Investigating Protein Sequence-structure-dynamics Relationships with Bio3D-web
Summary
A protocol for the online investigation of protein sequence-structure-dynamics relationships using Bio3D-web is presented.
Abstract
We demonstrate the usage of Bio3D-web for the interactive analysis of biomolecular structure data. The Bio3D-web application provides online functionality for: (1) The identification of related protein structure sets to user specified thresholds of similarity; (2) Their multiple alignment and structure superposition; (3) Sequence and structure conservation analysis; (4) Inter-conformer relationship mapping with principal component analysis, and (5) comparison of predicted internal dynamics via ensemble normal mode analysis. This integrated functionality provides a complete online workflow for investigating sequence-structure-dynamic relationships within protein families and superfamilies.
Introduction
The protein data bank (PDB) now contains more than 120,000 protein structures – many of which are of the same protein family but resolved under different experimental conditions. These multiple structures represent an invaluable resource for understanding the intricacies of protein form and function. For example, the rigorous comparison of these structure ensembles can reveal important molecular mechanisms 1,2,3 and inform on conformational dynamics involved in processes including ligand binding, enzymatic catalysis and bi-molecular recognition 4,5,6,7. New insights can often be obtained from the detailed large-scale analysis of the sequence, structure and dynamics of protein families. However, this typically requires considerable bioinformatics and computer programming expertise together with familiarity with the protein systems under study. For example, software packages such as Bio3D, ProDy and Maven require programing in R, python and Matlab, respectively 8,9,10. Conversely, online tools for analysis of structural flexibility are generally limited to the investigation of individual structures 11,12. An exception in this regard is the recently developed WebNM@ server, which allows for the comparison of flexibility patterns obtained from normal mode analysis (NMA) of several pre-aligned user specified structures13. However, this server lacks an automated procedure for the identification of structures for comparison, their alignment or further analysis beyond NMA. Another recent contribution is the online PDBFlex database, which presents pre-computed analysis of PDB structures sharing 95% or higher sequence identity14. However, analysis of more diverse structure sets is not currently available.
We have previously presented Bio3D-web – an easy to use web application for the analysis of protein sequence-structure-dynamic relationships15. Bio3D-web is unique in providing easy to use integrated functionality for the identification, comparison and detailed analysis of large homologous structure sets online. Here we present a detailed protocol for the online investigation of protein sequence-structure-dynamics relationship using Bio3D-web. Bio3D-web provides a variety of functions to support the five major steps of data analysis shown in Figure 1 and discussed in detail below. These steps constitute a workflow that spans from query sequence or structure input, through multiple levels of sequence-structure-dynamic analysis, to summary report generation. Results are available immediately through extensive in-browser visualization and plotting devices, as well as through downloading result files in commonly used formats. In addition to a convenient easy to use dynamic interface for exploring the effects of parameter and method choices, Bio3D-web also records the complete user input and subsequent graphical results of a user's session as a sharable reproducible report in PDF, DOC and HTML formats. User sessions may be saved and reloaded at future times and complete results downloaded and further interpreted by the Bio3D R package on a user's local machine.
Bio3D-web is powered by the Bio3D R package for analysis of biomolecular structure, sequence and molecular simulation data 8,16. In particular, Bio3D algorithms for rigid-core identification 8, superposition, principal component analysis (PCA) 8, and ensemble normal mode analysis(eNMA) 16 form the basis of the application. We also utilize Bio3D protocols that depend on pHMMER 17 for the identification of related protein structures, and MUSCLE 18 for multiple sequence alignment. Structure and sequence annotations are derived via Bio3D utilities from the RCSB PDB 19 and PFAM databases 20. Bio3D-web can be run from our online server or installed locally on any computer running R. Bio3D-web is open to all users and is provided free of charge under a GPL-3 open-source license from: http://thegrantlab.org/bio3d/webapps
Protocol
NOTE: A typical Bio3D-web session proceeds through five consecutive and dependent steps (see Figure 1 for a schematic representation). Each step is implemented as a consecutive navigation tab of the web application namely SEARCH, ALIGN, FIT, PCA, and eNMA.
