Designing, Packaging, and Delivery of High Titer CRISPR Retro and Lentiviruses via Stereotaxic Injection
Summary
The CRISPR/Cas9 system offers the potential to make targeted genome editing accessible and affordable to the scientific community. This protocol is intended to demonstrate how to create viruses that will knockout a gene of interest using the CRISPR/Cas9 system, and then inject them stereotaxically into the adult mouse brain.
Abstract
Replication defective lentiviruses or retroviruses are capable of stably integrating transgenes into the genome of an infected host cell. This technique has been widely used to encode fluorescent proteins, opto- or chemo-genetic controllers of cell activity, or heterologous expression of human genes in model organisms. These viruses have also successfully been used to deliver recombinases to relevant target sites in transgenic animals, or even deliver small hairpin or micro RNAs in order to manipulate gene expression. While these techniques have been fruitful, they rely on transgenic animals (recombinases) or frequently lack high efficacy and specificity (shRNA/miRNA). In contrast, the CRISPR/Cas system uses an exogenous Cas nuclease which targets specific sites in an organism’s genome via an exogenous guide RNA in order to induce double stranded breaks in DNA. These breaks are then repaired by non-homologous end joining (NHEJ), producing insertion and deletion (indel) mutations that can result in deleterious missense or nonsense mutations. This manuscript provides detailed methods for the design, production, injection, and validation of single lenti/retro virus particles that can stably transduce neurons to express a fluorescent reporter, Cas9, and sgRNAs to knockout genes in a model organism.
Introduction
To study the basis of normal physiology and disease pathology, there is a need to precisely manipulate gene expression in model organisms. For mammalian model organisms, this is largely centered on the creation and development of transgenic mice wherein a genetic element of interest is flanked by sites recognized by a recombinase. This can result in a site specific manipulation of these flanked genes. While this has been a successful strategy, it is time and resource intensive; for example, creating a triple transgenic animal that would express a floxed gene, Cre recombinase, and a Cre reporter gene requires multiple matings and validation. In contrast, the stereotaxic injection of replication defective viral particles encoding a fluorescent protein and the recombinase into a floxed gene animal does not require complex genotyping or breeding strategies1. Further, if a fluorescent protein and Cre expressing virus is co-injected with a second virus encoding a different fluorescent protein, then this provides a within-tissue control for the targeted genetic manipulation. While this strategy still requires the use of knock-in animals, virally mediated RNA based strategies circumvent the need for transgenic animals. For example, stereotaxic injection of replication-deficient viruses that encode a fluorescent protein and a short hairpin RNA (shRNA) can use the cell's endogenous RNAi machinery to result in a potent reduction of the transcript of a gene of interest. However, shRNA strategies produce subtle gene knock-downs often resulting in modest cellular phenotypes2. While a knock-down may be more physiologically relevant for heterozygous gene dysfunction, its decreased robustness compared to a knock-out is not ideal for phenotypic discovery of novel genes.
A third technique that has recently emerged, the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat)/Cas9 (CRISPR-associated protein 9) system, relies on the expression of both a small exogenous RNA and a DNA cutting enzyme. The CRISPR/Cas9 system was adapted from the prokaryotic immune system which evolved a method of identifying foreign, invading DNA from viruses and targeting it for degradation via the Cas9 enzyme3,4. This powerful genome editing technique can be used for creating targeted deletions, insertions, and mutations; and the following protocol will outline how to make deletions in a gene of interest in order to knockout its expression in vivo. The Cas9 enzyme must be expressed with a guide RNA homologous to the region of interest and contiguous with a scaffold RNA. Knockout of a gene using this technique requires targeting Cas9 to a specific region in the genome using synthetic guide RNAs (sgRNA), and inducing double stranded breaks (DSBs) at a site of interest. These DSBs are then repaired by the endogenous cell-repair machinery via non-homologous end-joining (NHEJ) which lead to indels that may produce missense or nonsense mutations and can therefore create a loss of functional protein expression5. Because this system produces genomic alterations, it only requires the transient expression of the Cas9 and sgRNA. However, it is desirable for a stable fluorescent indicator to identify cells and their progeny manipulated in this manner.
Lenti- and retroviruses have the advantage of stably integrating DNA of interest into host cells which maintain long-term expression and are passed down to daughter cells during mitosis. This protocol describes the design and production of two types of replication defective, high titer retroviruses: the human immunodeficiency virus derived lentiviral particles (lentivirus) and those based on murine Maloney Leukemia virus (retrovirus). While both of these viruses are capable of supporting stable expressing of large transgenes, the retroviral particles can only integrate into the genome during cell division with the degradation of the nuclear envelope, and therefore can be used as a tool to label and birth-date cells6. While lentiviruses have a reputation for being relatively low titer7, this methodology, including the use of caffeine8 during viral collection, routinely produces titers of 109 and 1010 particles/ml. Another advantage of lenti- and retroviruses is the tolerance for very large inserts. The following collection of protocols outlines the procedure for designing a lenti or retrovirus encoding a fluorescent reporter, sgRNAs, and Cas9 to utilize the CRISPR/Cas9 system to modify DNA as well as express a fluorescent protein.
