High-throughput, Automated Extraction of DNA and RNA from Clinical Samples using TruTip Technology on Common Liquid Handling Robots
Summary
TruTip is a simple nucleic acid extraction technology whereby a porous, monolithic binding matrix is inserted into a pipette tip. Consequently, the sample preparation format is compatible with most liquid handling instruments, and can be used for many medium to high-throughput clinical applications and sample types.
Abstract
TruTip is a simple nucleic acid extraction technology whereby a porous, monolithic binding matrix is inserted into a pipette tip. The geometry of the monolith can be adapted for specific pipette tips ranging in volume from 1.0 to 5.0 ml. The large porosity of the monolith enables viscous or complex samples to readily pass through it with minimal fluidic backpressure. Bi-directional flow maximizes residence time between the monolith and sample, and enables large sample volumes to be processed within a single TruTip. The fundamental steps, irrespective of sample volume or TruTip geometry, include cell lysis, nucleic acid binding to the inner pores of the TruTip monolith, washing away unbound sample components and lysis buffers, and eluting purified and concentrated nucleic acids into an appropriate buffer. The attributes and adaptability of TruTip are demonstrated in three automated clinical sample processing protocols using an Eppendorf epMotion 5070, Hamilton STAR and STARplus liquid handling robots, including RNA isolation from nasopharyngeal aspirate, genomic DNA isolation from whole blood, and fetal DNA extraction and enrichment from large volumes of maternal plasma (respectively).
Introduction
Nucleic acid purification is necessary for most molecular diagnostics, research use only, and life science applications. Various approaches have emerged since the time of phenol/chloroform extractions, many of which are based on the fundamental principle of nucleic acid binding to silica in the presence of chaotropic salts 1. The extraction process has been streamlined and automated through the use of various bead- and membrane-based formats, with spin filters, magnetic beads and related approaches dominating the life sciences industry (see examples in 2-13). While effective, particles and membranes have known limitations when confronted with challenging clinical matrices. For example, membranes and bead-based columns are compliant, have small pore sizes (thus, high back pressures), and require some type of support in order to be processed by a centrifuge or vacuum system. The physical characteristics of membranes and bead columns result in significant fluidic resistance, which limits the type of samples that can be efficiently processed without clogging the consumable, and/or the total (input) sample volume that can be uni-directionally processed through the flow path. Conversely, magnetic particles must be distributed throughout the sample by agitation. The need to homogenously distribute magnetic particles within a solution limits the total input sample volume that can be processed with most magnetic bead consumables. Clinical sample attributes (such as viscosity or complexity) can lead to inefficient magnetic particle concentration on the side of a tube or rod. In addition, silica fine particulates can break off of the beads during the extraction process, losing their magnetization and contaminating the final sample.
The TruTip technology was developed to overcome some of these nucleic acid sample processing constraints and limitations 14. By embedding a porous monolith within a pipette tip, fluidic backpressure is lowered, which enables flow control by vacuum (i.e. pipette aspiration). This feature enables the extraction process and instrumentation required to purify nucleic acids from difficult sample types to be greatly simplified (Figure 1). The geometry and porosity of the monolith is tailored to minimize clogging, while the thickness of the monolith provides sufficient nucleic acid binding capacity for sample volumes ranging from 1.0 to 5.0 ml. Bidirectional flow during sample aspiration and dispensing allows for prolonged residence times between the sample extract and the binding monolith for efficient nucleic acid recovery and elution, and enables relatively large sample volumes to be processed that exceed the volume capacity of the pipette tip itself. We previously reported the development and application of a manual TruTip procedure for purifying influenza RNA from nasopharyngeal samples using a single- or multi-channel Rainin pipettor 15. Equivalent extraction efficiencies were obtained between automated QIAcube and manual TruTip methods at 106 gene copies influenza A per ml nasopharyngeal aspirate. The objective of this study was to demonstrate medium- and high-throughput, automated TruTip nucleic acid purification procedures for nasopharyngeal aspirate (NPA) and other clinically relevant sample types, using liquid handling robots commonly found in reference clinical laboratories.
Protocol
Three automated TruTip extraction protocols are described and demonstrated here: 1) medium-throughput RNA extraction from NPA on an Eppendorf epMotion 5070; 2) high-throughput genomic DNA extraction from low volumes of whole blood on the Hamilton STAR, and 3) selective isolation and enrichment of fragmented fetal DNA from large volumes of maternal plasma on the Hamilton STARplus. Automated scripts are available from Akonni and only high-level program descriptors are provided here. The extraction and elution reagents are automatically dispensed from bulk reagent troughs to 96-well plates by the automated script(s) before the clinical samples are processed.
1. Automated RNA Extraction from Nasopharyngeal Aspirate
The previously described manual TruTip method 15 is now adapted to run on an Eppendorf epMotion 5070 liquid handling robot, using a large pore TruTip matrix embedded in 1.0 ml Eppendorf pipette tips, a 2 ml deep-well plate (USA Scientific), Akonni TruTip extraction reagents, and NPA as the sample type. The epMotion 5070 liquid handling robot holds up to 8 tips simultaneously, so a baseline automated protocol is described for 8 parallel extractions. However, up to 24 samples can be processed during a single program in one deep-well 96-well sample plate. A separate epMotion program is available (and required) in order to process 16 or 24 samples. The protocol outlined below is for an 8 sample automated script.
Setup
- Bring nasopharyngeal samples to RT before starting the extraction.
- Dispense 100 μl nasopharyngeal aspirate plus 150 μl nuclease-free water into column 1 of the sample plate (Figure 2A).