1. Structure Search and Selection (SEARCH)
- Input structure
- Obtain the PDB ID of adenylate kinase (Adk), e.g. by searching the PDB [http://www.rcsb.org/pdb]. Alternatively, obtain the protein amino acid sequence of interest, e.g. from UniProt [http://uniprot.org].
- Enter the four character long PDB ID for Adk (e.g. 1AKE), or paste a protein sequence, to the text box in the "Input structure or sequence" panel.
- Hit selection
- Click the blue "Next" (Hit selection) button in the first panel or simply scroll down to panel B) "Hit selection" for further analysis.
- Make sure the "Limit total number of included structures" slider is set to its maximum value to include all structures above the cutoff.
- Lower the "Adjust the inclusion BitScore cutoff" to include more distantly related hits, or increase it to exclude.
- Optional hit filtering
- Click the blue "Next" (Hit selection) button in the first panel or simply scroll down to panel C) "Optional filtering of related structures for further analysis".
- Make sure the selected hits represent relevant structures by inspecting details of the table, e.g. PDB name, species, and bound ligands.
- Manually refine the selected subset of structures if needed by clicking the rows of the table.
NOTE: Rows highlighted with a blue color depict PDB IDs selected for further analysis in subsequent tabs.
2. Multiple Sequence Alignment Analysis (ALIGN)
- Click the ALIGN tab to perform sequence alignment of the selected structures from the SEARCH tab.
- Alignment summary
- Review the alignment summary in panel A) "Alignment summary". Make sure that the regions of interest are aligned and not masked by gaps in one or more structures.
- If needed, toggle the "Display alignment editing options" and remove unwanted PDB IDs, e.g. PDBs with missing residues.
- Sequence alignment analysis
- Click the blue "Next" (Analysis) button to perform sequence-based clustering analysis of the collected structures.
- Select the plot option Dendrogram. Adjust the Cluster into K groups slider to partition the structures into k groups.
- Optionally change the clustering method if desired by toggling the More clustering and output options checkbox.
- Residue conservation analysis
- Click the blue "Next" (Conservation) button to calculate the column-wise residue conservation.
- Select the Aligned structure sets to generate a plot of the residue conservation at each alignment position.
- Select Structures aligned with PFAM seed alignment to show conservation calculated with respect to the associated PFAM seed alignment containing representative members of the family.
- Sequence alignment display
- Click the blue "Next" (Alignment) button to show the full sequence alignment with the in-browser alignment visualization tool.
3. Structure Fitting and Analysis (FIT)
- Perform structure superimposition by entering the FIT tab.
- Structure superposition
- Toggle the "Show PDBs" checkbox to visualize the aligned protein structures in-browser.
- Make sure the protein structures are superimposed to corresponding and relevant regions by visual inspections. Click and drag the mouse over the structures to rotate, and scroll to zoom.
- Adjust the coloring of the structures by clicking on the "Color options". Coloring options include alignment position, structural variability per position, RMSD cluster groups, sequence cluster groups, aligned regions and secondary structure.
- Download the superposed structures as either conventional PDB files or as a single PyMOL session file for visualization in a specialized molecular viewer program.
- Structure analysis
- Click the blue "Next" (Analysis) button to perform structure-based clustering of the collected PDB structures.
- Toggle the RMSD Heatmap in the Plot options dropdown menu.
- Adjust the clustering options, including the clustering method itself, through toggling the "More clustering and output options" checkbox.
NOTE: Pairwise RMSD data can also be visualized as a dendrogram, a histogram or a heat map.
- Residue fluctuations
- Click the blue "Next" (RMSF) button to view the structural variability of each residue (shown as an RMSF plot) with major secondary structure elements shown in the marginal regions of the x-axis.
- Toggle the Show B-factors checkbox to overlay crystallographic B-factors of the reference structure onto the RMSF plot.
4. Principal Component Analysis (PCA)
- Perform principal component analysis by entering the "PCA" tab.
- Visualization of the principal components
- Toggle the "Show PC Trajectory" checkbox to visualize motions described by the PCs with the in-browser visualization tool.
- Make sure "Principal Component 1" is chosen from the first drop down menu.
- To visualize the motions described by other PCs, choose the desired PC from the "Choose Principal Component" drop down menu.