Mouse stereotaxic neurosurgery is a valuable method for injecting viruses in vivo to study morphology, function, and connectivity of infected neurons. Viral infection in neurons can be used to manipulate expression levels over an extended period of time, such as throughout development, and expression can be precisely controlled by the use of various drug inducible systems and specific Cre driven expression. This particular protocol explains how to inject a virus expressing an sgRNA and Cas9 to knockout a gene of interest in the brain of an adult mouse. Mice recover very quickly from this procedure and expression of the viral transgene can be seen within 48 hours post injection. However, fluorophore expression appears to increase over the course of weeks resulting in near maximal levels by 3 weeks post-infection. Mice that undergo viral stereotaxic injection can be used for behavior, electrophysiology, or morphological studies. Overall, the purpose of these procedures is to demonstrate how to knockout a gene in the adult mouse brain using stereotaxic surgery and a virus expressing a specific sgRNA and Cas9.
Protocol
Ethics Statement: All protocols were approved by the Dartmouth Institutional Biosafety and Institutional Animal Care and Use Committee review boards.
1. Protocol for Designing a Guide Strand (sgRNA) for CRISPR/Cas9 Retrovirus
NOTE: There are many non-profit websites that can be used to generate sgRNAs to target a gene of interest (https://benchling.com/ and http://crispr.mit.edu/). The goal of this protocol is to design and order single stranded oligos from a commercial vendor that are annealed to each other. This annealed oligo will be ligated into the PXL transfer vector. See supplemental video 1 for an example of designing sgRNAs using Benchling.
- Use a website dedicated to designing sgRNAs.
NOTE: For example, inputting a coding/exonic sequence from near the start of the gene of interest into the website will generate a 20 nucleotide sgRNA. Use the 20 nucleotide sgRNA sequence to design the sense and anti-sense oligos that will be ordered to create the sgRNA. - After generating the sgRNA, copy this sequence into a word processor. Add a G (guanine) to the beginning of the 20 nucleotide sgRNA sequence in the document if it does not already begin with one (i.e., G-20 nucleotide sgRNA sequence). This is necessary to ensure good transcription off the U6 promoter.
- Document the reverse complement of this now 20-21 nucleotide sequence. For the sense oligo, add "CACC" to the 5' end of the sequence in the document (i.e., CACC-G-20 nucleotide sgRNA sequence). This overhang will be used to ligate the sequence into the PXL vector.
- For the antisense oligo, add AAAC to the 5' end. Use this overhang to ligate the sequence into the PXL vector.
- Obtain the sense and antisense oligos. After receiving the oligos, make 100 μM stocks using DNAse free water. Mix together 10 µl each of the 100 μM sense and antisense oligos with 4 µl of 10x NEB Buffer 2, and 16 µl of water.
NOTE: They do not need page purification or 5'phosphorylation (as the BbsI overhangs are non-compatible cohesive ends).- Bring 200 ml of water to a boil in a 500 ml beaker, then float the tube containing this mixture in a foam "floater" holder. Allow the water to slowly cool from 95 °C for 2 hr at room temperature.
- Dilute the now annealed-oligo mix 1:1,000 in sterile water and use immediately in the following ligation or store the remaining reaction at -20 °C.
- Digest pXL, a PX330 vector derivative, with the BbsI restriction enzyme at 37 °C for 2 hr. Use a 40 µl reaction containing 2 µg of pXL, 4 µl of 10x NEB Buffer 2, 1 µl of Bbs1 and balance water. Subject the digested-pXL to routine gel purification using a commercial kit. Ensure that the yield is ~8.5 kB product.
- Using a commercial ligation kit, ligate 1 µl of the 1:1,000 annealed-oligos with 50 ng of digested and gel purified-pXL. Transform the ligation product into competent recombination deficient E. coli (NEB 5-alpha, subcloning efficiency). Screen the transformants for the presence of the correct guide by plasmid DNA sequencing.
- Digest the U6, guide strand, and RNA scaffold elements (sgRNA) out of pXL using BstB1 and Pac1 restriction enzymes and ligate into the fluorescent protein-T2A-Cas9 viral backbone digested with the same restriction enzymes. Transform the ligation product (NEB 5-alpha maximum efficiency). The fluorescent protein-T2A-Cas9 viral backbone plasmids are low copy plasmids and thus low copy protocols must be used.
NOTE: A second sgRNA can be similarly introduced by PacI digestion of another pXL guide strand plasmid and ligated into the Pac1 digested and calf-intestinal phosphatase treated viral backbone already containing the first guide strand. Phosphatase treatment of the viral transfer plasmid will help reduce the number of transformants from the self-ligation of plasmids not containing the second guide. - Sequence verify the final plasmid and maxi prep using the Nucleobond Xtra maxi prep kit and package it into a virus using the following "Protocol for Retro/Lentiviral Production — CaPO4 Method".
2. Prepare 293FT/293GP Cells for Transfection (Retro/Lentiviral Production — CaPO4 Method)
- (Day 1) Rapidly thaw 1 vial of cells per 10 cm cell culture plate in a 37 °C water bath. For lentiviral packaging, use 293FT cells. For retroviral packaging, use 293GP (gag/pol) cells.
- Pipette all of the thawed-cells from the cryo tube into a 15 ml conical tube and add 2 ml of pre-warmed CO2 equilibrated-complete Iscove's Modified Dulbecco's Medium.
- Centrifuge cells for 5 min at 500 x g to pellet. Aspirate the supernatant and resuspend the cell pellet in 10 ml of complete Iscove's Modified Dulbecco's Medium. Plate the cells onto a 10 cm cell culture dish. Incubate the cells overnight at 37 °C in a 5% carbon dioxide incubator.
