High-throughput Screening of Carbohydrate-degrading Enzymes Using Novel Insoluble Chromogenic Substrate Assay Kits
Summary
A high-throughput assay for enzyme screening is described. This multiplexed ready-to-use assay kit comprises of pre-chosen Chromogenic Polymer Hydrogel (CPH) substrates and complex Insoluble Chromogenic Biomass (ICB) substrates. Target enzymes are polysaccharide degrading endo-enzymes and proteases.
Abstract
Carbohydrates active enzymes (CAZymes) have multiple roles in vivo and are widely used for industrial processing in the biofuel, textile, detergent, paper and food industries. A deeper understanding of CAZymes is important from both fundamental biology and industrial standpoints. Vast numbers of CAZymes exist in nature (especially in microorganisms) and hundreds of thousands have been cataloged and described in the carbohydrate active enzyme database (CAZy). However, the rate of discovery of putative enzymes has outstripped our ability to biochemically characterize their activities. One reason for this is that advances in genome and transcriptome sequencing, together with associated bioinformatics tools allow for rapid identification of candidate CAZymes, but technology for determining an enzyme’s biochemical characteristics has advanced more slowly. To address this technology gap, a novel high-throughput assay kit based on insoluble chromogenic substrates is described here. Two distinct substrate types were produced: Chromogenic Polymer Hydrogel (CPH) substrates (made from purified polysaccharides and proteins) and Insoluble Chromogenic Biomass (ICB) substrates (made from complex biomass materials). Both CPH and ICB substrates are provided in a 96-well high-throughput assay system. The CPH substrates can be made in four different colors, enabling them to be mixed together and thus increasing assay throughput. The protocol describes a 96-well plate assay and illustrates how this assay can be used for screening the activities of enzymes, enzyme cocktails, and broths.
Introduction
Techniques for mining genomes and metagenomes have developed rapidly in recent years, and so have medium- and high-throughput strategies for cloning and expressing recombinant enzymes. Furthermore, bioinformatic resources and associated depositories, such as (CAZy)1,2 have expanded greatly. However, there are considerable challenges inherent in the exploitation of microbial enzyme diversity for industrial purposes and the empirical determination of enzyme activities has now become a serious bottleneck. For example, it is estimated that, using current methods, we can safely predict the activities of no more than 4% of the proteins within the CAZy database. Although numerous methods are available for monitoring enzyme activities they all have some limitations. Well-established techniques based on chromatography combined with mass spectrometry are available for assessing the oligomeric fragments of glycosyl hydrolase (GH) activities3,4. However, these approaches are labor intensive and generally low-throughput. Methods based on the measurement of reducing sugars such as the dinitrosalicylic acid5 and Nelson-Somogyi6 assays are widely used for assessing GH activities. However, these assays have limited throughput and can be prone to side-reactions. Individual chromogenic polysaccharide substrates, such as azurine cross-linked (AZCL) are widely used for determination of enzyme activities, but purchasing all of the substrates separately and manually distributing the substrate powders within the assay plate can be cumbersome and costly7.
We have developed a new generation of chromogenic polymer hydrogel (CPH) substrates based on chlorotriazine dyes that, when used in conjunction with a 96-well filter plate, form a high-throughput assay system. Additional Insoluble Chromogenic Biomass (ICB) substrates were developed which provide information about substrate availability within complex polymer mixtures, such as those that exist in lignocellulosic biomass. Each substrate can be produced in one of four colors, and different colored substrates can be combined in a single well. In this protocol is shown that this methodology can be applied to a wide variety of polysaccharides and proteins and the potential for screening GHs, lytic polysaccharide monooxygenases (LPMOs) and proteases. Specific protocols are provided for the use of 96 well plates and representative results illustrate the high efficiency of the CPH and ICB substrate kits as tools for enzyme screening.
One significant advantage of the assay kits described, regardless of the substrate, is that the kits are ready to use within 15 minutes, after the activation step. This eliminates the need for time-consuming assembly of the assay from raw substrate materials as it is the case with some other methods7. The CPH and ICB substrates have excellent storage (at least one year at room temperature), pH and temperature stability 8 and require no specialized equipment or training. The CPH or ICB assays are based on 96-well filter plate within which the reaction with the enzyme is conducted. If the enzyme is active with a given substrate, soluble dyed oligomers are generated, producing a colored supernatant which can then be filtered into a regular clear-well 96-well plate using a vacuum manifold or a centrifuge 8.
