Generering CRISPR / Cas9 Mediated monoallelisk Sletninger til Studieinformationen Enhancer funktion i mus embryonale stamceller
Summary
Experimental validation of enhancer activity is best approached by loss-of-function analysis. Presented here is an efficient protocol that uses CRISPR/Cas9 mediated deletion to study allele-specific regulation of gene transcription in F1 ES cells which contain a hybrid genome (Mus musculus129 x Mus castaneus).
Abstract
Enhancers control cell identity by regulating tissue-specific gene expression in a position and orientation independent manner. These enhancers are often located distally from the regulated gene in intergenic regions or even within the body of another gene. The position independent nature of enhancer activity makes it difficult to match enhancers with the genes they regulate. Deletion of an enhancer region provides direct evidence for enhancer activity and is the gold standard to reveal an enhancer’s role in endogenous gene transcription. Conventional homologous recombination based deletion methods have been surpassed by recent advances in genome editing technology which enable rapid and precisely located changes to the genomes of numerous model organisms. CRISPR/Cas9 mediated genome editing can be used to manipulate the genome in many cell types and organisms rapidly and cost effectively, due to the ease with which Cas9 can be targeted to the genome by a guide RNA from a bespoke expression plasmid. Homozygous deletion of essential gene regulatory elements might lead to lethality or alter cellular phenotype whereas monoallelic deletion of transcriptional enhancers allows for the study of cis-regulation of gene expression without this confounding issue. Presented here is a protocol for CRISPR/Cas9 mediated deletion in F1 mouse embryonic stem (ES) cells (Mus musculus129 x Mus castaneus). Monoallelic deletion, screening and expression analysis is facilitated by single nucleotide polymorphisms (SNP) between the two alleles which occur on average every 125 bp in these cells.
Introduction
Transkriptionelle regulatoriske elementer er afgørende for spatio-temporale finjustering af genekspression under udvikling 1 og modifikation af disse elementer kan resultere i sygdom på grund af afvigende genekspression 2. Mange sygdom-associerede områder identificeret ved genom bred forening undersøgelser er i ikke-kodende områder og har træk af transkriptionelle forstærkere 3-4. Identificering enhancere og matche dem med de gener, de regulerer er kompliceret, da de ofte er placeret adskillige kilobaser væk fra generne, de regulerer og kan aktiveres i en vævsspecifik måde 5-6. Enhancer forudsigelser er almindeligt baseret på histon modifikation mærker, mediator-cohesin komplekser og binding af celletype-specifikke transkriptionsfaktorer 7-10. Validering af forudsagte enhancere er oftest sker gennem en vektor baseret assay, ved hvilken enhancer aktiverer ekspression af et reportergen 11-12. Disse data giver valuable information om de lovgivningsmæssige potentiale formodede forstærkersekvenser men afslører ikke deres opgaver i deres endogene genomiske kontekst eller identificere de gener, de regulerer. Genom redigering tjener som et effektivt redskab til at studere funktionen af transkriptionelle regulatoriske elementer i deres endogene kontekst ved tab af funktion analyse.
Nylige fremskridt i genom redigering, nemlig CRISPR / Cas9 genom redigering system, lette efterforskningen af genom-funktion. Den CRISPR / Cas9 systemet er let at bruge og kan tilpasses til mange biologiske systemer. Den Cas9 protein er målrettet til et specifikt sted i genomet af en vejledning RNA (gRNA) 13. Den SpCas9 / gRNA komplekset scanner genomet for sit mål genomiske sekvens, som skal være 5 'til en protospacer tilstødende motiv (PAM) sekvens, NGG 14-15. Baseparring af gRNA til dens mål, en 20 nukleotid (nt) sekvens komplementær til gRNA, aktiverer SpCas9 nukleaseaktivitet resulterer i en dobbele streng brud (DSB) 3 bp opstrøms for PAM-sekvensen. Specificitet opnås gennem fuldstændig baseparring i gRNA frø region, 6-12 nt siden af PAM; omvendt ikke stemmer overens 5 'af frø sædvanligvis tolereres 16-17. Den indførte DSB kan repareres enten af ikke-homolog ende sammenføjning (NHEJ) DNA-reparation eller homologi rettet reparation (HDR) mechanisms.NHEJ DNA-reparation skaber ofte insertion / deletion (indels) af et par bp på målstedet, der kan forstyrre den åbne læseramme (ORF) af et gen. For at generere større deletioner i genomet to gRNAs, som flankerer regionen af interesse, kan anvendes 18-19. Denne fremgangsmåde er særligt anvendelig for studiet af transkriptionelle enhancere grupperet i locus kontrol regioner eller super-enhancere, der er større end konventionelle forstærkere 9,18,20-22.