- (Day 2) 24 hr after plating, change media on the plate by aspirating the existing media and adding 10 ml of pre-warmed Iscove's Modified Dulbecco's Medium to the plate.
- (Day 3-4) 24-48 hr after the media change and once the cells become confluent, split the cells to a confluency of 2.5-3.0 x 106 cells/plate (10 cm plate).
- To split the cells, aspirate the media and wash the plate with 5 ml of PBS. Add 1 ml of 0.25% trypsin to the plate and incubate at 37 °C until the cells lift off the plate. Add 0.5 ml of Iscove's Modified Dulbecco's Medium to neutralize the 0.25% trypsin reaction and pipette the cells into a 1.5 ml tube.
- Spin the cells at 500 x g for 5 min. Resuspend the cells in 1 ml of Iscove's Modified Dulbecco's Medium. Dilute 10 µl of cells in 90 µl of PBS. Count the cells using either a hemocytometer or automated cell counter. Re-plate 2.5-3.0 x 106 cells/plate with complete Iscove's Modified Dulbecco's Medium.
NOTE: Cells will be ~50% confluent 24-34 hr after plating. Transfect the cells when they are ~50% confluent.
3. (Day 5) CaPO4 Transfection and Viral Particle Collection
- Change the media 2 hr prior to transfection by aspirating the existing media and adding 10 ml of pre-warmed Iscove's Modified Dulbecco's Medium. Ensure that there is exactly 10 ml of media in the 10 cm plate.
- Prepare the transfection reagents for 2 dishes using 2, 5 ml round bottom polystyrene tubes. Label the first tube "DNA" and the second tube "2X HBS". Adjust the concentration of DNA to 1μg/μl in Tris-EDTA at pH 7.4.
- For lentiviruses, slowly add 20 μl of transfer vector (the viral construct of interest), 13 μl of CMVdelta8.9, 9 μl of VSV-g , 860 μl of molecular biology grade H2O, and 100 μl of 2.5 M CaCl2 to the first tube ("DNA") while continuously tapping on the tube to mix.
- For retroviruses, omit the CMVdelta8.9 plasmid. Add 20 µl Transfer Vector, 15 μl VSV-g, 860 μl Molecular Biology Grade H2O, and 100 μl 2.5 M CaCl2 to the tube labeled "DNA".
- Add 1 ml of 2x HEPES-buffered saline (pH 7.0; this pH is absolutely critical) to the tube labeled "2X HBS".
- Slowly add the 1 ml contents of the "DNA" tube to the "2X HBS" tube, one drop at a time. Continuously tap the "2X HBS" tube with the index or middle finger while adding the contents of the "DNA" tube. Observe apparent CaPO4 vesicles after each drop. Incubate the tube in the dark for 30 min at room temperature.
- Add 1 ml of the transfection in slow droplets to each 10 cm cell plate, then incubate overnight at 37 °C.
- (Day 6) Replace media with 8 ml of Iscove's Modified Dulbecco's Medium + 0.5% FBS and 40 mg Caffeine/100 ml media. If the transfer vector contains a fluorescent marker, then fluorophore expression indicates a successful transfection.
- (Day 7) Collect the media containing the viral particles by using a 10 ml serological pipette and dispense into a 50 ml conical tube. Store the 50 ml conical tube at 4 °C. Add 8 ml of 0.5% FBS media to each plate.
- (Day 8) Again, collect the media containing the viral particles and combine with the previous day's harvest in the 50 ml conical tube.
4. Concentration and Purification of the Virus
- Make a 5x polyethylene glycol 6000 solution by adding 40% polyethylene glycol 6000 and 1.5 M NaCl to ddH2O. Autoclave the solution on the liquid cycle for 45 min at 121 °C. Slowly mix the solution when cooling.
NOTE: The solution will start cloudy and then become clear as it cools. - Centrifuge the 50 ml conical tube containing both collections of viral supernatant at 2,000 x g for 10 min in order to pellet insoluble material. Purify the viral media by filtering through a 0.45 µm low protein binding syringe filter (PES or PVDF).
- Add 5x polyethylene glycol 6000 solution to media (The final concentration should be 8% polyethylene glycol 6000 and 0.3 M NaCl). Mix by inverting the tube several times (do not vortex). Incubate the viral containing polyethylene glycol solution at 4 °C for 12 or more hr, remixing occasionally.
- (Day 9) Centrifuge the viral containing polyethylene glycol solution at 2,500 x g for 45 min. Remove and discard the supernatant and spin again for 2 min. Again, remove and discard the supernatant.
- Resuspend the pellet by adding 320 µl of sterile phosphate-buffered saline (the 320 µl is 1/100th of the original volume of viral containing media collected) and incubate overnight at 4 °C. Optionally, resuspend the pellet at room temperature on a rocker for 30 min.
- After re-suspending the pellet, aliquot the virus (5-10 µl per 0.5 ml tube) and freeze aliquots at -80 °C (freeze thawing or storage at 4 °C will dramatically reduce the titer).
- Titer the viruses using standard protocols, i.e., perform a dilution series on a 6-well plate of HEK293T cells and manually count fluorescent colonies 48 hr later.
