Visual and Microscopic Evaluation of Streptomyces Developmental Mutants
Summary
Here we present protocols for novice researchers to initiate phenotyping for the pharmacologically important bacterial genus Streptomyces.
Abstract
Streptomycetes are filamentous soil bacteria belonging to the phylum Actinobacteria that are found throughout the world and produce a wide array of antibiotics and other secondary metabolites. Streptomyces coelicolor is a well-characterized, non-pathogenic species that is amenable to a variety of analyses in the lab. The phenotyping methods described here use S. coelicolor as a model streptomycete; however, the methods are applicable to all members of this large genus as well as some closely related actinomycetes. Phenotyping is necessary to characterize new species of Streptomyces identified in the environment, and it is also a vital first step in characterizing newly isolated mutant strains of Streptomyces. Proficiency in phenotyping is important for the many new researchers who are entering the field of Streptomyces research, which includes the study of bacterial development, cell division, chromosome segregation, and second messenger signaling. The recent crowdsourcing of antibiotic discovery through the isolation of new soil microbes has resulted in an increased need for training in phenotyping for instructors new to the field of Streptomyces research and their college or high school students. This manuscript describes methods for bacterial strain propagation, storage, and characterization through visual and microscopic examination. After reading this article, new researchers (microbiology education laboratories and citizen scientists) should be able to manipulate Streptomyces strains and begin visual characterization experiments.
Introduction
Streptomycetes are Gram-positive, filamentous soil bacteria known for their ability to produce a variety of secondary metabolites, including over two-thirds of the commercially available antibiotics, as well as anti-tumor, anti-HIV, and anti-parasitic drugs1. S. coelicolor is the most genetically characterized member of the genus2,3 and is the species used in the methods described here. S. coelicolor has a complex life cycle that begins with germination of a single spore, progressing to an extensive, branching vegetative mycelium that grows into the agar medium. As the life cycle proceeds, aerial filaments are formed that break the surface tension of the substrate mycelium and are finally divided into long chains of cells that are ultimately converted into mature, grey-pigmented spores. Dispersion of these newly formed spores constitutes the beginning of the next life cycle4.
Because of its complex pattern of differentiation, S. coelicolor serves as an excellent model for the study of bacterial development. Historically, mutations result in blocks for two major stages of development and produce distinct visual phenotypes. The bld (bald) mutants are blocked for aerial mycelium formation and result in the lack of a fuzzy aerial mycelium imparting a "bald" colony appearance. Mutants inhibited in spore formation and maturation are referred to as whi (white) mutants because they typically fail to produce wild-type levels of the gray spore pigment and the aerial mycelium remains white. Other interesting mutants are inhibited in antibiotic production, cell division, chromosome segregation, or other important processes4,5.
Despite the discovery of many developmental genes in Streptomyces species, there are many more believed to exist based on the lack of saturation of mutant screens. Our laboratories continue to identify new developmental genes using a mini-transposon system that we have constructed. Novel mutant strains isolated in random mutagenesis experiments with our transposon undergo phenotypic screening to identify the possible role of each new gene discovered6,7. The methods for bacterial phenotyping described here are relevant to Streptomyces mutants isolated by transposon mutagenesis8,9,10,11,12,13 and other random methods, such as chemical and ultra-violet (UV) mutagenesis14, as well as the directed construction of mutations such as gene deletions using recombineering15,16 or CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) genome editing technology17,18,19 and point mutations20.
As antibiotic resistance among pathogens becomes increasingly prevalent, the need for new antibiotics becomes increasingly urgent21,22. The "Small World Initiative" (or SWI) is an effective Science, Technology, Engineering and Mathematics (STEM) learning23 and research strategy24 to combat antibiotic resistance through the crowdsourcing of college students, and more recently high school students. Supported courses work includes identifying new soil microbes that produce novel antibiotics (http://www.smallworldinitiative.org). It is believed that a major source of undiscovered antibiotics will continue to be Streptomyces species found in a wide range of soil and water habitats25,26,27,28,29,31. Recently, our laboratory and the work of others have discovered and characterized signaling genes in Streptomyces species that regulate morphology and development, including antibiotic production7,32,33. Changes in expression of these genes result in a change in the amount and timing of antibiotics produced. Skilled phenotyping of new species and new antibiotic-producing mutants will continue to be important. As new instructors and their students become major contributors to the field of drug discovery, training in bacterial phenotyping is necessary for the success of these novice individuals. Furthermore, these experiments are tractable for high school STEM or university microbiology education laboratories. They represent a demonstration of basic microbial genetic principles in a teaching lab setting.