The substrates are dyed with chlorotriazine dyes which absorb in the visible spectrum (VIS) range and individual colors (red, blue, yellow and green) can be resolved using linear regression if different CPH substrates of different colors are mixed in a single well, and the enzyme acts on more than one substrate. The resulting plate with the supernatants can be measured using a standard microtiter-plate reader capable of measuring absorbance in the VIS range. Mixing different substrates with different colors in one well increases the throughput of the assay system, to a total of 384 experiments in a 96-well plate (4 different substrates of different colors per well).
CPH substrates provide a valuable tool for assessing the specific activity of an enzyme while ICB substrates are used to evaluate the capacity of an enzyme to digest a component within the context of complex substrate mixtures that enzymes usually encounter within biomass. Although ICB substrates do not provide information about individual enzyme specificities, they are nonetheless useful tools for assessing the commercial performance of enzymes, cocktails or broths.
Protocol
1. Chromogenic Assay with CPH Substrates in a 96-well Plate Format
- Activation of the assay kit plate
- Activate the 96-well filter (containing blue CPH-xylan) assay kit plate (List of Materials) by adding 200 µl activation solution (obtained with the kit) into each well, followed by 10 min incubation at room temperature without agitation.
- Apply vacuum using a vacuum manifold (with the spacer block inside and any standard, transparent 96-well plate as a collection plate) to remove extant activation solution. It is also possible to use a centrifuge at 2,700 x g for 10 min for this step instead of the vacuum manifold.
- Wash the CPH substrates by adding 100 µl sterile water and apply vacuum (or centrifugal force) to remove the stabilizer. Repeat this step two more times and the plates are now ready to use.
- Enzyme reaction
NOTE: Always include buffer alone as a negative control and if possible previously characterized enzymes as positive controls. Use a statistically appropriate number of replicates. The CPH substrates are stable between pH 3.0 to 10.0 and the total volume of buffer end enzyme solution in each well should not exceed 180 µl. Plant extract or culture broth can also be used instead of purified enzyme solution.- Add 150 µl 100 mM sodium acetate buffer, pH 4.5 and 5 µl endo-cellulase solution (to a final concentration of 1 U/ml) to each well of the assay kit plate.
- Put the product plate (a clear-well plate compatible with the microtiter-plate reader) underneath the assay kit plate to collect any potential leakage from the reaction plate during shaking.
- Incubate the assay kit plate at 25 °C for 30 min in a horizontal shaker at 150 rpm.
NOTE: Mixing the reaction in the assay kit plate during the incubation is crucial for achieving a consistent and reproducible result. CPH substrates are stable up to 90 °C. The incubation time should be increased up to 24 hr when testing culture broths containing unknown enzyme concentrations with CPH substrates. Note that appropriate incubation times depend on the activity of the enzyme(s), but in general if there is no detectable activity within 24 hr, it is likely that the enzyme will not degrade the tested substrate. An active enzyme is degrading the insoluble chromogenic polysaccharides of the CPH substrate into soluble chromogenic oligosaccharides, which are visible as colored supernatant. - Place the clean product plate inside the vacuum manifold with the spacer block inside.
- Place the assay kit plate on top and apply vacuum (maximum negative pressure of -60 kPa). It is also possible to use a centrifuge at 2,700 x g for 10 min.
NOTE: The filtrate containing the colored oligosaccharides as reaction product is now in the product plate for further analysis8.
- Detection and Quantification
- Check that the volume of liquid in each well of the product plate is approximately the same by visual inspection.
- Read the absorbance of the collection plate at 595 nm for blue CPH-xylan using a plate reader.
- When doing data analysis, subtract the buffer-only negative control values from the values from the wells where an enzyme was added. Calculate the mean value and the standard error of means (SEM) from the replicate wells8.
2. Chromogenic Assay with ICB Substrates in a 96-well Format
- Enzyme reaction
- Add the 150 µl 100 mM sodium acetate buffer, pH 4.5 and 5 µl 31 U/ml endo-xylanase solution to each well of the assay kit plate containing red ICB-wheat straw (final enzyme concentration in the well: 1 U/ml).
NOTE: The assay kit plates (96-well filter plates containing ICB substrates) are manufactured as described in literature (see List of Materials). The ICB substrates are stable in buffers with a pH range between pH 3.0 to 10.0. Always include buffer alone as a negative control, commercial enzymes as a positive control and use a statistically appropriate number of replicas.- Do not activate the ICB substrates like the CPH substrate plate, but remove the stabilizer by washing three times with 100 µl water followed by vacuum filtration or centrifugation.
- Put the product plate underneath the substrate plate to collect any potential leakage from the substrate plate during shaking.
- Incubate the reaction at 25 °C shaking at 150 rpm for 2 hr.