Monoallelisk sletninger er en værdifuld model til at studere cis -forordning af transskription. Den observerede Change i udskrift niveau efter monoallelisk sletning af en forstærker korrelerer til den rolle, som forstærker i genregulering uden de forstyrrende effekter, der kan opstå, når transkription af begge alleler påvirkes potentielt påvirke cellulære fitness. Evaluering reduceret ekspression er vanskeligt dog uden evnen til at skelne mellem den udgår af vildtype allelen. Endvidere genotypebestemmelse deletioner på hver allel uden evnen til at skelne mellem de to alleler er udfordrende, især for store deletioner af> 10 kb til 1 Mb 23, i hvilken det er vanskeligt at amplificere hele vildtype region ved PCR. Anvendelsen af F1 ES celler frembragt ved krydsning Mus musculus 129 med Mus castaneus tillader de to alleler, der skal differentieres ved allel-specifik PCR 18,24. Den hybride genomet i disse celler letter allel-specifik deletion screening og ekspressionsanalyse. I gennemsnit er der en SNP hver 125 bp mellem disse to genomer, Hvilket giver fleksibilitet i primer design for ekspression og genotypebestemmelse analyser. Tilstedeværelsen af én SNP kan påvirke primer smeltetemperatur (T m) og målrette specificitet i real-time kvantitativ PCR (qPCR) amplifikation tillader diskrimination af de to alleler 25. Endvidere et mismatch i 3'-enden af primeren i høj grad påvirker evnen af DNA-polymerase til at forlænge fra primer forhindrer amplifikation af det uønskede allel target 26. Beskrevet i det følgende protokol er anvendelsen af F1 ES celler til allelspecifikke enhancer deletioner af mere end 1 kb og efterfølgende ekspression analyse under anvendelse CRISPR / Cas9 genom redigering system (figur 1).
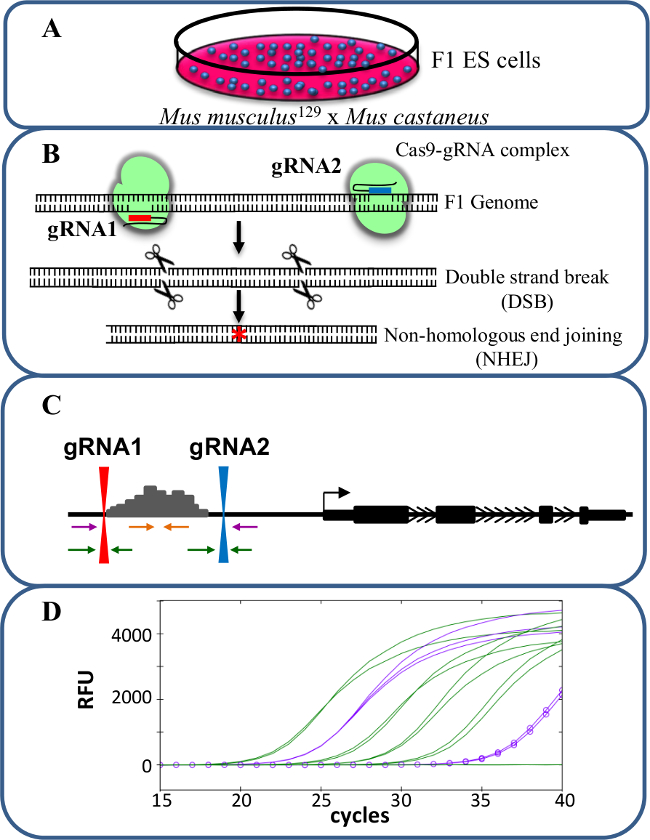
Figur 1. Enhancer sletning brug CRISPR / Cas9 at studere cis -regulation af genekspression. (A) F1 ES celler frembragt af en krydsning mellem Mus musculus 129 og Mus castaneus anvendes til at give mulighed for allel specifik deletion. (B) To guide RNA'er (gRNA) anvendes til at inducere et stort Cas9-medieret deletion af enhancerregionen. (C) Primersæt bruges til at identificere store mono- og bi-allele deletioner. De orange primere er de indvendige primere, de lilla primere er de udvendige primere og de grønne primere de gRNA flankerende primere. (D) Ændringer i genekspression overvåges ved hjælp af allel-specifik qPCR. RFU betegner relative fluorescensenheder. Klik her for at se en større version af dette tal.
