), where:
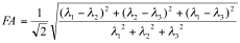
Transverse (λ⊥=(λ2+λ3)/2) and longitudinal (λll=λ1) components of the diffusion tensor were obtained as described elsewhere 21. Once the maps are constructed, ROIs are drawn on T2 weighted MRI overlaid with color encoded λ1 directionality maps. Examples of the regions chosen for analysis in the HIV mouse model are displayed in Figure 1. - Spectroscopic Analysis: Quantitation of the metabolic compounds contributing to the peaks of brain 1H MRS is determined using one of a variety of curve fitting methods. A variety of curve fitting techniques have been developed. In our laboratory, we employ a time domain fitting method (QUEST)22 in the jMRUI23 signal processing package that is a linear combination of individual metabolite spectra contributing to the final spectrum. We use a basis set of 22 individual metabolites as potential contributing factors. The basis spectra are simulated and checked using spectra of solutions of individual metabolites. An example of a curve fit result from a single spectrum is shown in Figure 2.
6. Representative Results
Examples of DTI and 1HMRS are shown in figures 1 and 2. Additional examples of 1H MRS24-26 and DTI27 results can be seen in our previous publications. Examples of preinjection T2* weighted MRI with an overlay of the location of labeled cells in yellow is shown in Figure 3. The mouse had labeled monocytes derived macrophages injected into the tail vein. Five days later, T2* weighted MRI was acquired and processed as described, above. The mouse was prepared by injection of HIV infected human macrophages into the brain, which is seen as a line of detected mouse monocytes derived macrophages. Further examples of both detection of labeled cells and coregistration with histology can be seen in our previous publications10,12.
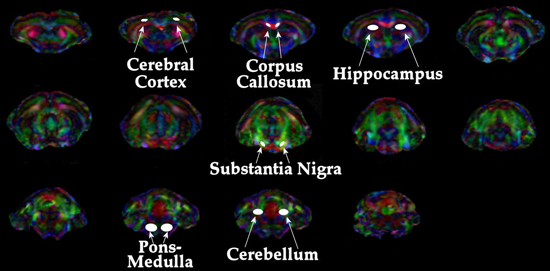
Figure 1. Depiction of regions analyzed for DTI metrics.
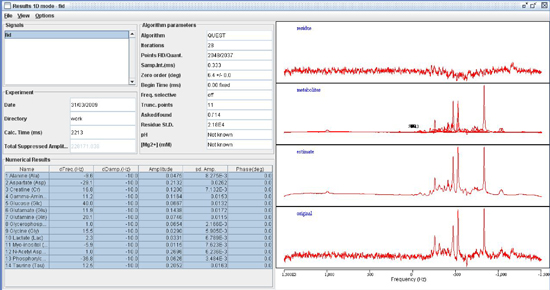
Figure 2. Spectroscopic fitting using QUEST in the jMRUI signal processing suite.
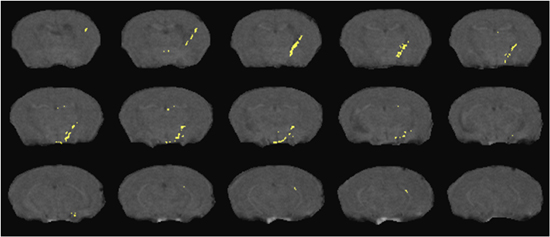
Figure 3. Detection of SPIO labeled cells migrating from the peripheral blood into a brain region with focal encephalitis. Cell positions (yellow) overlay representative slices from a T2* weighted MRI acquisition as detailed in the text.
Discussion
The accurate registration of histology with in-vivo imaging results is a critical step in the development of imaging biomarkers for detection and staging of neuronal disease. Some imaging metrics are likely to be correlated with gross morphological changes including changes in magnetic relaxation properties of tissue used for detecting the presence of white matter disease and cancers. Other more subtle methods, such as DTI, are likely to detect early cellular changes that may not be detectable as histological changes caused by the disease do not appear until later the stages of disease. Still other markers, such as spectroscopic markers, may be indicators of early and reversible changes, which precede even the subtlest cellular alterations.