- Place the sample plate into position B1 on the epMotion Worktable (Figure 2B).
- Place pipette tips, TruTips and 30 ml reagent troughs onto their respective epMotion Worktable positions (Figure 2B).
- Open the Eppendorf epBlue software, select the Run file provided by Akonni for 8 samples, and load the method by clicking the RUN button on the RUN tab.
- Under Level Sensor Settings, select Levels and Tips, and click the RUN button.
- Input the sample volume into the software and click RUN.
- The epMotion script will prompt the user to add extraction and elution reagents to the reagent reservoirs located at position B2 on the Worktable. Add the recommended volumes of each reagent to the respective trough. For 8 samples, the minimum reagent volumes are:
| Reagent | Volume (ml) | Trough Position |
| 95% ethanol | 3.5 | 2 |
| Wash Buffer D | 9.0 | 3 |
| Wash Buffer E | 9.0 | 4 |
| Elution Buffer A2 | 1.3 | 5 |
| Lysis and Binding Buffer D | 11.0 | 6 |
- Input the reagent volumes into the Table presented by the epMotion software during the prompt from Step 8 above. The input value should reflect the actual volume of buffer dispensed by the user into each respective reagent reservoir, and must be greater than or equal to the minimum volumes noted above. Incorrect volume entries could result in incorrect volumes delivered by the epMotion hardware to each tube or well in the 96-well plate(s).
Automated Program
- Select RUN to start the automated method. The automated script will move through the following steps without user intervention:
Sample Lysis and Reagent Distribution:- Dispense 375 μl Lysis Buffer D into column 1 and mix for 10 cycles (apirate + dispense = 1 cycle). This step starts the lysis incubation process while the remaining reagents are aliquotted.
- Dispense 1.6 ml Wash Buffer D into column 2.
- Dispense 1.6 ml Wash Buffer E into column 3.
- Dispense 50 μl Elution Buffer A into column 4.
- Pause for 6 min to complete the 10 min sample incubation in Lysis Buffer D.
- Add 375 μl ethanol to each well in column 1, mixing each sample with ethanol through 10 pipetting cycles.
- Load 8 TruTips from position A2 on the Worktable, and begin the extraction process outlined in Figure 1.
- Aspirate and dispense sample/lysis buffer/ethanol mixture from column 1 of the sample plate for seven cycles to bind the nucleic acid to the TruTip monolith. Although sample flow through the TruTip may vary (due to differences in clinical sample viscosity), nucleic acid yield for these sample was not affected by flow variation. Options for improving sample flow are described in the Discussion.
- Move TruTips to sample plate column 2, and cycle Wash Buffer D 5x over the monolith to remove residual lysis buffer and sample matrix.
- Move TruTips to sample plate column 3, and cycle Wash Buffer E 5x over the monolith to remove proteins and other contaminants from the bound nucleic acid.
- Move TruTips to the empty reagent reservoir position 1 (in Worktable location B2) and cycle 80x (at a fast flow rate) to dry the monolith. It is important to thoroughly dry the TruTip, as residual solvents in eluted nucleic acid preparations will negatively affect enzymes such as reverse transcriptase and Taq DNA polymerase.
- Move TruTips to sample plate column 4 and cycle 5x in Elution Buffer A. The extracted and purified nucleic acid is now in elution buffer in sample plate column 4 wells.
- Eject TruTips into the epMotion waste bin.
- When the program is finished, manually remove the sample plate from the instrument and transfer the purified nucleic acid to new tubes for long-term storage or further use. Advanced epMotion users can add instructions to the Run file to transfer eluted samples into separate storage tubes or PCR plates, if desired.
The program for 16 total samples will repeat steps 10.1 through 10.13 using sample plate columns 5-8. For the 24-sample program, steps 10.1 through 10.13 are repeated 2x using sample plate columns 5-8 and 9-12, respectively.
2. 96-well Genomic DNA Extraction from Whole Blood
A Hamilton STAR liquid handling robot is used to demonstrate automated extraction of 96 samples simultaneously from whole blood. The Hamilton STAR differs from the epMotion system in that an optional heater/shaker unit is available on the deck, which is important for enzymatic digestion of certain clinical matrices, such as whole blood. Because the system can be fitted with a 96-channel pipette head, there is a dedicated 96-well plate for each of the TruTip steps and reagents.
Setup
- Turn on the STAR instrument and computer.
- Open the Hamilton Run Control software.
- Open the Run file provided by Akonni for 96 samples.
- Place labware onto the STAR deck as shown in Figure 3.
- Dispense reagents into their corresponding troughs (volumes denote the minimum required to process 96 samples):
| Reagent | Volume (ml) | Trough Position |
| Lysis and Binding Buffer F | 75 | 5 |
| 95% ethanol | 100 | 6 |
| Wash Buffer J | 175 | 7 |
| Wash Buffer K | 175 | 8 |
| Elution Buffer A2 | 12 | 9 |
| Proteinase K (20 mg ml-1) | 8 | 15 |
6. Allow samples to equilibrate to RT.
7. Place the sample tubes in the Sample Carrier racks (deck position 4 in Figure 3). Place Sample 1 in the rear of the far left carrier and move sequentially down each carrier with Sample 96 ending in the front right position.
Automated Program
- Select the PLAY button in the upper left of the Run file window and input the number of samples being processed. The automated script will then move through the following steps without user intervention:
Sample Lysis and Reagent Distribution- Transfer 200 μl from each sample tube to the incubation plate at position 14 on the heater/shaker module (Figure 3).
- Dispense 80 μl proteinase K into each sample well of the incubation plate.