Protocol
Representative Results
Discussion
CRISPR / Cas9 medieret genom redigering teknologi giver en enkel, hurtig og billig metode til genom modifikation. Metoden beskrevet her for at generere og analysere monoallelisk forstærker sletning til funktionel forstærker karakterisering udnytter SNPs i F1 museceller. Fordelene ved denne type fremgangsmåde er: 1) monoallelisk enhancer deletioner ikke producerer forstyrrende effekter, som opstår, når et kritisk enhancer udgår fra begge alleler, dvs. en stor reduktion i protein-niveauer i det regulerede g…
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We would like to thank all the members of the Mitchell lab for helpful discussions. This work was supported by the Canadian Institutes of Health Research, the Canada Foundation for Innovation and the Ontario Ministry of Research and Innovation (operating and infrastructure grants held by JAM).
Materials
| Phusion High-Fidelity DNA Polymerase | NEB | M0530S | high fidelity DNA polymerase used in gRNA assembly |
| Gibson Assembly Master Mix | NEB | E2611L | |
| gRNA_Cloning Vector | Addgene | 41824 | A target sequence is cloned into this vector to create the gRNA plasmid |
| pCas9_GFP | Addgene | 44719 | Codon-optimized SpCas9 and EGFP co-expression plasmid |
| AflII | NEB | R0520S | |
| EcoRI | NEB | R3101S | |
| Neon Transfection System 100 µL Kit | Life Technologies | MPK10096 | Microporator transfection technology |
| prepGEM | ZyGEM | PT10500 | genomic DNA extraction reagent |
| Nucleo Spin Gel & PCR Clean-up | Macherey-Nagel | 740609.5 | |
| High-Speed Plasmid Mini Kit | Geneaid | PD300 | |
| Maxi Plasmid Kit Endotoxin Free | Geneaid | PME25 | |
| SYBR select mix for CFX | Life Technologies | 4472942 | qPCR reagent |
| iScript cDNA synthesis kit | Bio-rad | 170-8891 | Reverse transcription reagent |
| 0.25% Trypsin with EDTA | Life Technologies | 25200072 | |
| PBS without Ca/Mg2+ | Sigma | D8537 | |
| 0.5M EDTA | Bioshop | EDT111.500 | |
| HBSS | Life Technologies | 14175095 | |
| 1M HEPES | Life Technologies | 13630080 | |
| BSA fraction V (7.5%) | Life Technologies | 15260037 | |
| Max Efficiency DH5α competent cells | Invitrogen | 18258012 | |
| FBS | ES cell qualified | FBS is subjected to a prior testing in mouse ES cells for pluripotency | |
| DMSO | Sigma | D2650 | |
| Glutamax | Invitrogen | 35050 | |
| DMEM | Life Technologies | 11960069 | |
| Pencillin/Streptomycin | Invitrogen | 15140 | |
| Sodium pyruvate | Invitrogen | 11360 | |
| Non-essential aminoacid | Invitrogen | 11140 | |
| β-mercaptoethanol | Sigma | M7522 | |
| 96-well plate | Sarstedt | 83.3924 | |
| Sealing tape | Sarstedt | 95.1994 | |
| CoolCell LX | Biocision | BCS-405 | alcohol-free cell freezing container |
| CHIR99021 | Biovision | 1748-5 | Inhibitor for F1 ES cell culture |
| PD0325901 | Invivogen | inh-pd32 | Inhibitor for F1 ES cell culture |
| LIF | Chemicon | ESG1107 | Inhibitor for F1 ES cell culture |
Riferimenti
- Sagai, T., Hosoya, M., Mizushina, Y., Tamura, M., Shiroishi, T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 132 (4), 797-803 (2005).