Biodistribution can be determined non-invasively using a variety of methods. The primary non-invasive methods are positron emission tomography (PET), single photon emission computed tomography (SPECT), optical imaging, and MRI. Nuclear medicine based imaging (PET and SPECT) have been used over the years for many biodistribution, but these methods are limited by the half life of the radiotracers used for labeling the compounds or nanomaterials, especially for PET tracers. Optical imaging can be used for small rodents but cannot be translated to human use except for regions easily accessible such as surface tumors due to light absorption and light scattering. In addition, it is difficult to quantify the optical signals for these same reasons. MRI uses persistent tags such as SPIO that can be tracked in the body over a period of weeks. This, too, must be used with caution, as the label can be transferred to different cells or be reabsorbed by the body.
Detection specificity for SPIO in MRI can be provided by a variety of methods. Detection methods, which provide positive as well as negative signals, are used for improving specificity of the MRI for detecting the presence of SPIO in tissue. The subtraction method used in this work has been used by others, as well28. Other approaches include off resonance detection29-31, phase sensitive imaging that produces a particular pattern near SPIO voids32, and zero echo time image that uses T1 weighting to produce a positive signal intensity in the region of SPIO33. The advancement of these methods for improving quantitation of label, sensitivity and specificity is an area of active research today.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The work was supported by grants 1K25MH089851, 1P01DA028555-01A1, 2R01 NS034239, 2R37 NS36126, P01 NS31492, P20RR 15635, P01MH64570, and P01 NS43985 from the National Institutes of Health. The authors thank Ms. Robin Taylor for critical reading of the manuscript and outstanding graphic and literary support. The authors would also like to thank Erin McIntyre, Melissa Mellon, and Lindsay Rice for their technical support.
Materials
| Material Name | Tipo | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| Isoflurane | ||||
| Medical Oxygen | ||||
| Isoflurane vaporizer | ||||
| Rodent gas anesthesia mask | ||||
| MRI compatible Stereotactic head holder | ||||
| Syringe | ||||
| Polyethylene catheter tubing | ||||
| Non-magnetic needle | ||||
| Eye lubricant | ||||
| Gauze | ||||
| Tape | ||||
| Perfusion media | ||||
| OCT compound for embedding tissue | ||||
| MRI system | ||||
| Digital Camera | ||||
| Tissue Sectioning Cryostat |
Riferimenti
- Nowacek, A., Gendelman, H. E. NanoART neuroAIDS and CNS drug delivery. Nanomedicine (Lond). 4, 557-574 (2009).
- Nowacek, A., Kosloski, L. M., Gendelman, H. E. Neurodegenerative disorders and nanoformulated drug development. Nanomedicine (Lond). 4, 541-555 (2009).
- Nowacek, A. S., Miller, R. L., McMillan, J. NanoART synthesis, characterization, uptake, release and toxicology for human monocyte-macrophage drug delivery. Nanomedicine (Lond). 4, 903-917 (2009).
- Herbein, G., Varin, A. The macrophage in HIV-1 infection: from activation to deactivation. Retrovirology. 7, 33-33 (2010).
- Qian, B. Z., Pollard, J. W. Macrophage diversity enhances tumor progression and metastasis. Cell. 141, 39-51 (2010).
- Nathan, C., Ding, A. Nonresolving inflammation. Cell. 140, 871-882 (2010).
- McNeill, E., Channon, K. M., Greaves, D. R. Inflammatory cell recruitment in cardiovascular disease: murine models and potential clinical applications. Clin Sci (Lond). 118, 641-655 (2010).
- Bondeson, J. Activated synovial macrophages as targets for osteoarthritis drug therapy. Curr Drug Targets. 11, 576-585 (2010).
- Benoit, L. A., Holtzman, M. J. New immune pathways from chronic post-viral lung disease. Ann N Y Acad Sci. 1183, 195-210 (2010).
- Beduneau, A., Ma, Z., Grotepas, C. B. Facilitated monocyte-macrophage uptake and tissue distribution of superparmagnetic iron-oxide nanoparticles. PLoS One. 4, e4343-e4343 (2009).
- Dou, H., Grotepas, C. B., McMillan, J. M. Macrophage delivery of nanoformulated antiretroviral drug to the brain in a murine model of neuroAIDS. J Immunol. 183, 661-669 (2009).
- Liu, Y., Uberti, M. G., Dou, H. Ingress of blood-borne macrophages across the blood-brain barrier in murine HIV-1 encephalitis. J Neuroimmunol. 200, 41-52 (2008).
- Hetherington, H. P., Chu, W. J., Gonen, O., Pan, J. W. Robust fully automated shimming of the human brain for high-field 1H spectroscopic imaging. Magn Reson Med. 56, 26-33 (2006).