- Change the coloring of the trajectory from the "Color options" drop down menu.
- Choose "Variability per position" from the "Color options" to color by displacement magnitude.
- Click the "Download PDB trajectory" button in the "Principal Component Visualization" panel to obtain a trajectory view of the motion described by the PCs.
- Click the button "Download PyMOL" session file to generate a PyMOL session file giving the motions as a vector field.
- Conformer analysis
- Project the individual structures onto two selected PCs by clicking on the blue "Next" (Plot) button.
- Make sure "PC on X-axis" is set to 1, and "PC on Y-axis" to 2. To project the structures onto other PCs, adjust the PC numbering accordingly.
- Choose "Cluster by PC Subspace" to color the structures in the plot by PC-based clustering; "RMSD" to color by "RMSD-based" clustering; and "Sequence" to color by sequence based clustering.
- Click on any individual points in the plot to label the structures. Alternatively, highlight one or more structures in the table "PCA conformer plot annotation" below the plot.
- Slide the PCs in subspace slider to including more/less PCs for the clustering algorithm.
- Residue contributions
- Calculate the residue contributions to the individual PCs by clicking the blue "Next" (Residue contributions) button.
- Plot the contributions for additional PCs by including the PC number in the "Choose Principal Component" text box.
- Toggle the "Spread lines" checkbox avoid plotting the residue contributions on top of each other.
- Toggle off the "Multiline plot" checkbox to plot the residue contributions in separate plots.
- Toggle the "Show RMSF" to include the RMSF values (from the FIT tab).
5. Ensemble Normal Mode Analysis (eNMA)
- Click the eNMA tab to initiate normal modes (NMs) calculation.
- Filter structure
- Adjust the number of structures by lowering or increasing the "Cutoff" for structure inclusion/exclusion.
- Click the green "Run Ensemble NMA" to start the NMA calculation.
- Normal modes visualization
- Scroll down to the second panel of the eNMA tab (Normal Modes Visualization) for visualization of the NMs.
NOTE: By default, the NM with the highest overlap (similarity) to PC-1 is displayed in the visualization window. - To visualize the motions described by other NMs or other PDB structures, choose the desired NM and structure from the "Choose Mode" and "Show NMs for structure" drop down menus, respectively.
- Scroll down to the second panel of the eNMA tab (Normal Modes Visualization) for visualization of the NMs.
- Residue fluctuations
- Click the blue "Next" (Fluctuations) button to calculate the residue-wise fluctuations of structures selected for eNMA.
- Toggle the "Cluster by RMSD" to color the fluctuation profiles by RMSD-based clustering.
- Toggle the "Cluster by RMSIP" to color the fluctuation profiles by RMSIP-based clustering.
- Toggle the "Spread lines" checkbox to plot the grouped fluctuation profiles apart from each other.
- Comparing NMA and PCA
- Click the blue "Next" (PCA-vs-NMA) button to calculate the similarity between the individual NMs and PCs.
- Select a PDB ID from the "Compare NMs of structure" drop down menu to calculate the similarity between the NMs of this structure to the PCs calculated in the PCA tab.
- Overlap analysis
- Click the blue "Next" (Overlap analysis) button to calculate the overlap between calculated NMs and the structure difference vector between two selected structures.
- Select a 'reference' PDB ID from the "Compare NMs of structure" drop down menu and or one or more PDB IDs in the structure table for the pairwise comparison with the reference PDB.
- Clustering analysis
- Click the blue "Next" (Clustering) button to perform structure clustering based on pair-wise NM similarity (RMSIP).
Representative Results
Adenylate kinase (Adk) is a ubiquitous enzyme that functions to maintain the equilibrium between cytoplasmic nucleotides essential for many cellular processes. Adk operates by catalyzing the reversible transfer of a phosphoryl group from ATP to AMP. This reaction is accompanied by well-studied rate limiting conformational transitions 3,21. Here we analyze all currently available Adk structures with Bio3D-web to reveal detailed features and mechanistic principles of these essential transitions.