NOTE: An active enzyme is degrading the insoluble chromogenic polysaccharides in the ICB substrate into soluble oligosaccharides, which are visible as colored supernatant. ICB substrates are stable up to 90 °C. The incubation time should be increased up to 24 hr if non-purified enzymes such as culture broths are used. - Place the product plate inside the vacuum manifold with the spacer block inside.
- Place the assay kit plate on top and apply vacuum (maximum negative pressure of -60 kPa) or use a centrifuge to filtrate the product from the assay kit plate into the well of the product plate.
NOTE: The filtrate containing the colored oligosaccharides as reaction product is now in the collection plate for further analysis8.
- Add the 150 µl 100 mM sodium acetate buffer, pH 4.5 and 5 µl 31 U/ml endo-xylanase solution to each well of the assay kit plate containing red ICB-wheat straw (final enzyme concentration in the well: 1 U/ml).
- Detection and quantification
- Check that the volume of liquid in each well of the collection plate is approximately the same by visual inspection.
- Read the absorbance of the collection plate at 517 nm for red ICB-wheat straw using a plate reader.
- When doing data analysis — subtract the buffer — only negative control values from the values from the wells where an enzyme was added. Calculate the mean value and the standard error of means (SEM) from the replicate wells8.
NOTE: In case of screening unknown enzymes, we suggest making a dilution series in order to obtain more detailed data about the dynamic range of enzyme's activity.
Representative Results
The high-throughput and multiplexing capacity of this assay is based on insoluble chromogenic polymer (or protein) hydrogel (CPH) substrates arranged in 96-well filter plates. Enzymes as well as negative controls are added to the assay kit plate (Figure 1A) and the enzymes degrade the corresponding substrate producing a colored supernatant (Figure 1B). After the reaction is finished, the supernatant is transferred into a clear-well product plate and the absorbance can be measured directly using a spectrophotometer suitable for 96-well plates (Figure 1C).
An example of a dose response of CPH-arabinoxylan to xylanase at different concentrations of the enzyme (0.00 – 0.75 U/ml) is shown in Figure 1D where the decreasing enzyme concentration can be observed visually. A more detailed spectrophotometric quantification can be used to plot the absorbance versus enzyme concentration (Figure 1E). The signal intensity corresponds to the enzyme activity. The reproducibility of the assay is shown by the error bars (standard error of mean, SEM, of three replicas). More detailed experiments on the reproducibility of this assay are published elsewhere8.
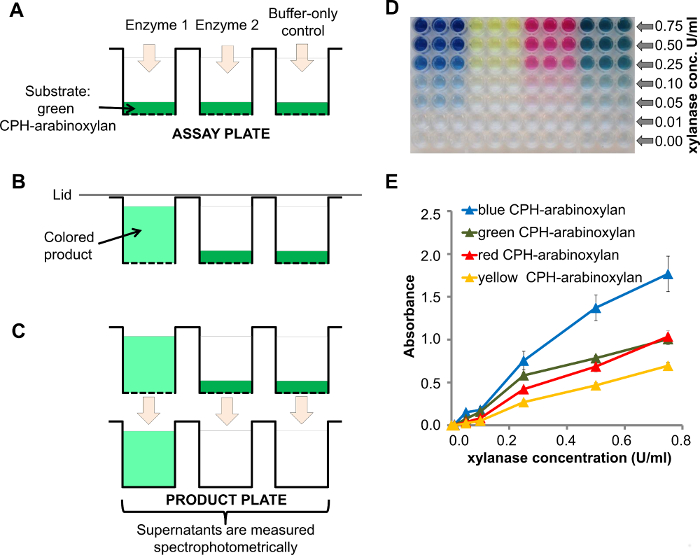
Figure 1. Xylanase treatment of CPH-arabinoxylan. A) A scheme of the assay kit plate with the CPH substrate (e.g., CPH-arabinoxylan) loaded into 96-well filter plate wells just before the addition of enzymes 1 and 2 and the buffer-only control (enzyme 1 had endo-xylanase activity); B) Degradation of CPH-arabinoxylan by enzyme 1 produced a colored supernatant; C) Upon vacuum-assisted filtration of the supernatants in to the product plate, the absorbance is measured spectrophotometrically; D) product plate containing the reaction products after treatment of CPH-arabinoxylan in 4 different colors with different concentrations of endo-β-1,4-xylanase in 100 mM sodium acetate buffer, pH 4.5 for 60 min at room temperature; E) Quantification of the reaction products from D using spectrophotometry. Please click here to view a larger version of this figure.