Protocol
1. Prepare Instruments, Culture Media, Solutions, and Petri Dishes
- Autoclave toothpicks in a 50 mL beaker covered with aluminum foil to keep sterile.
- Sterilize all dry goods that will be used (tips, sticks, glassware, etc.) in an autoclave.
NOTE: Caution! Follow all safety directions for the autoclave model used. Autoclaves pose a risk for explosions, and burns from steam and contact with hot surfaces, materials, and media. For autoclave safety, view "Proper Use of Autoclaves"34. - Prepare the MS agar (Mannitol Soya Flour (SFM))14 to grow cultures.
- Add 5 g of soya flour and 5 g of agar to 1 L flask.
- Dissolve 5 g of mannitol in 250 mL of tap water and dispense into the flask containing the soy flour and agar.
- Add a foam plug and foil to the opening of the flask and autoclave at 121 °C, 15 psi for at least 30 min.
CAUTION: This medium easily boils over. Use a flask that can hold at least four times the actual volume of MS agar being autoclaved. A standard pressure cooker can be used in place of an autoclave with similar safety considerations. Alternatively, a microwave oven may be used to sterilize media and materials if access to an autoclave is not possible35,36. High school teachers may also want to collaborate with local colleges or universities to access autoclaved supplies. - Cool media until comfortable enough to handle. Be careful to not cool it for too long because agar solidifies at temperatures below 50 °C.
- Pour MS agar from the flask into sterile Petri dishes until the bottom of each Petri dish is approximately half full (approximately 25 mL of media per Petri dish). Allow to solidify before using agar plates. Store agar plates in a plastic bag at room temperature until needed.
NOTE: Agar plates may also be stored in a refrigerator, especially if antibiotics are needed in the media.
2. Streak Streptomyces onto Plates for Propagation
- Streak Streptomyces strains onto MS agar plates to single colonies using a standard method, such as the quadrant streak method as shown in Sanders, 201237.
NOTE: An inoculating loop or sterile toothpicks may be used. Optional: Other media are commonly used for Streptomyces species, such as R2YE and minimal media14. - Incubate plates at 30 °C.
- Restreak plates by picking a single colony every 4–6 days. As colonies age they become prone to random mutation.
3. Create Glycerol Mycelial Stocks for Bacterial Storage
NOTE: Mycelial stocks may be created for all Streptomyces strains, including whi and bld strains, and other developmental mutants that are unable to complete sporulation.
- Macerate 15–20 colonies in 200–500 µL of 20% glycerol using a sterile wooden dowel applicator.
- Transfer the slurry of macerated colonies to a 1.5 mL freezer culture tube, using a micropipette or sterile transfer pipette.
NOTE: The basics of micropipette use are covered elsewhere38. - Add enough 20% glycerol to make the final volume 1 mL and gently mix. Store samples at -80 °C.
NOTE: Caution! Use cryogenic safety gloves and a lab coat to protect skin from contact with extreme cold. Alternatively, a -20 °C freezer may be used with potential reduction in long term storage time. Avoid excessive freezing and thawing to extend viability.
4. Create Glycerol Spore Stocks for Strains Capable of Sporulation
NOTE: Spore stocks are preferable for long-term viability but are only feasible for strains that are capable of completing sporulation.
- Spread 100 µL of macerated material (from step 3.1) on an agar plate to obtain a confluent lawn of growth37.
NOTE: Spread plating is shown in Sanders37 (Modification: A turntable is not required.). The plate can be turned with one hand while using the other hand to move the spreader in a gentle back and forth motion across the agar media.- Pipet 100 µL of macerated mycelium to the center of an MS agar plate.
- Sterilize a glass or metal spreading tool by partially submerging in 70% ethanol and passing the spreader once slowly through a Bunsen burner flame to speed the evaporation of the ethanol.
CAUTION: Ethanol is highly flammable. Do not place the flaming spreader into ethanol or allow the flaming drop of ethanol to fall into the ethanol dish. Alternatively, use a disposable sterile cotton swab to spread the bacteria on the plate, which requires no ethanol or flame. - Turn the MS agar plate with one hand, while using a back and forth motion to inoculate the plate with the sterile spreader.
- Incubate for 5–7 days at 30 °C until the strain is well-sporulated.
NOTE: Incubation times will vary depending upon the species and strain.
- Add 2 mL of sterile saline (0.8% NaCl) to the center of a confluent lawn of Streptomyces cells that has undergone sporulation.