We can begin our Bio3D-web analysis of Adk by entering the RCSB PDB code of any known Adk structure. For example, entering the PDB ID 1AKE in panel A of the SEARCH tab returns 167 sequence similar structures from which the top 26 are automatically selected for further analysis (see panel B). The annotation presented in panel C indicates that these selected structures are all from E. coli, were solved by x-ray diffraction in a range of space groups; have a resolution range of 1.63 to 2.8 Å, and were co-crystalized with a range of different ligands (including no ligands, AMP, ADP, MG and the inhibitor AP5). Note that additional annotation details can be displayed by clicking on "Show/Hide Columns" option in panel C.
Multiple sequence alignment is performed upon entering the ALIGN tab. The first panel of the ALIGN tab displays a summary of the alignment providing details on the number of sequence rows (equivalent to the number of PDB structures), as well as the number of positions (i.e. alignment columns). This includes a specification of the number of gap and non-gap containing columns. The figure on the right hand side of the first row provides a schematic representation of the sequence alignment. Here the grey areas represent non-gap positions, while white areas in the alignment correspond to gaps. A representation of the sequence conservation is shown above the alignment with red areas indicating well-conserved positions, and white indicating less conserved. Note that the sequences in this figure are ordered based on their similarity provided by the clustering dendrogram on the left hand side. The second panel of this tab further facilitates clustering of the selected PDBs based on their pair-wise sequence similarity, which can be visualized either as a dendrogram or a heat map. By default, a dendrogram (or tree diagram) representing the arrangement of clusters is shown. The y- axis of the dendrogram represents the distance (in terms of sequence identity) between the clusters.
Structure superposition is performed automatically upon entering the FIT tab. The superimposed structures, displayed interactively in panel A, indicate the presence of a relatively rigid core region (encompassing residues 1-29, 68-117, and 161-214; see the 'optional core and RMSD details' panel at the bottom of the FIT tab for details). Two more variable nucleotide-binding regions (residues 30-67 and 118-167) are also clearly visible (Figure 2). RMSD-based clustering groups these structures into two distinct conformations.
Clicking on the PCA tab more clearly shows the relationship between the structures in terms of the displacements of these regions that effectively close over the bound nucleotide species in related structures (Figure 2B and 2C). The majority of structures are in the 'closed' form (blue in Figure 2C) and are associated with a bound ligand or inhibitor. In contrast more 'open' conformations are nucleotide and inhibitor free. This is consistent with the extensive body of research on Adk structure and dynamics indicating that an open configuration of these regions is required for nucleotide binding and a closed conformation for efficient phosphoryl transfer and suppression of detrimental hydrolysis events. It is notable that a single PC captures 97% of the total mean square displacement in this Adk structure set and provides a clear and compelling description of the open to closed transition along with the individual residue contributions to this functional displacement (panel C of the app and Figure 2).
Visiting the NMA tab and increasing the number of structures considered for calculation (via decreasing the cutoff for filtering similar structures) indicates that open state structures display enhanced local and global dynamics in comparison to the closed form structures (Figure 2D and panel C of app). Comparing PCA and NMA results for individual structures (panel D) indicates that the first mode of all open form structures displays a relatively high overlap to PC1 (with a mean value of 0.37 ± 0.04). In contrast, closed form structures display lower values (with a mean of 0.30 ± 0.01). RMSIP values for open form structures (0.62 ± 0.003) are also higher than those of closed structures (0.56 ± 0.008). In addition, overlap analysis shows that the first modes of the open state are in good agreement with the conformational change that describes the difference of the open and closed states (panel E). Clustering based on RMSIP values again displays a consistent partitioning of open and closed state structures (panel F).
Collectively these results indicate the existence of two major distinct conformational states for Adk. These differ by a collective low frequency displacement of two nucleotide-binding site regions that display distinct flexibilities upon nucleotide binding.