There are different options for using this assay in enzyme screening. One option is to use a 96-well plate containing different polysaccharides for screening, e.g., a small number of (purified) endo-enzymes with unknown activity. In this case the result will show which polysaccharides are degradable by the target enzyme. To show this principle, an endo-cellulase was tested against different CPH substrates at 25 °C. Three different enzyme concentrations (0.5 U/ml, 1.0 U/ml and 5 U/ml) were incubated for 30 min. The result is clearly visible in the product plate (Figure 2A). The product sheet for this endo-cellulase provided by the supplier specifies side-activity for xyloglucan (tamarind), barley β-glucan, glucomannan, birchwood xylan and low side-activity for galactomannan. Consistent with this, activity additional to cellulase was found against CPH-β-glucan (barley), CPH-xyloglucan (tamarind), CPH-xylan (beechwood) and low activity against CPH-galactomannan (Figure 2B). Glucomannan was not tested. The same CPH substrates were digested with commercial available enzymes (three different enzyme concentrations: 0.1 U/ml, 0.5 U/ml and 1.0 U/ml) used as positive controls under the same conditions than the previous experiment. All substrates were degraded by the positive control enzyme and the signal intensity increased corresponding to higher enzyme concentration (Figure 2C).
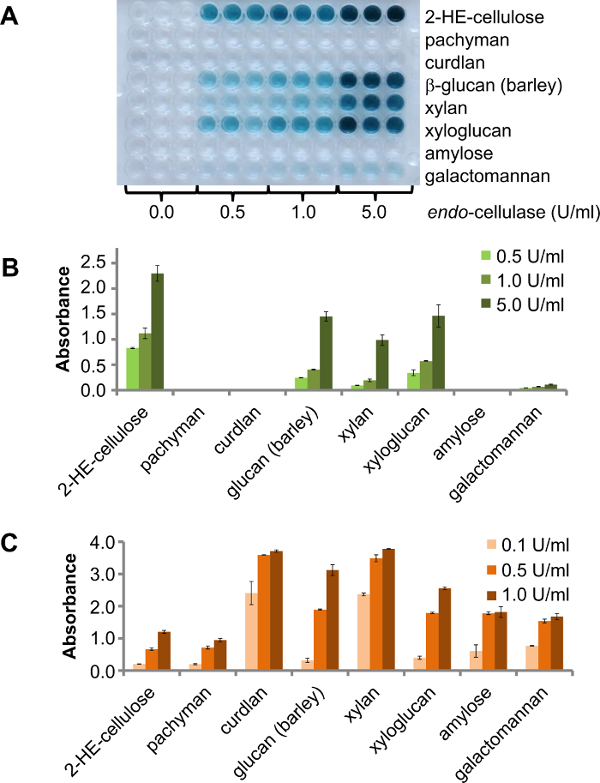
Figure 2. Eight different CPH substrates were incubated under agitation at 25 °C for 30 min. A) The product plate of different CPH substrates digested with an endo-cellulase, at different concentrations. B) Quantification of the activity and various side activity of the endo-cellulase. The error bars represent the standard error of mean of three replicas. C) Activity of different commercial enzymes to the corresponding CPH substrate (endo-cellulase and 2-HE-cellulose; E-LAMSE and CPH-pachyman, CPH-curdlan, CPH-β-glucan (barley); E-XYAN4 and CPH-xylan, E-XEGP and CPH-xyloglucan, E-BLAAM and CPH-amylose, E-BMACJ and CPH-galactomannan; all enzymes from Megazyme). The error bars represent the standard error of mean of two replicas. Please click here to view a larger version of this figure.
The insoluble chromogenic biomass (ICB) substrates are a useful addition to the chromogenic substrate repertoire because they retain in part the natural arrangement of polysaccharides in plant cell walls which are the major constituent of biomass. CPH and ICB substrates are used in our example to analyze the secreted enzymes of Phanerochaete chrysosporium when cultivated in liquid medium. The plate setup of the assay kit plate is shown in Figure 3A, with 19 CPH substrates and 5 ICB substrates (4 wells for each substrate, Figure 3B). P. chrysosporium was cultivated for three days and then the culture supernatant analyzed. Therefore, 125 µl 200 mM buffer was transferred to each well and 25 µl culture supernatant added. Three different pH conditions have been tested using sodium acetate buffer pH 4.0, sodium phosphate buffer pH 6.0 or pH 8.0 (Figure 3C). The plate was incubated shaking (150 rpm) at 25 °C for 2 hr.
The reaction products were transferred to the product plate (Figure 3D) and analyzed. P. chrysosporium produced enzymes for the degradation of various glucans, starch and xylans (Figure 3E). Lower signals could be detected for the hemicelluloses arabinan (sugar beet) and pectic galactan as well as for RGI (soybean). The enzymes produced were more active in acidic conditions (pH 4.0) than in neutral or slightly basic conditions (pH 8.0). Lower activity towards ICB substrates (Figure 3F) demonstrates that when the polysaccharides are in a more natural context, the efficiency of the enzyme is not the same as with a pure polysaccharide and that is why ICB substrates demonstrate a more realistic view on enzyme efficiency, if it was applied to raw or pre-treated plant material.
