映射的适体对ATP的结合部位使用MicroScale程序热泳
Summary
MicroScale Thermophoresis (MST) is a sensitive technology to characterize aptamer-target interactions. This manuscript describes an MST protocol to characterize aptamer-small molecule interactions.
Abstract
Characterization of molecular interactions in terms of basic binding parameters such as binding affinity, stoichiometry, and thermodynamics is an essential step in basic and applied science. MicroScale Thermophoresis (MST) is a sensitive biophysical method to obtain this important information. Relying on a physical effect called thermophoresis, which describes the movement of molecules through temperature gradients, this technology allows for the fast and precise determination of binding parameters in solution and allows the free choice of buffer conditions (from buffer to lysates/sera). MST uses the fact that an unbound molecule displays a different thermophoretic movement than a molecule that is in complex with a binding partner. The thermophoretic movement is altered in the moment of molecular interaction due to changes in size, charge, and hydration shell. By comparing the movement profiles of different molecular ratios of the two binding partners, quantitative information such as binding affinity (pM to mM) can be determined. Even challenging interactions between molecules of small sizes, such as aptamers and small compounds, can be studied by MST. Using the well-studied model interaction between the DH25.42 DNA aptamer and ATP, this manuscript provides a protocol to characterize aptamer-small molecule interactions. This study demonstrates that MST is highly sensitive and permits the mapping of the binding site of the 7.9 kDa DNA aptamer to the adenine of ATP.
Introduction
分子之间的相互作用是自然的基础。因此,科学家基础和应用研究的众多领域尝试了解不同种类的分子相互作用的基本原则。 MicroScale程序热泳(MST),科学家可以在溶液中进行快速,精确,成本效益和质量控制的分子间相互作用的特征,与缓冲区的自由选择。目前已经有超过1000出版物使用MST,2016年独自一人,描述不同类型的分析,包括图书馆放映,结合事件验证,竞争分析,和实验与多个合作伙伴的结合1-8。在一般情况下,台MST允许古典结合参数,如任何类型的分子相互作用的结合亲和力(1pM至毫米),化学计量,和热力学,研究。 MST的一大优点是研究独立的互动合作伙伴的大小结合事件的能力。即使CHAL目标小核酸适体之间有挑战性相互作用(15-30个核苷酸)和诸如小分子,药物,抗生素,或代谢产物可以定量。
当前国家的最先进的技术来表征适体靶相互作用或者是实验室强烈和高度复杂的或无法量化适配子的小分子的相互作用9,10。表面等离子体共振(SPR)为基础的试验11,12和真正的无标记热量的方法,如温滴定量热(ITC)13-15度洗脱16,平衡渗透17,18,在线探测19,凝胶-移位测定,stopped-溢流FL荧光光谱20,21,荧光各向异性(FA)22,23,单分子FL荧光成像24,25,和生物层干涉测量(BLI)26也无论是不精确或适体-小分子不相容互动。其他principa这些方法升问题是灵敏度低,高样品消耗,固定化,在表面上的质量传递的限制,和/或缓冲的限制。只有少数这些技术提供了聚集和吸附作用的综合控制。
台MST表示一个强大的工具为科学家克服这种限制,研究其它靶的适体和小分子27-29之间的相互作用,以及如蛋白质30-33。该技术依赖于分子通过温度梯度的运动。这种定向运动,被称为“热泳,”取决于大小,电荷,并且该分子34,35的水化外壳。配体分子的结合将直接改变这些参数中的至少一个,从而产生改变热泳迁移率。小尺寸的配体可能没有从绑定尺寸的变化对束缚态方面相当大的影响,但可以拥有博士上水合壳和/或电荷amatic影响。在分子与结合配偶体相互作用后的热泳运动的变化使得基本结合参数2,7,34,36,37的量化。