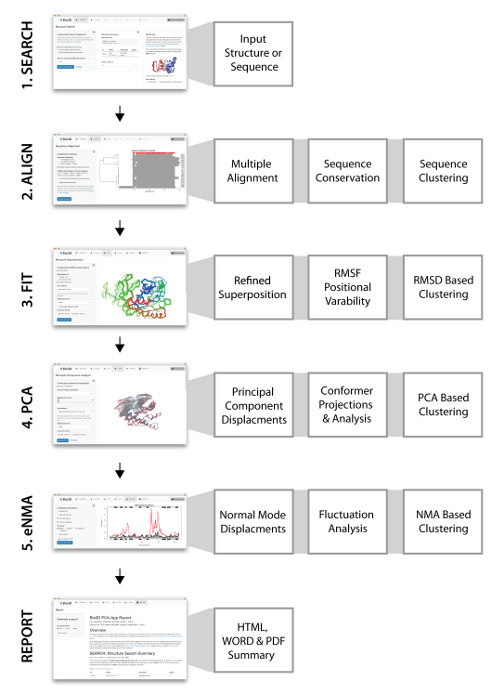
Figure 1: Bio3D-web overview with screen shots of the PCA and NMA tabs. Bio3D-web takes a user provided protein structure or sequence as input in the SEARCH tab (1). The server provides a list of related structures, which can be selected for further analysis. (2) The ALIGN tab provides sequence alignment and analysis of the structures selected in the SEARCH tab. (3) In the FIT tab all structures are superimposed and visualized in 3D together with the results of conventional pair-wise structure analysis. (4) Principal component analysis of the structure set is performed in the PCA tab to characterize inter-conformer relationships. (5) Normal mode analysis on each structure can be carried out in the eNMA tab to explore dynamic trends for the available structural states. Please click here to view a larger version of this figure.
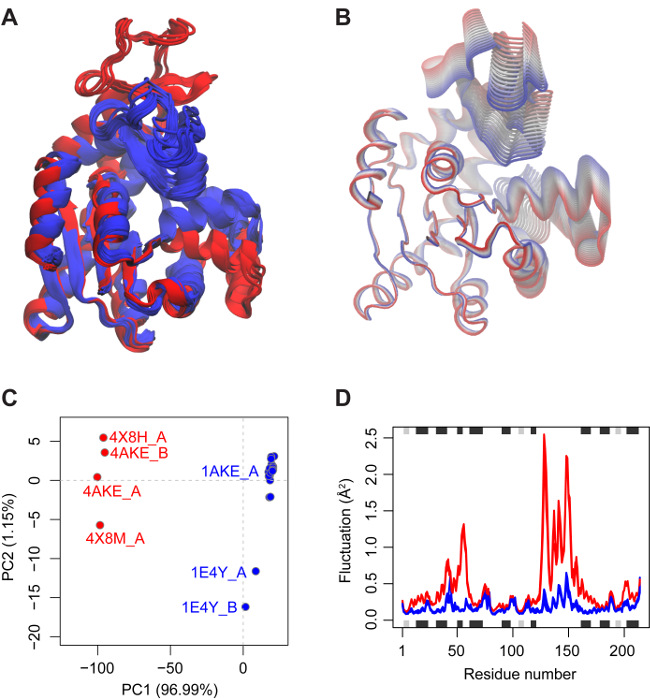
Figure 2: Results of Bio3D-web analysis of adenylate kinase. (A) Available PDB structures of adenylate kinase superimposed on the identified invariant core. Structures are colored according to RMSD-based clustering provided in the FIT tab. (B) Visualization of the principal components is available from the PCA tab to characterize the major conformational variations in the data set. Here, the trajectory corresponding to the first principal component is shown in tube representation showing the large-scale closing motion of the protein. (C) Structures are projected onto their two first principal components in a conformer plot showing a low-dimensional representation of the conformational variability. Each dot (or structure) is colored according to user specified criteria, in this case PCA-based clustering results. (D) Normal mode analysis in the eNMA tab suggests enhanced local and global dynamics for structures in the open state (red) in comparison to the closed form (blue) structures. Please click here to view a larger version of this figure.
Discussion
Bio3D-web can be used to interactively explore and map the structural, dynamic and functional states of proteins from available crystallographic structures. Furthermore, the NMA and PCA based clustering results, together with the annotations and sequence based analysis, can be particularly useful for selecting representative structures for more time-consuming analysis such as ensemble small molecule docking or molecular dynamics simulations. Bio3D-web thus facilitates advanced structural bioinformatics analysis for a broader range of researchers by reducing the required level of technical expertise. The current design of Bio3D-web emphasizes simplicity over exhaustive inclusion of the many analysis methods available in the full standalone Bio3D package. In many cases it is envisaged that researchers will use Bio3D-web to understand general trends in their protein family or superfamily of interest, which may then inform more specialized analyses. Bio3D-web is therefore designed to quickly explore biomolecular structure datasets and to act as a hypothesis-generating tool. We encourage users to further explore their data by providing example Bio3D code in the reproducible report that also stores all query details and analysis results.