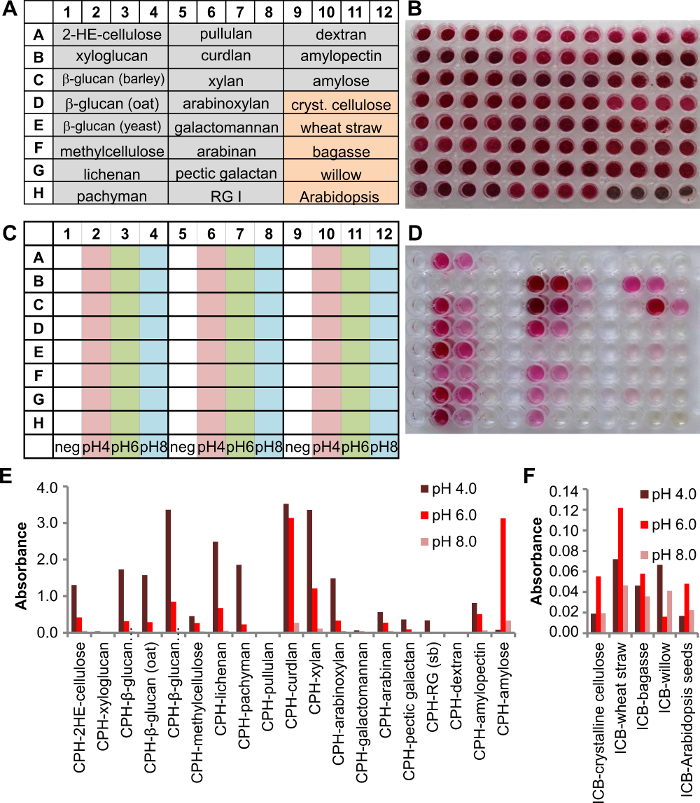
Figure 3. Screening of a culture supernatant from a 3-day old liquid culture of Phanerochaete chrysosporium using a multi substrate plate containing 19 CPH and 5 ICB substrates. A) A scheme of the plate setup with 4 wells for each individual substrate (grey background = CPH substrates, orange background = ICB substrates). B) Picture of the assay plate containing the substrate. C) Scheme showing the buffer conditions used in that experiment (200 mM sodium acetate pH 4.0, sodium phosphate pH 6.0 and sodium phosphate pH 8.0). D) picture of the product plate after 2 hr at 25 °C. E) Absorbances were detected at 517 nm and plotted for each individual CPH substrate and F) ICB substrate results for the respective enzymes. Please click here to view a larger version of this figure.
The chromogenic substrates can be also used to study synergistic effects by using a mixture of different colored CPH substrates in one well and analyzing the reaction supernatant after treatment using single enzymes or enzyme cocktails.
In the following example shown in Figure 4, red CPH-cellulose and yellow CPH-xylan substrates were mixed together at an approximately equal ratio in 96-well filter plate wells. Figure 4A shows the colored reaction products after 1 hr treatment at room temperature with no enzyme (control), cellulose cel (2 U/ml), xylanase xyl (1 U/ml) and a mixture of both enzymes (3 replicates for each approach) in 100 mM sodium acetate buffer pH 4.5. For analysis, the reaction product was quantified spectrophotometrically by scanning the absorbance spectrum from 350 nm to 700 nm (Figure 4B). Often visual inspection alone can give an indication of whether the enzyme is acting on one or multiple substrates, however recorded absorbance spectra stemming from different dyes can also be resolved using simple linear regression8 to give a more accurate indication of the extent of degradation of each substrate from the mixture.
Using CPH substrates as mixtures substantially adds to the throughput of the assay, enabling screening against up to 4 different substrates in one experiment (one well). In the example shown are four different substrates used: blue CPH-β-glucan (barley), yellow CPH-xylan (beechwood), green CPH-amylose and red CPH-pectic galactan (lupin). The layout of the substrate plate is shown in Figure 4C and a picture of the assay plate in Figure 4D. The reaction was performed in 100 mM sodium acetate buffer pH 4.5 for 30 min at 25 °C and 150 rpm. First single enzymes with increasing enzyme concentration were tested with the corresponding CPH substrate (Figure 4E) and the colored supernatant was received as expected in the product plate (Figure 4F, row 1A – 12D). Row E contained the two different CPH substrates yellow CPH-xylan and blue CPH-β-glucan, which were degraded with different ratios of the corresponding enzymes endo-xylanase and endo-glucanase. After the reaction, the result is visible in the product plate: the color of the reaction product was darker green-blue, when more endo-glucanase was present (Figure 4F, 4E-6E) and turned into a lighter yellow-green, when the endo-xylanase concentration increased (Figure 4F, 10-12E). The same is seen in row F, where the two substrates red CPH-pectic galactan and yellow CPH-xylan were degraded with endo-galactanase and endo-xylanase. All four different colored CPH substrate were present (Figure 4F, 1G-12F) and single enzymes degraded the appropriate CPH substrate and by adding additional enzymes a combination of the colored reaction products was received.