- Dispense 600 μl Lysis Buffer F into each well of the incubation plate.
- Mix the solution for 10 cycles, and then incubate for 20 min at 70 °C and 500 rpm. The 70 °C set temperature results in an ~60 °C sample temperature within the deep well plate, which is within the range of proteinase K activity. While the samples are incubating, the liquid handling system continues operating by dispensing reagents into their corresponding plates and wells:
- 100 μl Elution Buffer A into each well of the deep well plate at position 13.
- 800 μl ethanol into each well of the deep well plate at position 10.
- 1.6 ml Wash Buffer K into each well of the deep well plate at position 12.
- 1.6 ml Wash Buffer J into each well of the deep well plate at position 11.
- After the 20 min incubation, the sample mixture is transferred from the incubation plate to the deep well plate at position 10, and mixed through 12 pipetting cycles.
- Eject reagent tips into the waste bin.
Extraction
This portion of the gDNA blood procedure is very similar to the epMotion influenza protocol, except for the composition of wash reagents and cycle numbers. The Hamilton TruTips tips are black, so the flow of liquids through the TruTip is not visible.
- Load 96 TruTips from deck position 3.
- Aspirate and dispense the sample/lysis buffer/ethanol mixture in position 10 for 10 cycles to bind nucleic acids to the TruTip monolith.
- Move TruTips to position 11 and cycle Wash Buffer J 5x over the monolith to remove residual lysis buffer and sample matrix.
- Move TruTips to position 12 and cycle Wash Buffer K 5x to remove proteins and other contaminants from the bound nucleic acid.
- Cycle the TruTip 40x at high speed to air dry.
- Move TruTips to position 13 and cycle 5x in Elution Buffer A2. The extracted and purified nucleic acid is now in elution buffer in the deep well plate.
- Eject TruTips into the waste bin.
When the program is finished, remove the Elution Plate from the instrument and transfer the extracted samples to the appropriate tubes for storage or downstream applications.
3. DNA Extraction from Large Volume Plasma Samples
The Hamilton STARplus instrument is used to demonstrate an automated protocol for extracting freely circulating fetal DNA from 5 ml of maternal plasma. The STARplus system can support two automated pipette channel arms, one with 8 x 5 ml channels and one with 8 x 1 ml channels. These arms can operate in parallel for staggered processing in batches of 8 samples each. A 5 ml TruTip is used for the initial large-volume extraction, and a 1 ml TruTip is used for size separation and further concentration of the extracted nucleic acid.
Set-up
- Turn on the STARplus instrument and computer.
- Open the Hamilton Run Control software.
- Open the Run file provided by Akonni for 8 large volume plasma samples.
- Place labware onto the STARplus deck as shown in the Figure 4.
- Dispense reagents into their corresponding reservoirs:
| Reagent | Volume (ml) | Trough Position |
| CN-W1 | 17 | 5A |
| CN-W2 | 17 | 5B |
| CN-W4 | 21 | 5C |
| Proteinase K (20 mg ml-1) | 5 | 6A |
| EBB | 17 | 6B |
| EBA2 | 5 | 6C |
| CN-W3 | 5 | 6D |
| CN-B2 | 5 | 6E |
| CN-B3 | 5 | 6F |
| CN-L1 | 52 | 7 |
| CN-B1 | 175 | 8 |
- Allow sample to equilibrate to RT.
- Place the sample tubes in the Sample Carrier racks (deck position 3 in Figure 4). Place Sample 1 in the rear and move sequentially toward the front of the deck.
Automated Program
Because of the large input sample volume, lysis and homogenization steps must be performed off of the Hamilton STARplus instrument in a water bath. Steps requiring user intervention within the automated protocol are indicated with an asterisk (*) at the beginning of the sentence, and bold type.
Sample Lysis and Reagent Distribution: The sample is incubated with proteinase K and lysis buffer to homogenize the sample and release the DNA.
- Select the PLAY button in the upper left of the Run file window. Input the number of samples being processed, the location of pipette tips on the deck, and the location of TruTips on the deck.
- The automated script adds 615 μl proteinase K, 5 ml plasma, and 6.2 ml Lysis Buffer CN-L1 to each 50 ml conical tube, and will then PAUSE.
- *Remove the 50 ml conical tubes from the sample deck, vortex for 30 sec on high speed, and incubate off-line for 30 min at 60 °C in a water bath or heat block. After the conical tubes are removed from the Hamilton deck, RESUME the automated script to continue dispensing reagents into their respective plates and wells (Figure 4B and 4C):
- 2 ml CN-W1 to every other well in position 9 column 1.
- 2 ml CN-W2 to every other well in position 9 column 2.
- 2 ml CN-W4 to every other well in position 9 column 3.
- 250 μl EBA2 to every other well in position 9 columns 4 and 5.
- 495 μl CN-B2 to every other well in position 10 column 1.
- 1 ml EBB to every other well in position 10 column 2.
- 500 μl CN-W3 to every other well in position 10 column 3.
- 500 μl CN-W4 to every other well in position 10 column 4.
- 50 μl EBA2 to every other well in position 10 column 5.
Because the 5 ml channels are too wide to use adjacent wells for each sample, the automated program dispenses reagents into every other well of the deep well plate in deck position 9. The mechanical arm used for the 1 ml channels also requires the use of every other well in reagent plates for the exclusion and concentration steps of the protocol.
After dispensing reagents, the program will PAUSE.
- *After the 30 min, 60 °C incubation, place the 50 ml conical tubes on ice for 5 min.