如在图1A中所描绘的MST装置由红外激光聚焦到使用相同的光学元件作为用于荧光检测的玻璃毛细管内的试样。而激光建立温度梯度(2-6℃的ΔT)可监控的蛋白质经由色氨酸6或荧光标记的相互作用配偶3,8的固有FL荧光的热泳的运动。在空间,ΔT,所产生的温度差会导致在升高的温度下的区域耗尽或分子的积累,这可通过索瑞量化系数音响cient(S T)的 :
G“/>
C 热表示在被加热区域的浓度,和c 冷是在初始冷区域中的浓度。
如图1B所示 ,一个典型的MST试验结果在MST运动轮廓(时间跟踪),由不同的相位,这可以通过它们各自的时间尺度来分离的。初始荧光在第一5秒中不存在温度梯度的测量来定义的精确开始荧光和检查漂白或photoenhancement。温度跳跃(T-跳转)表示相,其中热泳动前的荧光变化。荧光这个初始下降取决于uorophore量子产率FL的热依赖性变化。热泳相位如下,其中达到荧光减少(或增加),由于分子直至稳态分布的热泳运动。如在图1B中所示的激光被关闭后,可以观察到的反向TJump和FL uorescent分子伴随背面扩散。为了获得基本的绑定参数,相互作用伙伴不同的摩尔比进行了分析和比较。通常情况下,16个不同的比例进行了研究一个MST实验,而光可见分子被保持恒定,并且与未标记的配体的增加量供给。两个结合配偶之间的相互作用诱导的热泳的变化,并因此在规范化FL荧光,女范数 ,这是因为以下计算:
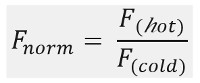
˚F 热和F 冷代表在MST痕迹德科幻奈德时间点的平均荧光FL强度。结合亲和力(K D或EC 50值)可以通过CURV计算易连接( 图1C)。
总体来看,MST是一个强大的工具来研究任何种类的分子间的相互作用。这份手稿提供了一个协议来表征小分子三磷酸腺苷(ATP 0.5 kDa的)之间的相互作用充满挑战和25-nt的单链DNA短适体DH25.42(7.9 kDa的)。在手稿的过程中,在ATP分子的适体的结合位点被映射下来到腺嘌呤基团的ATP。
Protocol
Representative Results
Discussion
质量控制:
非特异性粘着/样品材料的表面,以及聚集效应的吸附,对亲和数据的质量有巨大影响。但是,只有少数国家的最先进的技术提供准确,快速的选择,监督和避免这些影响。台MST提供了检测和帮助克服这些问题,从而允许技术设置的逐步优化集成质量控制。上粘附和荧光效果的重要信息可以从毛细管扫描和毛细管形状(步骤5.2和7.5)被提取,而聚集/沉淀的效果(步骤…
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
作者没有确认。
Materials
| Aptamer binding buffer | 20 mM Tris pH7.6; 300 mM NaCl; 5 mM MgCl2; 0.01%Tween-20 | ||
| Fluorescently labeled ATP aptamer | IDT, Leuven, Belgium | sequence: DH25.42 50-Cy5-CCTGGGGGAGT-ATTGCGGAGGAAGG-3 | |
| ATP | Sigma Aldrich, Germany | A2383 | 10 mM stock solutions stored at – 20 °C |
| ADP | Sigma Aldrich, Germany | A2754 | 10 mM stock solutions stored at – 20 °C |
| AMP | Sigma Aldrich, Germany | A2252 | 10 mM stock solutions stored at – 20 °C |
| Adenine | Sigma Aldrich, Germany | A8626 | 10 mM stock solutions stored at – 20 °C |
| SAM | Sigma Aldrich, Germany | A7007 | 10 mM stock solutions stored at – 20 °C |
| dATP | Sigma Aldrich, Germany | 11934511001 | 10 mM stock solutions stored at – 20 °C |
| CTP | Sigma Aldrich, Germany | C1506 | 10 mM stock solutions stored at – 20 °C |
| GTP | Sigma Aldrich, Germany | G8877 | 10 mM stock solutions stored at – 20 °C |
| Monolith NT.115 | NanoTemper Technologies, Munich, Germany | MO-G008 | Blue/Red Channel MST device with standard detector, Monolith NT115 pico is MST device with high sensitivity detector |
| Monolith NT.115 capillaries Standard | NanoTemper Technologies, Munich, Germany | MO-K002 | |
| Eppendorf PCR tubes | Eppendorf, Germany | 30124537 | |
| Monolith control software. 2.1.33, pre-installed on the device | NanoTemper Technologies, Munich, Germany | ||
| MO.affinity analysis v2.1.1 | NanoTemper Technologies, Munich, Germany | ||
| Kaleidagraph 4.5.2 | Synergy Software |
Riferimenti
- Linke, P., et al. An Automated Microscale Thermophoresis Screening Approach for Fragment-Based Lead Discovery. J Biomol Screen. 21 (4), 414-421 (2015).