- Hasan, K. M., Parker, D. L., Alexander, A. L. Comparison of gradient encoding schemes for diffusion-tensor MRI. J Magn Reson Imaging. 13, 769-780 (2001).
- Hasan, K. M., Basser, P. J., Parker, D. L., Alexander, A. L. Analytical computation of the eigenvalues and eigenvectors in DT-MRI. J Magn Reson. 152, 41-47 (2001).
- Neeman, M., Freyer, J. P., Sillerud, L. O. A simple method for obtaining cross-term-free images for diffusion anisotropy studies in NMR microimaging. Magn Reson Med. 21, 138-143 (1991).
- Uberti, M. G., Boska, M. D., Liu, Y. A semi-automatic image segmentation method for extraction of brain volume from in vivo mouse head magnetic resonance imaging using Constraint Level Sets. J of Neurosci Methods. 179, 338-344 (2009).
- Liu, Y., Uberti, M. G., Dou, H., Mosley, R. L., Gendelman, H. E., Boska, M. D. An Image Warping Technique for Rodent Brain MRI-Histology Registration Based On Thin-Plate Splines with Landmark Optimization. Proc Soc Photo Opt Instrum Eng. 7259, 72592-K-72597-K (2009).
- Uberti, M. G., Liu, Y., Dou, H., Mosley, R. L., Gendelman, H. E., Boska, M. Registration of in vivo MR to histology of rodent brains using blockface imaging. Proc Soc Photo Opt Instrum Eng. 7262, 726213-726213 (2009).
- Basser, P. J., Mattiello, J., LeBihan, D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B. 103, 247-254 (1994).
- Hasan, K. M., Narayana, P. A. Retrospective measurement of the diffusion tensor eigenvalues from diffusion anisotropy and mean diffusivity in DTI. Magn Reson Med. 56, 130-137 (2006).
- Ratiney, H., Sdika, M., Coenradie, Y., Cavassila, S., Ormondt, D. v. a. n., Graveron-Demilly, D. Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed. 18, 1-13 (2005).
- Naressi, A., Couturier, C., Castang, I., Beer, R. d. e., Graveron-Demilly, D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 31, 269-286 (2001).
- Boska, L. e. w. i. s., Destache, T. B., J, C. Quantitative 1H magnetic resonance spectroscopic imaging determines therapeutic immunization efficacy in an animal model of Parkinson’s disease. J Neurosci. 25, 1691-1700 (2005).
- Mosley, R. L., Benner, E. J., Kadiu, I. Neuroinflammation, Oxidative Stress and the Pathogenesis of Parkinson’s Disease. Clin Neurosci Res. 6, 261-281 (2006).
- Nelson, J. A., Dou, H., Ellison, B. Coregistration of quantitative proton magnetic resonance spectroscopic imaging with neuropathological and neurophysiological analyses defines the extent of neuronal impairments in murine human immunodeficiency virus type-1 encephalitis. J Neurosci Res. 80, 562-575 (2005).
- Boska, H. a. s. a. n., Kibuule, K. M., D, . Quantitative diffusion tensor imaging detects dopaminergic neuronal degeneration in a murine model of Parkinson’s disease. Neurobiol Dis. 26, 590-596 (2007).
- Liu, W., Frank, J. A. Detection and quantification of magnetically labeled cells by cellular MRI. Eur J Radiol. 70, 258-264 (2009).
- Balchandani, P., Yamada, M., Pauly, J., Yang, P., Spielman, D. Self-refocused spatial-spectral pulse for positive contrast imaging of cells labeled with SPIO nanoparticles. Magn Reson Med. 62, 183-192 (2009).
- Stuber, M., Gilson, W. D., Schar, M. Positive contrast visualization of iron oxide-labeled stem cells using inversion-recovery with ON-resonant water suppression (IRON). Magn Reson Med. 58, 1072-1077 (2007).
- Suzuki, Y., Cunningham, C. H., Noguchi, K. In vivo serial evaluation of superparamagnetic iron-oxide labeled stem cells by off-resonance positive contrast. Magn Reson Med. 60, 1269-1275 (2008).
- Kim, Y. B., Bae, K. H., Yoo, S. S., Park, T. G., H, P. a. r. k. Positive contrast visualization for cellular magnetic resonance imaging using susceptibility-weighted echo-time encoding. Magn Reson Imaging. 27, 601-610 (2009).
- Zhou, R., Idiyatullin, D., Moeller, S. SWIFT detection of SPIO-labeled stem cells grafted in the myocardium. Magn Reson Med. 63, 1154-1161 .