- *Return the 50 ml conical tubes to their original positions within the sample carrier rack at deck position 4, and RESUME the automated script.
- Add 12 ml Binding Buffer CN-B1 to each sample tube and mix 10x.
Large Volume Extraction: 5 ml TruTips are used for extracting total DNA from the lysed plasma sample.
- Pick up 5 ml TruTips from position 2 for the large-volume nucleic acid extraction.
- Cycle the sample mixture15 times in the 50 ml conical tube, starting at the bottom of the tube and moving 3 mm higher after each pipetting cycle. This step binds the total nucleic acid to the TruTip monolith.
- Move TruTips to the deep well plates at position 6 column 1, and cycle 1x in Wash Buffer CN-W1.
- Move TruTips to position 9 column 2 and cycle 1x in Wash CN-W2.
- Move TruTips to position 9 column 3 and cycle 2x in Wash CN-W4.
- Move TruTips to position 9 column 4 and cycle 40x at high speed to dry binding matrix.
- Move TruTips to position 9 column 5 and cycle 10x to elute the bound nucleic acids from the 5 ml TruTips. This is large-volume elution #1.
- Move TruTips to column 6 and repeat the step with the second aliquot of elution buffer. This is large-volume elution #2.
- Transfer elution #2 into position 9 column 5 to combine it with elution #1, and discard the TruTips.
Exclusion and Concentration: The high molecular weight DNA is removed from the extracted sample, and the remaining DNA is isolated and concentrated.
- Add combined eluant from step 22 to position 10 column 1 and mix thoroughly 10x.
- Pick up 1 ml TruTips from position 13 and cycle 20x to bind the high molecular weight DNA to the monolith.
- Move the TruTips to position 10 column 2 and cycle 5x to rinse the tip and remove high molecular weight DNA. The TruTips are retained and placed back in the tip rack at position 13.
- With reagent tips from position 12, add 575 μl of Binding Buffer CN-B3 to the sample in position 10 column 1 and mix 10x.
- Pick up the TruTips from step 25, return to position 10 column 1, and cycle 20x to bind the remaining DNA from the sample to the 1 ml TruTip.
- Move TruTips to position 10 column 4 and cycle 1x in Wash CN-W3 to remove any remaining inhibitors.
- Move the TruTips to position 10 column 5 and cycle 1x in Wash CN-W4 to rinse residual salts from CN-W3.
- Raise TruTips over position 10 column 5 and cycle air through the tips 35x to dry the monolith.
- Move TruTips to position 10 column 6 and cycle 10x in EBA2 to elute the purified, size-selected and concentrated nucleic acid.
- Discard TruTips.
- Transfer the eluted sample from column 6 to 1.5 ml tubes in position 11. Extracted samples are ready for storage or downstream processing.
Representative Results
Real-time PCR data for influenza RNA extraction from NPA are shown in Figure 5A. Estimated nucleic acid extraction efficiency is similar to results obtained with the manual version of the protocol 15. A linear response in average Ct values is observed between 104 and 106 gene copies ml-1 amended influenza (R2 = 0.99 and 0.98 for influenza A and B, respectively), with standard deviations in average Ct values less than 1 cycle. The absence of cross-contamination with the epMotion system is demonstrated in Figure 5B, where 12 positive NPA samples containing 106 gene copies ml-1 NPA were interspersed with 12 buffer blanks. The average Ct for the positive controls was 30.16 ± 0.14, and all buffer blanks were negative. The total sample processing time is 16, 28 and 40 min for 8, 16 and 24 samples, respectively. Because a typical clinical nasopharyngeal aspirate or swab will contain 107 gene copies ml-1 (assuming 1,000 virions per TCID50 16 and > 104 TCID50 ml-1 influenza 17), the automated epMotion protocol is expected to be effective on a majority of clinical NPA specimens.
Given the range of molecular tests performed on human genomic DNA, the primary objective of nucleic acid extraction from whole blood is to produce contaminant-free, high molecular weight genomic DNA. The automated protocol for 96 samples is completed within 1 hr, which is an improvement over other automated systems (e.g. Promega MagneSil = 90 min and Qiagen QIAamp DNA Blood BioRobot MDx system = 2.5 hr). Figure 6A shows the UV/Vis absorbance profiles for 45 positive blood samples processed simultaneously with 45 reagent blanks on the Hamilton STAR protocol, with an average A260/280 ratio of 1.96 and average A260/230 ratio of 1.93. An A260/280 ratio between 1.7-2.0 and A260/230 ratio >1.7 are generally indicative of very pure DNA, free of residual salts, proteins or solvents, and acceptable for most downstream molecular applications. The 1% agarose gel in Figure 6B shows that the resulting gDNA is of high molecular weight (> 24 kb), with minimal shearing. The average nucleic acid yield from the full set of 45 positive samples is 5.26 ± 0.46 μg human DNA per 200 μl whole blood, based on the Life Technologies Quantifiler Human DNA Quantification Kit. Cross-contamination studies similar to those shown in Figure 5B were performed with negative control buffer blanks interspersed with the positive samples and extracted in parallel; all blanks were again negative by PCR (not shown). The purified gDNA is also suitable for numerous other PCR-based analyses (not shown).