5. Testing Efficacy of the CRISPR Virus
NOTE: Sequence clones using the following steps to test for production of double-strand breaks repaired by NHEJ using the mouse Neuro2A cells. This has the advantage over surveyor assays in that it can be used to determine the percentage of cells that have been modified and the nature of the indels resulting from NHEJ.
- Coat a 3.5 cm cell culture dish with a gelatinous protein mixture such as matrigel diluted 1:50 in Iscove's Modified Dulbecco's Medium and incubate for 30 min at 37 °C. Aspirate the gelatinous protein mixture and plate the Neuro2A cells.
- After the Neuro2A cells reach 50% confluence, add the CRISPR Lentivirus or Retrovirus at a multiplicity of infection of 10 (i.e., 10 viral particles per cell).
NOTE: Neuro2A cells are not as amiable to infection as are HEK293 cells, thus, a single infection will not result in 100% of the cells being infected with the lentivirus. Further, the retrovirus only infects dividing cells and therefore will also not result in 100% infection. In order to achieve a near 100% rate of infection, addition of virus can be repeated on multiple days, splitting the cells as needed. Alternatively, fluorescent positive cells can be isolated using fluorescence-activated cell-sorting. The most effective way to achieve a 100% infection rate with a lentiviruses is to perform a single infection followed by FACS isolation. For a retrovirus, perform 4-rounds of infection 24 hr apart and follow by FACS isolation to ensure 100% infection. - After infecting the cells for 1 week, expand the cells to near confluence on a 10 cm plate, aspirate the media, and wash the plate with 5 ml of phosphate-buffered saline. Add 1 ml of 0.25% trypsin and incubate at 37 °C until the cells lift off the plate. Add 0.5 ml of Iscove's Modified Dulbecco's Medium to neutralize the 0.25% trypsin reaction and pipette the cells into a 1.5 ml tube and pellet the cells at 500 x g for 5 min.
- To isolate DNA, add 100 µl of 50 mM potassium hydroxide, re-suspend the cells, and incubate at 95 °C for 5 min. Neutralize the solution using 10 µl of 1 M Tris, pH = 8.0.
- PCR amplify the region flanking the genomic region targeted by the CRISPR sgRNA using ~300 ng of the DNA isolated in step 5.5 using a high fidelity PCR master mix4.
- Run the PCR reaction on a 2.5% agarose gel and gel purify the appropriately sized fragment using a gel purification kit such as a gel DNA recovery kit. Ligate into a PCR cloning vector such as pGem-t-easy and transform into competent cells9.
- 24 hr later, add 2 ml of LB broth into a 15 ml round-bottomed tubes as well as the appropriate antibiotic (ampicillin, neomycin, etc.). Inoculate the tube with an individual colony by using a sterile pipette tip or loop to select a single colony from the LB-plate. Expand and grow the colony by shaking the tube at 250 RPM and 37 °C for 24 hr. Repeat for as many colonies as desired.
- Using a mini-prep kit, isolate DNA from the E. coli and sequence with primers designed to amplify the area of the gene targeted by the sgRNA in order to analyze the Cas9 targeted region of the mouse genome.
6. Stereotaxic Injection Protocol for the Adult Mouse
- Prepare for Surgery
NOTE: It is important to maintain sterile conditions during survival surgeries. This is accomplished in this protocol by heat sterilizing the surgery tools, using betadine to sterilize the injection site, and adding antibiotic ointment to the incision site after it is closed. It is also important to use sterile gloves as well as a thoroughly sterilized, dedicated surgery area.- Heat sterilize surgery tools prior to use by either autoclaving or a using hot bead sterilizer. Prepare recovery chamber and surgery area by turning on heating pads to maintain body temperature during surgery and recovery. Assemble the drill used to create holes in the skull for injection.
- Load 4 µl of virus into injection needle by withdrawing the virus directly from the sterile PCR tube. Briefly remove the viral aliquot from ice. Maintain the virus in the syringe at room temperature for no more than 1 hr prior to injection.
7. Stereotaxic Injection
- Confirm that the ventilation to the surgery suite is open to ensure proper air flow. Prepare the mouse for surgery by anesthetizing with 4% isoflurane in an induction chamber. Following anesthesia, shave the head to prepare the injection site.
- Place the mouse in the stereotaxic instrument by using tweezers to move tongue down and to the side. Insert the bite bar into mouth until teeth drop into the slot, then secure the ear bars. Ensure that the body of the mouse is on the heating pad and the nose is situated in the nose cone. Direct 4% isoflurane into the nose cone. Confirm anesthesia by pinching the foot with tweezers and ensuring that there is no startle reflex.
- Apply artificial tears to lubricate the eyes. Finish preparing the injection site by swabbing the shaved head with alternating rounds of povidone-iodine and lidocaine.
- Using a scalpel, cut small incision along center of scalp. Dry the skull with a swab and hydrogen peroxide if necessary to help visualize bregma.
- Using a dissecting scope, locate bregma on the skull and place the drill tip on bregma. Zero (or record) the digital stereotactic coordinates on the x, y, and z planes.
- In order to ensure that the head is level on the rostral to caudal y-axis, place the drill bit on lambda and level the head so that the z-coordinate is roughly equal at both bregma and lambda.
- In order to ensure that the head is level on the x-axis, place the drill bit to y = 1/2 lambda. Ensure that the z-coordinate is equal at 1 mm on each side (x = +/-1 mm) of the sagittal suture. Adjust the skull if there is a difference in the z-coordinate at 1mm to the left and right of lambda.