In the representative example protocol above, we show the capability of Bio3D-web to reveal the structural features of functional conformational transitions of Adk. Additional applications of Bio3D-web include structural and dynamics analysis of user-uploaded PDB structures. For example, the user can upload new structures or indeed protein sequences for analysis. The analysis steps mentioned earlier, especially the eNMA step, can reveal both local and global trends in protein motions, with collective motions being of functional significance. Comparison with apo structures can also reveal characteristics of unbound to bound conformational transitions. Additional examples of application to a range of different protein families are provided online.
Although all proteins are flexible and dynamic entities, not all proteins have atomic resolution structures available in a range of different states (e.g. active and inactive states). Our view of protein structure space is thus a limited one and hence the insight obtained from tools such as Bio3D-web is necessarily also limited for certain proteins. However, with current technological advances and new initiatives for structural genomics the protocol presented here will increasingly become an important route for gaining insight into important structure-function relationships. A critical step, which is particularly important when analyzing more distantly related proteins, is the potential emergence of alignment errors in the ALIGN tab. Alignment errors will inevitably occur when sequence similarity drops below 30% and the user must in such cases double check and correct the sequence alignment in the ALIGN tab. Alignment errors will possibly result in incorrect superimposed structures in the FIT tab and mask the most relevant conformational variations for the subsequent PCA. In addition, the user should be aware of missing residues in the selected PDB structures as in the current implementation PCA can only be performed on protein residues in which all structures have their corresponding carbon alpha atom resolved. Consequently, if a selected PDB has unresolved residues for a particular region of the protein this region will be omitted from PCA.
Bio3D-web is currently limited to the analysis of single chain PDB structures. Consequently, functional motions occurring at the quaternary level cannot be explored using the current protocol. Although we are currently developing new algorithms to include such analysis in Bio3D-web, the only current option is through conventional Bio3D use.
Bio3D-web is the only online application that makes it possible to query and identify structure sets, interpret their patterns of sequence and structural variability, and extract mechanistic information from both analysis and prediction of their structural plasticity. A wide range of molecular visualization tools and online servers enable researchers to explore and analyse individual biomolecular structures. However, existing tools for analysis of the sequence, structure and dynamics of large heterogeneous protein families often require significant computational expertise and typically remain accessible only to users with relevant programming skills. For example, the Bio3D package requires R 8, ProDy requires python and Maven requires Matlab knowledge 9,10. Bio3D-web in contrast requires no programming knowledge and thus increases the accessibility and decreases the entry barrier to performing advanced comparative sequence, structure and dynamics analysis. Furthermore, the preparation, curation, annotation and clean-up of molecular structures that is frequently necessary for efficient analysis is included with the Bio3D-web service. Additionally, the restriction to performing such analysis on capable computational resources is alleviated by our server instance that enables large-scale analysis of many structures that can be initiated and controlled from any modern web browser.
Open development of Bio3D-web is on-going (see https://bitbucket.org/Grantlab/bio3d). We continue to add new analysis functionality and improve existing methods. Future development will focus on the addition of distance matrix based PCA and torsional PCA, more extensive sequence conservation approaches that include a phylogenetic component, ensemble binding site identification, and new approaches for dynamic network analysis across protein families. In this respect the current web application represents the starting point for many other collaborative structural bioinformatic analysis workflows by enabling reproducible and shareable steps on user defined experimental structure sets. We also plan future support of reconstructed biological unit coordinate sets in addition to individual and multiple chains from the asymmetric unit of PDB structures. Additional features will include enhanced saving and loading of collaborative workspaces together with an undo possibility.