Real-time results from eight replicate samples of a pooled maternal plasma sample processed with the large-volume TruTip procedure are shown in Figure 7. The full protocol (including off-line proteinase K incubation) is finished in approximately 2.5 hr, similar to the Qiagen manual Circulating Nucleic Acids Kit (no comparable automated extraction kits are yet available). The average Ct values over all replicates are 34.58 ± 0.66 and 29.76 ± 0.50 for fetal male (CHY) and total (CH1) DNA, respectively, which demonstrates excellent repeatability of the automated extraction method. The concentration of fetal DNA within the total DNA pool (in genome equivalents), is calculated based on fit point analysis comparison to standards, with the resulting average % fetal DNA across all samples of 2.8%. The actual % fetal DNA for this sample is unknown because samples were pooled before performing the extraction. Fetal DNA composition in non-pooled plasma samples extracted using the TruTip method are typically 1.5-fold higher compared to results with a Qiagen Circulating Nucleic Acids Kit (not shown).
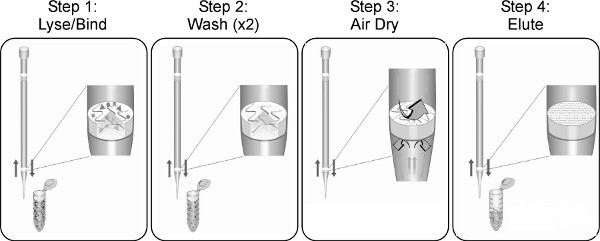
Figure 1. The TruTip extraction process and work flow, regardless of the liquid handling system. Other sample preparation or liquid handling steps can be incorporated into automated routines depending on the capabilities of the specific liquid handling instrument and software.
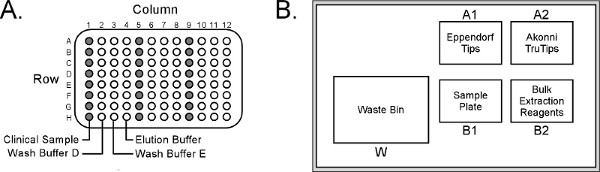
Figure 2. (A) Eppendorf epMotion 5070 sample plate layout. (B) Arrangement of reagents/consumables on the Worktable. The sample plate can be configured for up to 24 samples (columns 1, 5 and 9, respectively), although the epMotion will only process a maximum of 8 samples simultaneously. Click here to view larger figure.
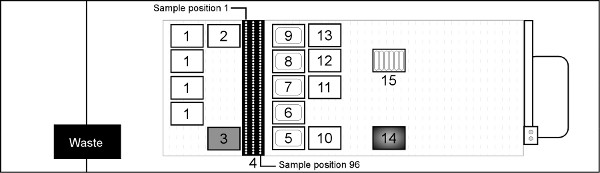
Figure 3. Hamilton STAR deck layout for purifying genomic DNA from whole blood (not to scale). Deck Position 1 = Hamilton 1 ml filtered tips; 2 = Hamilton 1 ml non-filtered tips; 3 = Akonni/Hamilton 1 ml LPT 2 mm TruTips; 4 = input blood sample carriers (blood collection tubes or microcentrifuge tubes); 5-9 = 290 ml reagent troughs for Lysis Buffer F, Ethanol, Wash Buffer J, Wash Buffer K and Elution Buffer A2, respectively; 10 = 96 deep well Binding plate; 11 = 96 deep well Wash J; 12 = 96 deep well Wash K; 13 = 96 deep well Elution plate; 14 = Hamilton HHS2 heater/shaker with Nunc 96 deep well Incubation plate; 15 = 50 ml reagent trough containing proteinase K. Click here to view larger figure.
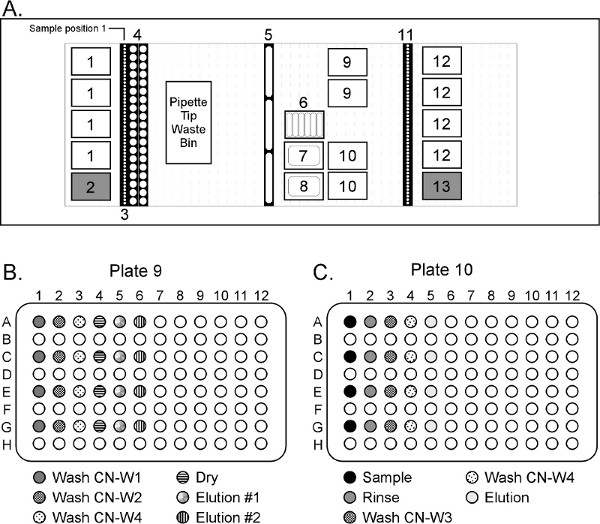
Figure 4. Hamilton STARplus deck layout for purifying DNA from large volume plasma samples (not to scale). The system is equipped with 8 x 5 ml channels and 8 x 1 ml channels (not shown in the deck layout). Deck Position 1 = Hamilton 4 ml filtered tips; 2 = Akonni/Hamilton 5 ml TruTips; 3 = source plasma samples; 4 = 50 ml conical tubes; 5 = 120 ml reagent troughs containing CN-W1, CN-W2 and CN-W4 reagents; 6 = low-volume reagent troughs containing proteinase K, CN-B2, CN-B3, EBA2, EBB and CN-W3 reagents; 7 = 290 ml reagent trough containing CN-L1 reagent; 8 = 290 ml reagent trough containing CN-B1 reagent; 9 = 96 deep well plates for Step 1; 10 = 96 deep well plates for Step 2; 11 = sample carriers for purified, final product; 12 = Hamilton 1 ml unfiltered tips; 13 = Akonni/Hamilton 1 ml LPT 4 mm TruTips. Click here to view larger figure.