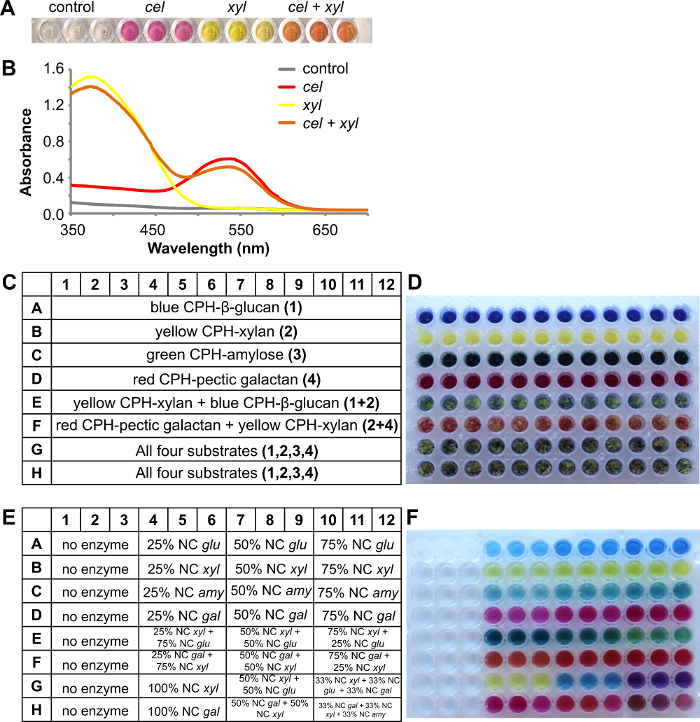
Figure 4. A combination of two different CPH substrates, red CPH-cellulose and yellow CPH-xylan, treated with different enzymes. A) Reaction supernatants after treatment of two substrates with endo-cellulase (cel) or endo-xylanase (xyl) or both enzymes. B) Absorbance spectra of the reaction supernatants. A combination of four different CPH substrates treated with different enzymes. C) Scheme of the substrate plate containing the CPH substrates: blue CPH-β-glucan (barley), yellow CPH-xylan (beechwood), green CPH-amylose and red CPH-pectic galactan (lupin). D) Picture of the assay plate containing the different CPH substrates. E) Scheme of the product plate showing the ratio of the added enzymes (nominal concentrations (NC): glu = 1 U/ml endo-glucanase, xyl = 1 U/ml endo-xylanase, amy = 5 U/ml endo-amylase and gal = 0,5 U/ml endo-galactanase). F) Picture of the product plate after 30 min incubation at 25 °C. Please click here to view a larger version of this figure.
| Substrate | Source |
| CPH-2-hydroxyethylcellulose | N/A |
| (CPH-2-HE-cellulose) | |
| CPH-amylopectin | potato |
| CPH-amylose | potato |
| CPH-arabinan | sugar beet |
| CPH-arabinoxylan | wheat |
| CPH-casein | bovine milk |
| CPH-chitosan | animal origin |
| CPH-curdlan | Alcaligenes faecalis |
| CPH-dextran | Leuconostoc spp. |
| CPH-galactomannan | carob |
| CPH-laminarin | Laminaria digitata |
| CPH-lichenan | Icelandic moss |
| CPH-methylcellulose | N/A |
| CPH-pachyman | Poria cocos |
| CPH-pectic galactan | potato |
| CPH-pullulan | Aureobasidium pullulans |
| CPH-rhamnogalacturonan I (RG I) | potato |
| CPH-rhamnogalacturonan I (-Gal)* | potato |
| CPH-rhamnogalacturonan | soy bean |
| CPH-xylan | beechwood |
| CPH-xyloglucan | tamarind |
| CPH-β-glucan from barley | barley |
| CPH-β-glucan from oat | oat |
| CPH-β-glucan from yeast | yeast |
| ICB-Arabidopsis | Rosette leaves from Arabidopsis thaliana Col-0 (adult plant) |
| ICB-Arabidopsis seeds | Arabidopsis thaliana |
| ICB-bagasse | Saccharum officinarum (dried adult plant, stem and leaves) |
| ICB-crystalline cellulose (filter paper) | commercial Whatman 3MM Chr Chromatography paper |
| ICB-fenugreek seeds | Trigonella spp. seeds |
| ICB-hemp | Cannabis spp. (dried adult plant, stem and leaves) |
| ICB-lupin seeds | Lupinus angustifolius seeds |
| ICB-pollen P. pratense | Phleum pratense pollen |
| ICB-spruce | Picea spp. (milled tree trunk) |
| ICB-tobacco | leaves from Nicotiana benthamiana (young plant) |
| ICB-wheat straw | Triticum spp. (dried adult plant, stem and leaves) |
| ICB-willow | Salix spp. (dried adult plant, milled tree trunk) |
| ICB-Sorghum | Sorghum spp. (leaves from adult plant) |
| *(β-1,4-D-galactan side chains removed with endo-β-1,4-D-galactanase) | |
Table 1. List of available Chromogenic Polymer Hydrogel (CPH) and Insoluble Chromogenic Biomass (ICB) substrates.