- Place the drill over the desired coordinates. For the dentate gyrus, use the coordinates y = -1.9 from bregma and x = +/- 1.1. Before drilling, lower the isoflurane to 2%, then slowly and carefully, drill through the skull.
- After drilling all the holes, affix the filled-syringe onto the stereotaxic instrument. Visually center the syringe over the hole and zero the z coordinate at the skull.
- Slowly lower the syringe to deepest z-depth. For the dentate gyrus, the z depths are -2.5, -2.4, and -2.3. Begin injection at a rate of 0.25 µl/min using a stereotaxic injector.
- After the injection is complete at lowest Z-depth, wait 1 min, then raise to the next coordinate and begin injection again. Continue this pattern until all z-injection coordinates are injected. Wait 2 min before removing the syringe after the last injection.
NOTE: No adverse effects have been noted on the tissue by injecting up to 2 µl of virus per hemisphere in the mouse brain.
- After the injection is complete at lowest Z-depth, wait 1 min, then raise to the next coordinate and begin injection again. Continue this pattern until all z-injection coordinates are injected. Wait 2 min before removing the syringe after the last injection.
- Repeat injection for other bore holes.
- After injection, remove the mouse from the stereotaxic instrument and suture the scalp with 6-0 silk sutures. Apply Lidocaine and an anti-bacterial cream to the wound.
- Inject 0.8-1.0 ml saline + Ketoprofen (3-5 mg/kg) via I.P. route to manage pain. Then place mouse into heated recovery chamber. Do not leave the animal unattended until it has regained sufficient consciousness to maintain sternal recumbency. Do not return the animal to the company of other animals until it has fully recovered.
- In the days following the surgery, weigh the mouse daily and provide soft food and treats. Check wound and note the general condition/demeanor.
- Following stereotaxic injection, the mice can be euthanized for slice prep electrophysiology1 or perfused and their brains removed, sliced, and stained via immunohistochemistry for analysis10.
Representative Results
Following the "Protocol for Designing a Guide Strand (sgRNA) for CRISPR/Cas9 Retrovirus", oligos targeting a particular sequence are inserted into the PXL cloning vector downstream of the hU6 promoter and downstream of a guide RNA scaffold using the BbsI cloning sites (Figure 1, step 1). This sgRNA is then excised from PXL and inserted into the pRubi backbone using the BstBI and PacI sites (Figure 1, Step 2). Finally, another sgRNA cloned into PXL can be placed downstream of the first guide in pRubi-Guide1 (Figure 1, Step 3), targeting another area of the gene and increasing the chances of a knockout via NHEJ. Verification of the correct constructs should be determined by sequence analysis. Once this construct is made, it can be packaged into a virus following the "Protocol for Retro/Lentiviral Production- CaPO4 Method". Successful packaging is confirmed by infection of virus into HEK293 cells in order to titer the virus. If there is no fluorophore expression then there was likely an error during packaging of the virus.
Figure 2 is a representative result of 2 retroviruses, one expressing GFP and the other expressing mCherry, co-injected into the dentate gyrus of neonatal mouse (7 days-old) and imaged 21 days post-injection. Labeling of neurons with mCherry or GFP allows morphological assessment of various genetic manipulations in the same tissue, where one virus can express a CRISPR/Cas9 mediated KO and the other a control virus, expressing solely a fluorophore. Stereotaxic injection allows precise anatomical selectivity as demonstrated by the discrete infection of the intended coordinates, the dentate gyrus. When analyzing brain sections for infection, it is important to keep surrounding tissue until it is determined that the correct anatomical region was infected. If there is no sign of infection, then it is possible that the injection occurred in a neighboring region and can be identified in surrounding sections. It can also be helpful to locate the needle track to find the exact injection region. VSVg pseudotyped viruses rarely spread out of the margins of the dentate gyrus when injected in vivo, and tend to spread along the rostral/caudal axis infecting cells along the entire dentate gyrus, as analyzed by 3D reconstruction (Figure 3).
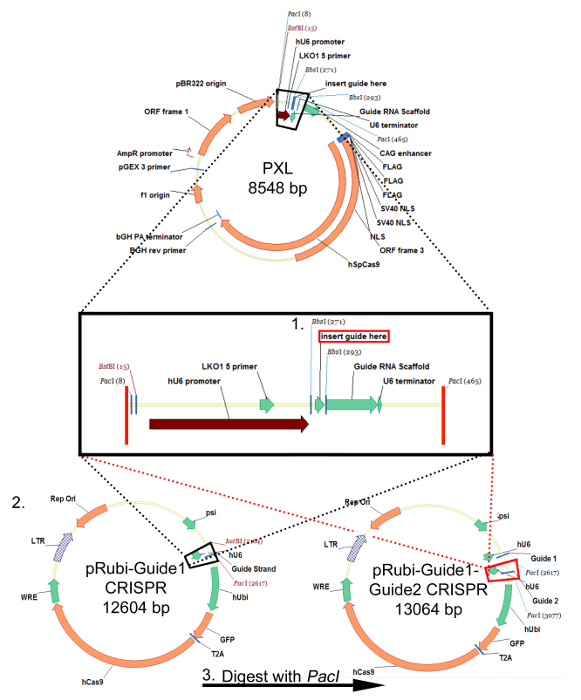
Figure 1: Cloning strategy for the retroviral pRubi-Guide1-Guide2-CRISPR plasmid. This strategy is identical for the FU-based lentiviral plasmids. sgRNA oligos are annealed and inserted into PXL using the BsbI cloning sites. After sequencing to ensure that the sgRNA is successfully inserted into PXL, digest the plasmid with BstBI and PacI. The insert that is dropped out (black box) is then cloned into the viral back bone (black dotted lines) pRubi-Guide1 CRISPR. A second sgRNA can also be inserted into PXL and digested out using the PacI enzyme. This is then cloned into the pRubi-Guide1 CRISPR vector (red dotted lines) using the PacI site. The resultant plasmid then contains both guide strands as well as the necessary viral elements, promoters, and fluorophores. Please click here to view a larger version of this figure.