- Kleinjan, D. A., Lettice, L. A. Long-range gene control and genetic disease. Adv Genet. 61, 339-388 (2008).
- Visel, A., Rubin, E. M., Pennacchio, L. A. Genomic views of distant-acting enhancers. Nature. 461 (7261), 199-205 (2009).
- Maurano, M. T., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 337 (6099), 1190-1195 (2012).
- Heintzman, N. D., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 459 (7243), 108-112 (2009).
- Shen, Y., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 488 (7409), 116-120 (2012).
- Johnson, D. S., Mortazavi, A., Myers, R. M., Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 316 (5830), 1497-1502 (2007).
- Rhee, H. S., Pugh, B. F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 147 (6), 1408-1419 (2011).
- Whyte, W. A., et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153 (2), 307-319 (2013).
- Chen, C. Y., Morris, Q., Mitchell, J. A. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics. 13 (1), 152 (2012).
- Patwardhan, R. P., et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 30 (3), 265-270 (2012).
- Melnikov, A., et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 30 (3), 271-277 (2012).
- Jinek, M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Cong, L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339 (6121), 819-823 (2013).
- Mali, P., et al. RNA-guided human genome engineering via Cas9. Science. 339 (6121), 823-826 (2013).
- Hsu, P. D., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 31 (9), 827-832 (2013).
- Cho, S. W., et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24 (1), 132-141 (2014).
- Zhou, H. Y., et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 28 (24), 2699-2711 (2014).
- Fujii, W., Kawasaki, K., Sugiura, K., Naito, K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 41 (20), e187 (2013).
- Tuan, D. Y., Solomon, W. B., London, I. M., Lee, D. P. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human ‘beta-like globin’ genes. Proc Natl Acad Sci U S A. 86 (8), 2554-2558 (1989).
- Amano, T., et al. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 16 (1), 47-57 (2009).
- Li, Y., et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 9 (12), e114485 (2014).
- Canver, M. C., et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 289 (31), 21312-21324 (2014).
- Mlynarczyk-Evans, S., et al. X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 4 (6), e159 (2006).
- Lefever, S., Pattyn, F., Hellemans, J., Vandesompele, J. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin Chem. 59 (10), 1470-1480 (2013).
- Huang, M. M., Arnheim, N., Goodman, M. F. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 20 (17), 4567-4573 (1992).
- Keane, T. M., et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 477 (7364), 289-294 (2011).
- Yalcin, B., et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 477 (7364), 326-329 (2011).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6 (5), 343-345 (2009).
- Gibson, D. G., Smith, H. O., Hutchison, C. A., Venter, J. C., Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 7 (11), 901-903 (2010).
- Ding, Q., et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 12 (4), 393-394 (2013).
- Basu, S., Campbell, H. M., Dittel, B. N., Ray, A. Purification of specific cell population by fluorescence activated cell sorting (FACS). J Vis Exp. (41), (2010).
- Forlenza, M., Kaiser, T., Savelkoul, H. F., Wiegertjes, G. F. The use of real-time quantitative PCR for the analysis of cytokine mRNA levels. Methods Mol Biol. 820, 7-23 (2012).
- Wu, J. H., Hong, P. Y., Liu, W. T. Quantitative effects of position and type of single mismatch on single base primer extension. J Microbiol Methods. 77 (3), 267-275 (2009).
- Sanyal, A., Lajoie, B. R., Jain, G., Dekker, J. The long-range interaction landscape of gene promoters. Nature. 489 (7414), 109-113 (2012).