Discussion
We are using a new generation of multi-colored CPH and ICB substrates that are based on chlorotriazine dyes (full list of substrates in Table 1) arranged in a custom designed commercial assay kit. Enzyme digestion of the substrates yields small, soluble, dyed products which are detectable in the assay solution and can be quantified using a plate reader9. This assay is designed for evaluation of endo-acting enzymes and the sensitivity of the assay is similar the one using azurine cross-linked (AZCL) substrates10, while other methods are available for exo-acting enzymes11,12. The limitation of this assay kit lies in the detection of endo-enzyme activity, as CPH as well as the ICB substrates are not degradable by exo-enzymes most likely due to steric hindrance arising from the dye and crosslinker molecules8.
The assay is performed in a 96-well format and individual reactions take place in the wells. The reaction has to be mixed in the plate to receive reproducible data. The resulting supernatants are filtered into a product plate where the absorbance of each well can be quantified using absorbance spectrometry. The basic principles and the layout of the assay are shown in Figure 1. The assay consists of an assay plate (a 96-well filter plate) with the substrates, and after incubation with enzymes, the supernatant is filtered through into a clear well plate and the absorbances are read providing a semi-quantitative measurement of enzyme specificity and activity. It has been shown, that this screening assay using CPH substrates can be also used in an agar plate format, where the soluble reaction products create a colored halo after overnight incubation.8
The assay kits can be used to screen purified enzymes and their potential side-activities as demonstrated in Figure 2. Side-activities can arise from a single enzyme and its promiscuous specificity but also from a fact that the sample analyzed is a mixture of different enzymes and their synergic effect needs to be studied. Additionally, as it has been shown in previous studies, that enzyme cocktails, gut microbiota13 and culture broths from fungi8 as well as endogenous plant enzyme and bacteria (unpublished data) can be used as an enzyme source.
ICB substrates address complex mixtures of cell wall components often encountered in industrial processes of biomass breakdown. These substrates are designed to evaluate polysaccharide availability and provide information about how to effectively optimize degradation cocktails for more efficient degradation output. As illustrated in Figure 3 CPH and ICB substrates can be used in enzyme screening side by side — revealing a wealth of information about enzyme specificity and activity in both the context of the preferred substrate (CPH) and a more natural complex containing other components in addition to the preferred substrate more closely mimicking a macromolecular assembly found in nature (ICB). The use of multiple colors allows for simultaneous detection of different enzyme activities against several substrates which increases the high-throughput and multiplexity of the assay. Spectra of different dyes can be resolved by simple linear regression and in most cases multi-substrate activity can be observed by visual inspection alone. A simulated example of such an experiment and its results is depicted in Figure 4.
This assay toolbox and the versatility of its application are very well suited for first-level screening of enzymes and culture broths with unknown activities. The most important aspects of this assay are its high-throughput nature, customizability, ease of use and flexibility. With that in mind, we believe that this novel set of tools will greatly improve and speed up enzyme screening processes in industrial as well as academic applications.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We would like to thank Prof. J. Paul Knox (University of Leeds, UK), who provided access to his laboratories for filming, and Susan E. Marcus for excellent technical assistance. JS acknowledges the WallTraC project (European Commission Seventh Framework Program (grant agreement no: 263916) and the project Biomass for 21st century (Innovation foundation Denmark; case no.: 103408). SKK is thanking the SET4Future project (Danish Strategic Research Council), the Bio-Value Strategic founded by the Danish Council for Strategic Research, the Danish Council for Technology and Innovation (Grant Case no.: 0603-00522B), and A Biology-driven Approach for Understanding Enzymatic Degradation of Complex Polysaccharide Systems (Grant Case no.: 107279) for funding. This paper reflects the authors’ views only. The European Union is not liable for any use that may be made of the information contain herein.