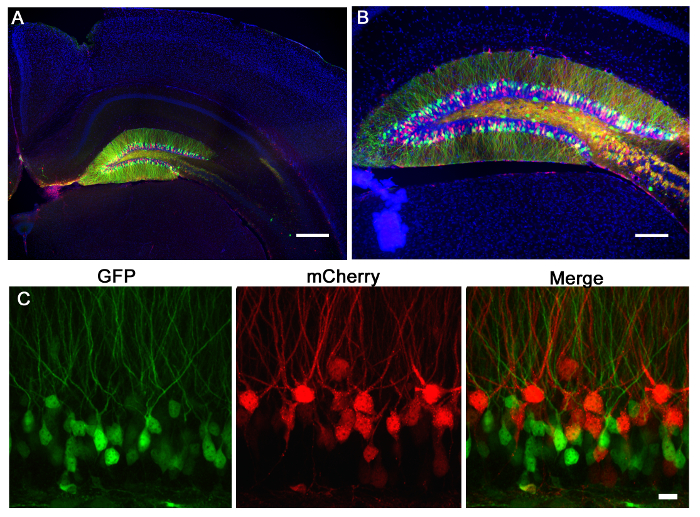
Figure 2: Retroviral injection of the mouse dentate gyrus. Retroviruses expressing mCherry (red) or GFP (green) were injected into the dentate gyrus of a p7 mouse. 21 days later the mice were perfused and the brains sectioned and stained for GFP and mCherry. (A) A 5X wide-field fluorescent image shows the precision of the dentate gyrus injection and the specificity of labeling dentate gyrus granule neurons. The morphology of the hippocampus can be seen via the Dapi (blue) staining. Scale bar measures 200 µm. (B) A 10X wide-field fluorescent image demonstrates that these high titer lentiviruses infect a large number of cells whose morphology can be accessed via fluorophore expression. Scale bar measures 100 µm. (C) Viruses expressing GFP or mCherry were co-injected into the dentate gyrus. Using a system of retroviruses, one can use one virus to make one genetic manipulation marked by GFP and another manipulation marked by mCherry, and then assess single or additive changes due to each virus. Scale bar measures 10 µm.
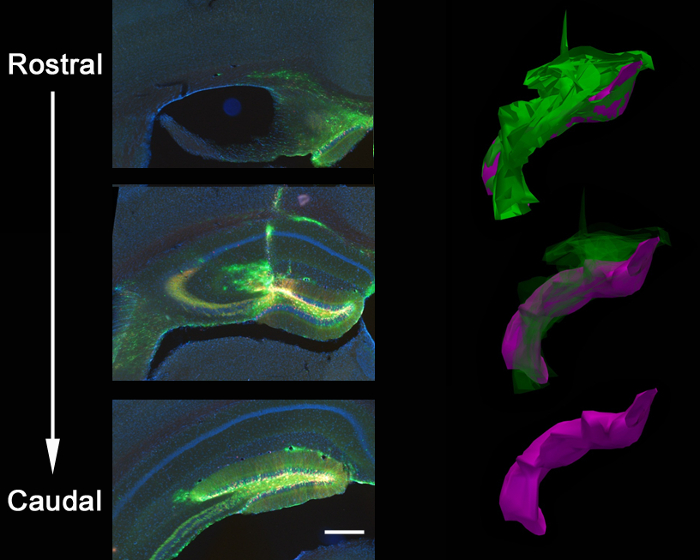
Figure 3: Anatomical spread of lentiviral injection. Stereotaxic coinjection of a GFP-shPten virus and an mCherry control virus into the brain of an adult Ptenloxp/+ mouse resulted in a viral spread along the entire rostral/caudal axis of the dentate gyrus of the hippocampus. This is shown in a 3D reconstruction of the extent of the injection in which closed contours of the viral spread were traced over 21 serial sections (Z = 50 μm/section) using reconstruction software. The contour tracings were then aligned to generate the 3D images for volume quantification. Total viral spread is shown in green (volume = 54,730,800 μm3) and dentate-localized spread is shown in purple (volume = 27,275,200 μm3). The virus spreads along the needle track and the corpus callosum at the intersection of the needle track in addition to filling the rostral/caudal axis of the dentate gyrus. Scale bar measures 200 µm.