- Jerabek-Willemsen, M., et al. MicroScale Thermophoresis: Interaction analysis and beyond. Journal of Molecular Structure. 1077, 101-113 (2014).
- Zillner, K., et al. Microscale thermophoresis as a sensitive method to quantify protein: nucleic acid interactions in solution. Methods Mol Biol. 815, 241-252 (2012).
- Zhang, W., Duhr, S., Baaske, P., Laue, E. Microscale thermophoresis for the assessment of nuclear protein-binding affinities. Methods Mol Biol. 1094, 269-276 (2014).
- Wienken, C. J., Baaske, P., Rothbauer, U., Braun, D., Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 1, 100 (2010).
- Seidel, S. A., et al. Label-free microscale thermophoresis discriminates sites and affinity of protein-ligand binding. Angew Chem Int Ed Engl. 51 (42), 10656-10659 (2012).
- Seidel, S. A., et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 59 (3), 301-315 (2013).
- Schubert, T., et al. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol Cell. 48 (3), 434-444 (2012).
- McKeague, M., Derosa, M. C. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012, 748913 (2012).
- Ruscito, A., DeRosa, M. C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front Chem. 4, 14 (2016).
- Chang, A. L., McKeague, M., Liang, J. C., Smolke, C. D. Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform. Anal Chem. 86 (7), 3273-3278 (2014).
- Chang, A. L., McKeague, M., Smolke, C. D. Facile characterization of aptamer kinetic and equilibrium binding properties using surface plasmon resonance. Methods Enzymol. 549, 451-466 (2014).
- Jing, M., Bowser, M. T. Methods for measuring aptamer-protein equilibria: a review. Anal Chim Acta. 686 (1-2), 9-18 (2011).
- Sokoloski, J. E., Dombrowski, S. E., Bevilacqua, P. C. Thermodynamics of ligand binding to a heterogeneous RNA population in the malachite green aptamer. Biochimica. 51 (1), 565-572 (2012).
- Burnouf, D., et al. kinITC: a new method for obtaining joint thermodynamic and kinetic data by isothermal titration calorimetry. J Am Chem Soc. 134 (1), 559-565 (2012).
- Mannironi, C., Scerch, C., Fruscoloni, P., Tocchini-Valentini, G. P. Molecular recognition of amino acids by RNA aptamers: the evolution into an L-tyrosine binder of a dopamine-binding RNA motif. RNA. 6 (4), 520-527 (2000).
- Jenison, R. D., Gill, S. C., Pardi, A., Polisky, B. High-resolution molecular discrimination by RNA. Science. 263 (5152), 1425-1429 (1994).
- Huizenga, D. E., Szostak, J. W. A DNA aptamer that binds adenosine and ATP. Biochimica. 34 (2), 656-665 (1995).
- Lee, E. R., Baker, J. L., Weinberg, Z., Sudarsan, N., Breaker, R. R. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 329 (5993), 845-848 (2010).
- Wickiser, J. K., Cheah, M. T., Breaker, R. R., Crothers, D. M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochimica. 44 (40), 13404-13414 (2005).
- Jucker, F. M., Phillips, R. M., McCallum, S. A., Pardi, A. Role of a heterogeneous free state in the formation of a specific RNA-theophylline complex. Biochimica. 42 (9), 2560-2567 (2003).
- Zhao, Q., Lv, Q., Wang, H. Aptamer fluorescence anisotropy sensors for adenosine triphosphate by comprehensive screening tetramethylrhodamine labeled nucleotides. Biosens Bioelectron. 70, 188-193 (2015).
- Zhang, D., et al. A sensitive fluorescence anisotropy method for detection of lead (II) ion by a G-quadruplex-inducible DNA aptamer. Anal Chim Acta. 812, 161-167 (2014).