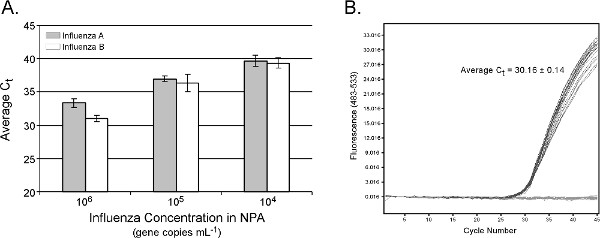
Figure 5. (A). Real-time PCR results from automated TruTip extraction of influenza virus added into nasopharyngeal aspirate (NPA). Input NPA volume = 100 μl, elution volume = 50 μl. Results are the average of 3 replicate extractions from 5 distinct NPA backgrounds (n = 15) per dilution level and influenza target. qPCR was performed on the LightCycler 480 system with assay conditions described previously 15. (B) No cross-contamination is detected when 12 positive NPA samples are interspersed with no template controls and subject to the automated extraction procedure. Click here to view larger figure.
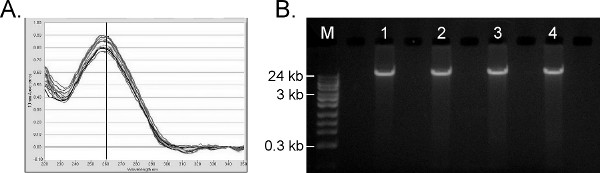
Figure 6. Results of human genomic DNA extraction from whole blood. A) UV-Visible traces from a NanoDrop 1000 (ThermoFisher) from 10 randomly selected replicates. B) 1% agarose gel of TruTip purified gDNA. M = Fisher 24 kb Max DNA Ladder. Lanes 1- 4 = ~100 ng purified gDNA from four randomly-selected replicates. Click here to view larger figure.
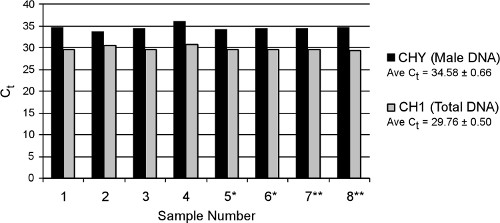
Figure 7. Real-time PCR results from eight replicate TruTip extractions of freely circulating DNA from plasma. Samples denoted by asterisks (* or **) were extracted on separate days. CHY quantifies male fetal DNA and CH1 quantifies total DNA present (fetal and maternal). qPCR was performed on a LightCycler 480 system (Roche) with previously published assays targeting CHY and CH1 18.
Discussion
The simplicity of the TruTip concept and workflow (Figure 1) make it readily adapted, automated, efficient and effective for a number of clinical sample matrices, input sample volumes, and liquid handling systems. It should be recognized, however, that every clinical sample is unique, and will vary one to the next in viscosity, particulates, mucus, surface contaminants, microbial, and/or human genetic backgrounds. Given expected variations in clinical sample composition and intended uses of an automated TruTip sample preparation protocol, it may therefore be necessary to modify certain steps in a TruTip procedure in order to achieve desired results for a specific sample type. Regardless of the sample type, however, TruTip parameters that typically have the most significant impact on nucleic acid purity and/or recovery include:
- Sample mixing and homogenization with lysis buffer (and alcohol). While TruTips have a relatively large pore size, sample homogenization and liquefaction is very important for efficient cell lysis, and subsequent binding steps to the TruTip monolith. With a homogenous and well-liquefied lysate, samples can move through the TruTip with higher flow rates, which reduces the overall sample processing time. The large volume plasma protocol demonstrates the potential for users to thoroughly homogenize and liquefy difficult samples (on-line or off-line), despite the large input sample volume.
- Flow rates. Slower flow rates during nucleic acid binding or elution typically result in higher nucleic acid yields, albeit at the expense of total processing time. Slower flow rates may also reduce the extent of DNA shearing.
- Cycle numbers. The optimum number of aspiration and dispense cycles is dependent upon sample type, total sample volume, and flow rates. Step 1 in Figure 1 is typically the point at which cycle numbers (and flow rates) may require some empirical optimization, with samples such as nasopharyngeal aspirate (Figure 5) representing one of the more challenging lysates to optimize due to the range of NPA viscosities from different patients.
- Drying. Complete drying of the TruTip monolith is imperative to prevent residual organic solvents from co-eluting with the purified nucleic acid sample and inhibiting downstream processes or tests. Because TruTip is not dried via centrifugation or vacuum filtration, it is important to maximize both the flow rate and cycle numbers during the drying step. Sometimes there is a residual droplet of wash solution on the terminus of TruTip after the drying cycles are completed. The Hamilton robot has the ability to perform a “tip touch” on the side of the well to release the droplet, thereby ensuring a solvent-free elution. The epMotion system does not have this feature, but a pre-rinse of the TruTip terminus in elution buffer can be programmed to achieve the same effect.
Because the geometry, pipette tip material, and attachment method to the robotic channel arms are unique for each instrument manufacturer, a different TruTip construct is required for each liquid handling system. The TruTip monolith dimensions (diameter, thickness, and pore size) do correlate with nucleic acid binding capacity (and elution efficiencies), as is expected for any solid-phase extraction technique. While thick (> 4 mm) matrices may be embedded into a 1 ml TruTip to increase nucleic acid binding capacity for large-volume samples and/or equalize the binding capacity across specific TruTip formats, there is a tradeoff between TruTip thickness and flow rates during the initial binding step (in the presence of crude lysates). Thus, it is sometimes advantageous to embed larger-diameter monoliths into larger-volume pipette tips for the initial steps of an automated protocol, (e.g. the 5 ml Hamilton/Akonni TruTips for large-volume extractions). Given the specific TruTip configurations dictated by the manufacturers of liquid handling robots, however, we do not necessarily expect TruTip nucleic acid yields to be identical across liquid handling platforms from different manufacturers, or across different TruTip sizes.