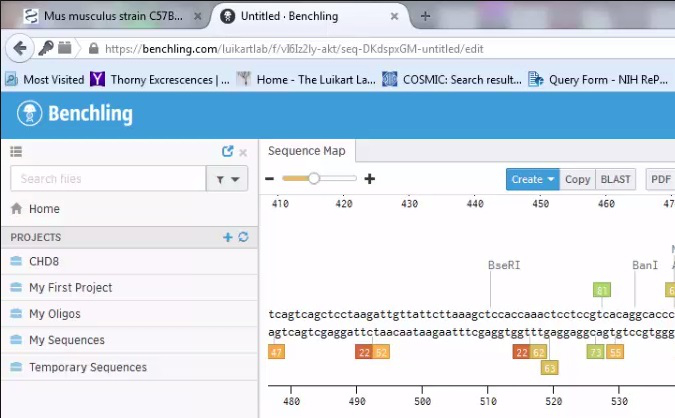
Supplemental Video 1. Design of sgRNAs to clone into retroviral and lentiviral backbone.
In this synthetic guide RNA (sgRNA) design example, the genomic sequence of mouse CHD8 is downloaded from NCBI. The start codon and exon structure are then visualized in Vector NTI. This allows us to copy the genomic region around the first coding exon and enter this sequence into Benchling. Benchling allows us to visualize all potential sgRNAs in the region. Further, after indicating the genomic region we have input, Benchling will show us the on-target and off-target scores for each guide RNA. The user can then select the guide RNA with the highest on- and off-target scores. Please click here to view this video. (Right-click to download.)
Discussion
There are a few critical steps that are important for successful viral packaging. Cell health is critical before and during the transfection, as unhealthy cells will greatly reduce the amount of virus produced. If the transfection and packaging are successful, then 100% of the cells should express the fluorophore and the cells should form a functional syncytium. In step 3.2.4, tapping the tube is necessary for high-titer transfection efficiently, and the pH of the HEPES-buffered saline must be exact. The maxi-preps that produce the plasmids necessary for viral packaging must be extremely pure. To this point, it is helpful to ethanol precipitate the final DNA elution and re-suspend in Tris-EDTA buffer. It is also very important to reduce the amount of serum to 2% or less in the media that the caffeine is added to on Day 6 (step 3.4) before viral collection. If the serum is not reduced, then the final purified virus will contain an undesirable amount of serum protein. The use of polyethylene glycol 6000 when precipitating the viral particles excludes the necessity of ultracentrifugation. It is also important to note that the Cas9 CRISPR containing viruses typically have a titer around 10 fold less than viruses solely containing a fluorophore.
For the stereotaxic surgery, the use of inhaled anesthesia allows rapid and precise control or the animal's consciousness compared to injectable anesthetics and allows anesthesia over a larger age range. It is highly important to keep the surgical instruments clean and sterile, and reproducible targeting requires precise positioning of the head. Make sure there is no pitching or rolling of the head in the stereotaxic instrument and that the skull feels firmly in place. It can be useful to allow the skull to dry in order to find the sutures to determine the stereotaxic coordinates. Also, the rate and volume for each stereotaxic coordinate should be empirically determined.
This technique is limiting in that the spread of a lenti- or retrovirus is restricted, especially when compared to adeno-associated viruses (AAVs).Therefore, these viruses are valuable when infecting a discrete brain region, but not for overall infection associated with AAVs used for behavioral analysis in animals. The use of caffeine in this protocol greatly increases the titer of these viruses, but they are still not as high as the titers achieved in AAV packaging. Also, stable integration is only an advantage of fluorophore expression, as CRISPR/Cas9 forms stable genomic edits even when transiently transfected and it is possible that on-going expression of the Cas9 and sgRNA may eventually produce off target effects. The transient expression the CRISPR/Cas9 system with AAVs is sufficient to produce genomic changes that are propagated throughout cell divisions, however, fluorophore expression will not be maintained.
Creation of lenti- and retroviruses utilizing the CRISPR/Cas9 system will impart the ability to target any novel gene in a wide variety of organisms. The efficiency of gene editing appears to be dependent on the sequence of the guide RNA targeting the Cas9 cleavage. It has been empirically determined that between 10% and 80% of clones contain indels after sequencing infected Neuro2A cells. It is presently unknown whether indel frequencies calculated in Neuro2A cells are reflective of those in neurons. Guide RNA design software such as Benchling now include an "on-target" score that may be able to predict the efficiency of a given target sequence. To what degree such "on-target" scores are reliable needs to be empirically determined in neurons and other cell-types as the CRISPR-Cas9 system becomes more widely implemented.
Lentivirus-based transgenic animal production has been variably successful with reports that the lentivirus-delivered transgenes become silenced11. CRISPR mediated gene editing of DNA may be passed through the germ line to generate whole animal models. Thus, stable genomic editing may be achievable despite the silencing of viral-delivered fluorophores and Cas9 transgenes. This may provide an efficient platform for targeted genomic alterations. The viral delivery of the CRISPR/Cas9 system, while not requiring transgenic organisms, is complementary to those techniques. For example, injecting such viral particles into a compound transgenic animal that inducibly expresses Cre and Cre dependent opto- or chemo-genetic transgenes should facilitate complex studies into the relationship between genetic manipulations and neuronal activity. A second example is to deliver these Cas9/sgRNA viral particles into a conditional knockout in an attempt to screen for gene-gene interactions. Finally, another exciting route of this research is the screening of phenotypes and therapeutic compounds in patient derived cells, which can be used to validate and discover genetic networks that are disrupted in various diseases.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the NINH grant R01MH097949 and the Autism Speaks Pilot Grant 7359 to BWL and the Norris Cotton Cancer Center Optical Imaging Shared Instrumentation Grant P30CA023108.