- Elenko, M. P., Szostak, J. W., van Oijen, A. M. Single-molecule imaging of an in vitro-evolved RNA aptamer reveals homogeneous ligand binding kinetics. J Am Chem Soc. 131 (29), 9866-9867 (2009).
- Elenko, M. P., Szostak, J. W., van Oijen, A. M. Single-molecule binding experiments on long time scales. Rev Sci Instrum. 81 (8), 083705 (2010).
- Zichel, R., Chearwae, W., Pandey, G. S., Golding, B., Sauna, Z. E. Aptamers as a sensitive tool to detect subtle modifications in therapeutic proteins. PLoS One. 7 (2), 31948 (2012).
- Baaske, P., Wienken, C. J., Reineck, P., Duhr, S., Braun, D. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew Chem Int Ed Engl. 49 (12), 2238-2241 (2010).
- Entzian, C., Schubert, T. Studying small molecule-aptamer interactions using MicroScale Thermophoresis (MST). Methods. 97, 27-34 (2016).
- Valenzano, S., et al. Screening and Identification of DNA Aptamers to Tyramine Using in Vitro Selection and High-Throughput Sequencing. ACS Comb Sci. 18 (6), 302-313 (2016).
- Jauset Rubio, M., et al. beta-Conglutin dual aptamers binding distinct aptatopes. Anal Bioanal Chem. 408 (3), 875-884 (2016).
- Breitsprecher, D., et al. Aptamer Binding Studies Using MicroScale Thermophoresis. Methods Mol Biol. 1380, 99-111 (2016).
- Stoltenburg, R., Schubert, T., Strehlitz, B. In vitro Selection and Interaction Studies of a DNA Aptamer Targeting Protein A. PLoS One. 10 (7), 0134403 (2015).
- Kinghorn, A. B., et al. Aptamer Affinity Maturation by Resampling and Microarray Selection. Anal Chem. 88 (14), 6981-6985 (2016).
- Duhr, S., Braun, D. Why molecules move along a temperature gradient. Proc Natl Acad Sci U S A. 103 (52), 19678-19682 (2006).
- Braun, D., Libchaber, A. Trapping of DNA by thermophoretic depletion and convection. Phys Rev Lett. 89 (18), 188103 (2002).
- Duhr, S., Arduini, S., Braun, D. Thermophoresis of DNA determined by microfluidic fluorescence. Eur Phys J E Soft Matter. 15 (3), 277-286 (2004).
- Jerabek-Willemsen, M., Wienken, C. J., Braun, D., Baaske, P., Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 9 (4), 342-353 (2011).
- He, K., Dragnea, V., Bauer, C. E. Adenylate Charge Regulates Sensor Kinase CheS3 To Control Cyst Formation in Rhodospirillum centenum. MBio. 6 (3), 00546 (2015).
- Brvar, M., et al. Structure-based discovery of substituted 4,5′-bithiazoles as novel DNA gyrase inhibitors. J Med Chem. 55 (14), 6413-6426 (2012).
- Pogorelcnik, B., et al. 4,6-Substituted-1,3,5-triazin-2(1H)-ones as monocyclic catalytic inhibitors of human DNA topoisomerase IIalpha targeting the ATP binding site. Bioorg Med Chem. 23 (15), 4218-4229 (2015).
- Jhaveri, S., Rajendran, M., Ellington, A. D. In vitro selection of signaling aptamers. Nat Biotechnol. 18 (12), 1293-1297 (2000).
- Khavrutskii, L., et al. Protein purification-free method of binding affinity determination by microscale thermophoresis. J Vis Exp. (78), (2013).
- Ramakrishnan, M., et al. Probing cocaine-antibody interactions in buffer and human serum. PLoS One. 7 (7), 40518 (2012).
- Chen, M., et al. Antiviral activity and interaction mechanisms study of novel glucopyranoside derivatives. Bioorg Med Chem Lett. 25 (18), 3840-3844 (2015).
- Wan, C., et al. Insights into the molecular recognition of the granuphilin C2A domain with PI(4,5)P2. Chem Phys Lipids. 186 (4,5), 61-67 (2015).
- Harazi, A., et al. The Interaction of UDP-N-Acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase (GNE) and Alpha-Actinin 2 Is Altered in GNE Myopathy M743T Mutant. Mol Neurobiol. , (2016).