Clinical samples (by definition) will contain significant quantities of human genomic DNA unless they are acquired from normally sterile sites (e.g. cerebral spinal fluid). Sometimes the human genomic DNA is desired (as in Figure 6), whereas in other applications the human DNA represents an unwanted genomic background (as for Figure 5). The presence of background DNA is usually not problematic as long as the total amount of nucleic acid in the sample does not exceed the binding capacity of the monolith, and the background DNA can serve as a carrier if the desired target nucleic acid is present in trace amounts. The objective of the high-volume plasma extraction protocol (Figure 7) is to isolate (fragmented) fetal DNA in the presence of a 10-20 fold excess of maternal DNA, which is similar to the sample preparation objective of infectious disease tests except that the sequences are highly congruent and can only be distinguished by highly specific molecular testing and/or size discrimination. In this case, total circulating DNA is isolated using a 5 ml TruTip, and subsequent high-molecular and low-molecular weight fetal DNA are separated through subsequent binding and elution to a 1 ml TruTip by altering the binding buffer conditions. Selective size separation and enrichment of target nucleic acids based on their binding and elution properties to a silica monolith is a significantly different mode of action than achieved by membranes or size exclusion spin columns. Size separation and enrichment of microbial DNA from human genomic DNA may be accomplished in future applications via customizing TruTip binding and elution buffers.
The automated protocols demonstrated here emphasize the utility of the TruTip monolith itself for processing diverse clinical samples, and how it can be adapted for large volumes and specific liquid handling robots. The streamlined methods typically result in faster extraction protocols compared to other automated systems. The simplicity of the TruTip technology, however, also affords some cost advantages for those interested in purchasing a new, automated nucleic acid purification system, because the primary hardware required for automating TruTip procedures is the pipette channel arm itself rather than magnetic rods, vacuum systems, or on-board centrifuges. Utilizing pre-filled reagent plates can also reduce space and required consumables, and double the throughput per run. Minimizing deck space with TruTip protocols also enables advanced users to integrate upstream or downstream automated processes with the TruTip. For example Hamilton’s easyBlood solution to fractionate whole blood can be incorporated with the automated TruTip extraction method, which would significantly streamline bio-banking processes. Post-extraction processes such as nucleic acid quantitation, normalization, PCR set-up, or DNA sequencing are also readily integrated with TruTip on the larger liquid handling platforms.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
Portions of this work were supported by the National Institutes of Health (NIH) under grant R 44 AI072784. We thank Dr. Kirsten St. George, Sara B. Griesemer, Daryl Lamson and Amy Dean of the Laboratory of Viral Diseases, Wadsworth Center, New York State Dept. of Health for quantified influenza virions and access to clinically-validated influenza real-time PCR assays.
Materials
| Reagent/Material | |||
| TruTip Influenza Extraction Kit (EPM TruTips) | Akonni Biosystems, Inc. | 300-11120 | |
| 95% Ethanol | Acros Organics/ThermoFisher Scientific | AC615110040 | |
| 99% Acetone | Sigma-Aldrich | 270725-4L | |
| DEPC-treated water | Life Technologies | AM9906 | |
| Reagent Reservoir, 30 ml | Eppendorf | 960050100 | |
| Deep well plate 96/2,000 μl | USA Scientific | 30502302 | |
| epT.I.P.S. Motion Filtertips, 1,000 μl | Eppendorf | 960050100 | |
| Equipment | |||
| epMotion 5070 System | Eppendorf | 5070 000.000 | |
| Dispensing tool TM1000-8 | 960001061 | ||
| Reservoir rack | 960002148 | ||
| Table 1. Reagents and equipment for automated RNA extraction from NPA. | |||
| Reagent/Material | |||
| TruTip gDNA Blood Extraction Kit (Hamilton TruTips) | Akonni Biosystems, Inc. | 300-20341 | |
| 95% ethanol | Acros Organics/ThermoFisher Scientific | AC615110040 | |
| Proteinase K | AMRESCO LLC | E195 | |
| 1 ml Hamilton filtered CO-RE 96 tip rack | Hamilton Robotics, Inc. | 235905 | |
| 1 ml Hamilton non-filtered CO-RE 96 tip rack | Hamilton Robotics, Inc. | 235904 | |
| 50 ml Reagent Trough | Hamilton Robotics, Inc. | 187297 | |
| Deep Well 2 ml plate | USA Scientific | 1896-2800 | |
| Nunc 96 DWP-2 ml | Thermofisher | 27874 | |
| Reagent Trough | Fisher | 14-222-412 | |
| Equipment | |||
| Hamilton STAR System | Hamilton Robotics, Inc. | 173027 | |
| 1 ml Independent Pipette Channels / Modular Arm | Hamilton Robotics, Inc. | 173081/173050 | |
| 1 ml 96-channel head | Hamilton Robotics, Inc. | 199090 | |
| Tip Carriers | Hamilton Robotics, Inc. | 182085 | |
| Sample Carriers/Inserts | Hamilton Robotics, Inc. | 173400/182238 | |