Materials
| List of Cell Culture Reagents | |||
| 293FT cell line | Life Technologies | R700-07 | For Lentivirus |
| 293GP cell line | Clontech | 631458 | For Retrovirus |
| Iscove's Modification of DMEM (IMDM) | Corning | 10-016-CV | Complete IMDM with 10% FBS, 1% NEAA, 1% L-Gln, and 1% P/S |
| Fetal Bovine Serum (FBS) | Corning | 35-011-CV | |
| MEM Nonessential Amino Acids (NEAA) | Corning | 25-025-CI | |
| L-Glutamine solution, 100X (L-Gln) | Corning | 25-005-CI | |
| Pennicillin/Streptomycin solution, 100X (P/S) | Corning | 30-002-CI | |
| Polystyrene 10cm plate | USA Scientific | CC7682-3394 | |
| Trypsin EDTA 1X | Corning | 25-053-CI |
|
| Reagent | Company | Catalog Number | Notes |
| List of Transfection Reagents | |||
| 5ml polystyrene tubes | Fisher Scientific | 352054 | |
| Calcium Chloride Dihydrate (CaCl2) | Fisher Scientific | C69-500 | Make a 2.5 M solution in ddH2O |
| Sodium Chloride (NaCl) | Fisher Scientific | S271-3 | |
| HEPES | Fisher Scientific | BP2939-100 | |
| Sodium phosphate dibasic | Fisher Scientific | S369-500 | |
| (Na2HPO4) | |||
| 2X HEPES Buffered Saline (HBS) | 500ml: 8.2g NaCl, 5.95g HEPES, 0.106g Na2HPO4, pH 7.01 (exact!) | ||
| filter and store at 4°C | |||
| Caffeine | Sigma-Aldrich | C0750-5G | |
| 0.22 µM syringe filter unit | EMD Millipore | SLGV033RS | |
| 0.45 µM syringe filter unit | EMD Millipore | SLHP033RS | |
| 60CC L/L Syringe | Med-Vet International | MV60CCLL | |
| 50 ml Conical Tube | Corning | 352098 | |
| polyethylene glycol 6000 (PEG 6000) | Millipore | 528877 | |
| (10X) Phosphate Buffered Saline (PBS) | National Diagnostics | CL-253 | |
| 0.5 ml microcentrifuge tubes | USA Scientific | 1605-0000 | |
| Matrigel | Fisher Scientific | CB-40230A | |
| 6-well plate | Fisher Scientific | 353046 | |
| Paraformaldehyde | Fisher Scientific | AC41678-5000 | |
| Donor Horse Serum | Cellgro | 35-030-CV | |
| TritonX-100 | Sigma-Aldrich | X100-500ML | |
| 10ml serological pipette | Fisher Scientific | 357551 | |
| anti-GFP, rabbit, 488 conjugate | Invitrogen | A21311 | |
| Reagent | Company | Catalog Number | Notes |
| Stereotaxic Surgery Reagents | |||
| Vet-Syringe vet use T.B. Syringe only 1cc luer slip T.B. 100/bx | Med-Vet International | 1CCVLS | |
| Isoflurane | |||
| Stainless Steel Scalpel Blades, #10, 100-pk | Med-Vet International | JOR580S | |
| artificial tear ointment | Med-Vet International | RXPARALUBE-O | |
| PVP PrepSolution | Med-Vet International | HPIV108208H | |
| normal saline | Med-Vet International | DYND500MLSH | |
| cotton tipped applicators | Med-Vet International | CTA6 | |
| Triple antibiotic ointment | Med-Vet International | RXTRIP-OI15 | |
| MONOJECT® Needles Soft Pack 25g x 5/8" | Med-Vet International | 25058 | |
| 6-0 silk sutures | Med-Vet International | MV-711 | |
Riferimenti
- Williams, M. R., DeSpenza, T., Li, M., Gulledge, A. T., Luikart, B. W. Hyperactivity of newborn Pten knock-out neurons results from increased excitatory synaptic drive. J Neurosci. 35 (3), 943-959 (2015).
- Luikart, B. W., et al. Pten knockdown in vivo increases excitatory drive onto dentate granule cells. J Neurosci. 31 (11), 4345-4354 (2011).
- Cong, L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339 (6121), 819-823 (2013).
- Ran, F. A., et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8 (11), 2281-2308 (2013).
- Sander, J. D., Joung, J. K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 32 (4), 347-355 (2014).
- Luikart, B. W., et al. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 6 (5), e19077 (2011).
- Nasri, M., Karimi, A., Allahbakhshian Farsani, M. Production, purification and titration of a lentivirus-based vector for gene delivery purposes. Cytotechnology. 66 (6), 1031-1038 (2014).
- Ellis, B. L., Potts, P. R., Porteus, M. H. Creating higher titer lentivirus with caffeine. Hum Gene Ther. 22 (1), 93-100 (2011).
- Fricano, C. J., et al. Fatty acids increase neuronal hypertrophy of Pten knockdown neurons. Front Mol Neurosci. 7, 30 (2014).
- Park, F. Lentiviral vectors: are they the future of animal transgenesis?. Physiol Genomics. 31 (2), 159-173 (2007).