Bio3D-web is an online application for interactive analysis of biomolecular structure data. Bio3D-web runs on any modern Web browser and provides functionality for: (1) The identification of related protein structure sets to user specified thresholds of similarity; (2) Their multiple alignment and structure superposition; (3) Sequence and structure conservation analysis; (4) Inter-conformer relationship mapping with principal component analysis, and (5) comparison of predicted internal dynamics via ensemble normal mode analysis. This integrated functionality provides a complete workflow for the investigation of sequence-structure-dynamic relationships within protein families and superfamilies. In addition to a convenient easy to use dynamic interface for exploring the effects of parameter and method choices, Bio3D-web also records the complete user input and subsequent graphical results of a user's session. This allows users to easily share and reproduce the sequence of analysis steps that created their results.Bio3D-web is implemented entirely in the R language and is based on the Bio3D and Shiny R packages. It can be run from our online server or installed locally on any computer running R. This includes local server installation to provide a customized multi user instance with access to priority structural datasets such as those common in the pharmaceutical industry. Full source code and extensive documentation is provided under a GPL-3 open-source license from: http://thegrantlab.org/bio3d/webapps
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Guido Scarabelli and Hongyang Li for extensive testing throughout development as well as the Bio3D user community and the University of Bergen structural bioinformatics workshop participants for feedback and comments that have improved this application.
Materials
| Bio3D-web | |||
| Web-site | http://thegrantlab.org/bio3d-web/ | ||
| Requirements | Web browser |
Riferimenti
- Kornev, A. P., Taylor, S. S. Dynamics-Driven Allostery in Protein Kinases. Trends Biochem. Sci. 40 (11), 628-647 (2015).
- Yao, X. -. Q., Grant, B. J. Domain-opening and dynamic coupling in the α-subunit of heterotrimeric G proteins. Biophys. J. 105 (2), L08-L10 (2013).
- Henzler-Wildman, K. A., et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 450 (7171), 838-844 (2007).
- Boehr, D., Nussinov, R., Wright, P. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5 (11), 789-796 (2009).
- Teilum, K., Olsen, J. G., Kragelund, B. B. Functional aspects of protein flexibility. Cell Mol Life Sci. 66 (14), 2231-2247 (2009).
- Henzler-Wildman, K., Kern, D. Dynamic personalities of proteins. Nature. 450 (7172), 964-972 (2007).
- Grant, B. J., Gorfe, A. A., McCammon, J. A. Large conformational changes in proteins: signaling and other functions. Curr. Opin. Struct. Biol. 20 (2), 142-147 (2010).
- Grant, B. J., Rodrigues, A. P. C., ElSawy, K. M., McCammon, J. A., Caves, L. S. D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 22 (21), 2695-2696 (2006).
- Bakan, A., Meireles, L. M., Bahar, I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 27 (11), 1575-1577 (2011).
- Zimmermann, M. T., Kloczkowski, A., Jernigan, R. L. MAVENs: motion analysis and visualization of elastic networks and structural ensembles. BMC Bioinformatics. 12 (1), 264 (2011).
- Yang, L. -. W., et al. oGNM: online computation of structural dynamics using the Gaussian Network Model. Nucleic Acids Res. 34, 24-31 (2006).
- Suhre, K., Sanejouand, Y. -. H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 32, W610-W614 (2004).
- Tiwari, S. P., et al. WEBnm@ v2.0: Web server and services for comparing protein flexibility. BMC Bioinformatics. 15 (1), 427 (2014).
- Hrabe, T., et al. PDBFlex: exploring flexibility in protein structures. Nucleic Acids Res. 44, D423-D428 (2016).
- Skjærven, L., Jariwala, S., Yao, X. -. Q., Grant, B. J. Online interactive analysis of protein structure ensembles with Bio3D-web. Bioinformatics. , (2016).
- Skjærven, L., Yao, X., Scarabelli, G., Grant, B. J. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinformatics. 15 (399), 1-11 (2014).
- Eddy, S. R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7 (10), (2011).
- Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792-1797 (2004).
- Berman, H. M. The Protein Data Bank. Nucleic Acids Res. 28 (1), 235-242 (2000).
- Finn, R. D., et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222-D230 (2014).
- Kerns, S. J., et al. The energy landscape of adenylate kinase during catalysis. Nat. Struct. Mol. Biol. 22 (2), 124-131 (2015).