| Plate Carriers | Hamilton Robotics, Inc. | 182090 | |
| Multiflex Carrier | Hamilton Robotics, Inc. | 188039 | |
| HHS2 Heater Shaker Unit | Hamilton Robotics, Inc. | 199033 | |
| Rack Carrier | Hamilton Robotics, Inc. | 188047 | |
| Table 2. Regents and equipment for 96-well genomic DNA extraction from whole blood. | |||
| Reagent/Material | |||
| TruTip R+D Circulating DNA Extraction Kit (Hamilton TruTips) | Akonni Biosystems, Inc. | Call to inquire | |
| 100% ethanol | Sigma-Aldrich | 459828-1L | |
| Isopropanol | Acros Organics/ThermoFisher Scientific | AC327270010 | |
| Proteinase K | AMRESCO LLC | E195 | |
| Filtered 4 ml Tips | Hamilton Robotics, Inc. | 184022 | |
| Unfiltered 1 ml Tips | Hamilton Robotics, Inc. | 235939 | |
| 96-Deep Well Plates | USA Scientific | 1896-2800 | |
| 50 ml Conical Tubes | Corning/ThermoFisher Scientific | 05-526B | |
| 50 ml Reagent Troughs | Hamilton Robotics, Inc. | 187297 | |
| 120 ml Reagent Troughs | Hamilton Robotics, Inc. | 182703 | |
| Large Volume 96-Pos Reagent Troughs | ThermoFisher Scientific | 14-222-412 | |
| Equipment | |||
| STARplus Autoload Workstation Base / Deck Module | Hamilton Robotics, Inc. | 173025/190012 | |
| 1 ml Independent Pipette Channels / Arm | Hamilton Robotics, Inc. | 173081/173052 | |
| 5 ml Independent Channel / Modular Arm | Hamilton Robotics, Inc. | 184090/173050 | |
| Plate Carriers | Hamilton Robotics, Inc. | 182090 | |
| Multiflex Carrier | Hamilton Robotics, Inc. | 188039 | |
| Rack Carrier for 50 ml Reagent Troughs | Hamilton Robotics, Inc. | 188047 | |
| 120 ml Reagent trough carrier | Hamilton Robotics, Inc. | 185290 | |
| Tip Carriers | Hamilton Robotics, Inc. | 182085 | |
| 50 ml Tube Carriers | Hamilton Robotics, Inc. | 182245 | |
| 24 Position Sample Carriers | Hamilton Robotics, Inc. | 173400 | |
| 32 Position Sample Carrier | Hamilton Robotics, Inc. | 173410 | |
| Table 3. Reagents and equipment for large volume DNA extraction from plasma. |
Riferimenti
- Boom, R., Sol, C. J. A., et al. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28, 495-503 (1990).
- Baker, M. P., Mitchell, A., et al. Isolation of genomic DNA from blood using a novel filter-based DNA purification technology. BioTechniques. 31, 142-145 (2001).
- Sinclair, B. To bead or not to bead: applications of magnetic bead technology. The Scientist. 12, 17-24 (1998).
- Dederich, D. A., Okwuonu, G., et al. Glass bead purification of plasmid template DNA for high throughput sequencing of mammalian genomes. Nucl. Acids Res. 30, e32 (2002).
- Levison, P. R., Badger, S. E., et al. Recent developments of magnetic beads for use in nucleic acid purification. J. Chromatogr. A. 816, 107-111 (1998).
- Hourfar, M. K., Schmidt, M., Seifried, E., Roth, W. K. Evaluation of an automated high-volume extraction method for viral nucleic acids in comparison to a manual procedure with preceding enrichment. Vox Sang. 89, 71-76 (2005).
- Perelle, S., Cavellini, L., et al. Use of a robotic RNA purification protocol based on the NucliSens easyMAG for real-time RT-PCR detection of hepatitis A virus in bottled water. J. Virol. Methods. 157, 80-83 (2009).
- Riemann, K., Adamzik, M., et al. Comparison of manual and automated nucleic acid extraction from whole-blood samples. J. Clin. Lab. Anal. 21, 244-248 (2007).
- Hukari, K. W., Shultz, M., Isely, N., Milson, R., West, J. A. A completely automated sample preparation instrument and consumable device for isolation and purification of nucleic acids. J. Lab. Autom. 16, 355-365 (2011).
- Kessler, H., Mühlbauer, G. Fully automated nucleic acid extraction. MagNA Pure LC. Clin. Chem. 47, 1124-1126 (2001).
- Miller, S., Seet, H., Khan, Y., Wright, C., Nadarajah, R. Comparison of QIAGEN automated nucleic acid extraction methods for CMV quantitative PCR testing. Am. J. Clin. Pathol. 133, 558-563 (2010).
- Dundas, N., Leos, N. K., Mitui, M., Revell, P., Rogers, B. B. Comparison of automated nucleic acid extraction methods with manual extraction. J. Mol. Diagn. 10, 311-316 (2008).
- Kruhøffer, M., Voss, T., et al. Evaluation of the QIAsymphony SP workstation for magnetic particle-based nucleic acid purification from different sample types for demanding downstream applications. J. Lab. Autom. 15, 41-51 (2010).
- Belgrader, P. Apparatus, system, and method for purifying nucleic acids. United States Patent. , (2010).
- Chandler, D. P., Griesemer, S. B., et al. Rapid, simple influenza RNA extraction from nasopharyngeal samples. J. Virol. Methods. 183, 8-13 (2012).
- Chan, K. H., Lai, S. T., et al. Analytical sensitivity of rapid influenza antigen detection tests for swine-origin influenza virus (H1N1). J. Clin. Virol. 45, 205-207 (2009).
- Chan, K. H., Lama, S. Y., et al. Comparative analytical sensitivities of six rapid influenza A antigen detection test kits for detection of influenza A. subtypes H1N1, H3N2 and. 38, 169-171 (2007).
- Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L., Quake, S. R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA. 105, 16266-16271 (2008).