Materials
| assay kit plates | Glycospot | customized assay kit plates | |
| activation solution | Glycospot | for activating CPH substrates | |
| 350 ml receiver plate spacer block for vacuum manifold | Pall Corporation | 5015 | spacer block |
| 96-well MultiScreen HV filter plate, 0.45 µm, clear, non-sterile | Millipore | MSHVN4510 | assay plate |
| 96-Well Microplates, Polypropylene | Greiner Bio-One | 651201 | collection plate after washing the substrates |
| Nunc™ MicroWell™ 96-Well Microplates | Thermo Scientific | 269620 | product plate |
| Diaphragm pump MZ 2 NT | Vacuubrand | 732000 | vacuum pump used with the vacuum manifold |
| Infors HT Ecotron | Infors HT | 4950132 (Buch & Holm) | horizontal shaker |
| SpectraMax M5 | Molecular Devices | 10067-750 (VWR) | 96-well plate absorbance reader |
| Vacuum manifold | Pall Corporation | 5017 | vacuum manifold |
| endo-cellulase (EGII) (Trichoderma longibrachiatum) | Megazyme | E-CELTR | cellulase [cel] |
| endo-β-1,4-mannanase (Cellvibrio japonicus) | Megazyme | E-BMACJ | mannanase [man] |
| endo-β-1,3-glucanase (Trichoderma spp.) | Megazyme | E-LAMSE | β-glucanase [glu] |
| endo-b-1,4-D-galactanase (Aspergillus niger) | Megazyme | E-EGALN | galactanase [gal] |
| endo-β-1,4-xylanase M4 (Aspergillus niger) | Megazyme | E-XYAN4 | xylanase [xyl] |
| endo-xyloglucanase (GH5) (Paenibacillus sp.) | Megazyme | E-XEGP | xyloglucanase [xg] |
| α-amylase (Bacillus licheniformis) | Megazyme | E-BLAAM | amylase [amy] |
Riferimenti
- Lombard, V., Ramulu, H. G., Drula, E., Coutinho, P. M., Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42 (Database issue), D490-D495 (2014).
- Cantarel, B. L., et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233-D238 (2009).
- Agblevor, F. A., Murden, A., Hames, B. R. Improved method of analysis of biomass sugars using high-performance liquid chromatography. Biotechnol. Lett. 26 (15), 1207-1211 (2004).
- Black, G. E., Fox, A. Recent progress in the analysis of sugar monomers from complex matrices using chromatography in conjunction with mass spectrometry or stand-alone tandem mass spectrometry. J. Chromatogr. A. 720 (1-2), 51-60 (1996).
- Miller, G. L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 31, 426-428 (1959).
- Somogyi, M. Notes on Sugar Determination. J. Biol. Chem. 195 (1), 19-23 (1952).
- Zantinge, J. L., Huang, H. C., Cheng, K. J. Microplate diffusion assay for screening of beta-glucanase-producing microorganisms. Biotechniques. 33 (4), 798 (2002).
- Kračun, S. K., et al. A new generation of versatile chromogenic substrates for high-throughput analysis of biomass-degrading enzymes. Biotechnol Biofuels. 8, (2015).
- Leemhuis, H., Kragh, K. M., Dijkstra, B. W., Dijkhuizen, L. Engineering cyclodextrin glycosyltransferase into a starch hydrolase with a high exo-specificity. J. Biotechnol. 103 (3), 203-212 (2003).
- Nyyssonen, M., et al. Coupled high-throughput functional screening and next generation sequencing for identification of plant polymer decomposing enzymes in metagenomic libraries. Front Microbiol. 4, 282 (2013).
- Sweeney, M. D., Xu, F. Biomass Converting Enzymes as Industrial Biocatalysts for Fuels and Chemicals: Recent Developments. Catalysts. 2 (2), 244-263 (2012).
- Biely, P., et al. Action of xylan deacetylating enzymes on monoacetyl derivatives of 4-nitrophenyl glycosides of beta-D-xylopyranose and alpha-L-arabinofuranose. J. Biotechnol. 151 (1), 137-142 (2011).
- Mackenzie, A. K., et al. A polysaccharide utilization locus from an uncultured bacteroidetes phylotype suggests ecological adaptation and substrate versatility. Appl Environ Microbiol. 81 (1), 187-195 (2015).