- Harvest spores by gently rubbing the surface of the mycelium with a sterile inoculating loop (or a cotton bud), slowly moving towards the edge of the lawn.
- Pipet the saline containing spores into a 15 mL conical tube. Vortex vigorously to break spore chains into individual cells.
- Centrifuge at room temperature for 5 min at approximately 2,500 x g.
- Pipet the supernatant and discard. Disperse the spore pellet by vigorous vortex agitation in the small amount of remaining liquid.
- Resuspend the spores in 1 mL of 20% glycerol by drawing the solution up and down with a pipette.
- Transfer the glycerol spore stock to a 1.5 mL freezer tube. Store at -80 °C.
CAUTION: Use cryogenic safety gloves and a lab coat to protect skin from contact with extreme cold. Alternatively, a -20 °C freezer may be used with potential reduction of viability in long term storage time. Avoid excessive freezing and thawing to extend viability.
5. Perform Mutagenesis
- Perform mutagenesis based on the desired outcomes as referenced in the Introduction and recently reviewed by Baltz, 201639.
NOTE: Some phenotyping experiments will not require mutagenesis, such as the intent to identify new species from the environment. Methods of random mutagenesis include the use of a transposon8,9,10,11,12,13, UV radiation, or chemicals14. Site-directed mutagenesis15,16,17,18,19,20 techniques can be used to create specific point mutations, deletions, or other types of mutations.
6. Compare and Record the Visual Appearance
- Take note of colony morphology for each mutant as compared to the wild-type parent strain after streaking all strains as in step 2. Compare the new isolate to that of a well-studied species, such as S. coelicolor or S. venezuelae, when characterizing new Streptomyces species. Note characteristics (Figure 1) such as basic shape, surface of colony (fuzzy, bald, wrinkled, etc.), opacity, elevation, and pigmentation (distinguish between vegetative mycelium, aerial mycelium, and/or surrounding medium).
- Label an MS agar plate and streak wild-type and mutant strains in wedge patterns (Figure 1). Be careful that no strain touches another strain to avoid cross contamination. Note the date and time that strains are streaked onto the plate. Incubate the plates at 30 °C.
- Place grown Petri dishes on colored or white paper to homogenize the background. Write on the paper the strain name, date, incubation temperature, and time from first plate streaking. Take digital picture(s) of the plate with the latter information written on the paper background so that confusion is minimized.
NOTE: The writing can be cropped from the photograph at a later time if it is to be used for figure preparation.
7. Perform Phase-Contrast Microscopy
- Sterilize tweezers by washing in 70% ethanol and then passing through a Bunsen burner flame to evaporate the ethanol. Allow to cool.
- Sterilize coverslips by washing in 70% ethanol and then allowing them to dry in a sterile Petri dish.
- Prepare a coverslip lift of the bacterial growth to be examined.
- Pick up a sterile coverslip with sterile tweezers and place the coverslip on an MS agar plate where bacterial growth is dense. Press on the back of coverslip gently with tweezers to ensure sufficient transfer of bacterial spores and aerial mycelium.
- Pick up the coverslip from the plate and place it so that the side with the cell material is facing toward the surface of a microscope slide, with a 15 µL drop of 50% glycerol prepared in water for mounting. Reduce air bubbles by placing the coverslip at a 45° angle to the slide and let it fall onto the 50% glycerol.
- (Optional) Seal with clear nail polish for preservation of the slide.
- Perform phase-contrast microscopy as described by Frohlich40 at various time intervals during the life cycles.
NOTE: Repeated imaging allows for a more complete analysis and the ability to detect delays in development. Independently repeat the observations to ensure accuracy and reproducibility.- Place the prepared slide on the microscope stage and add a drop of immersion oil to the center of the cover slip. Rotate the 100X phase objective in place and set the condenser turret to the proper "matching" phase setting. Focus the image using only the fine adjustment knob once the objective lens is in contact with the oil.
- Examine several fields of view to discern noticeable and consistent differences between mutant strains and the wild type, such as the ability to form spores, spore size and shape, and overall number of spores (see Figure 2).
NOTE: Alternatives to phase-contrast microscopy include differential interference contrast (DIC) microscopy40 and simple staining techniques for use with bright field microscopes41, such as crystal violet staining.
8. Perform Fluorescence Microscopy
- Sterilize tweezers by washing in 70% ethanol and then running through a Bunsen burner flame.
- Pick up a sterile coverslip with sterile tweezers and place on an MS agar plate where bacterial growth is evident. Touch the back of the coverslip gently with the tweezers to ensure sufficient transfer of bacterial spores and aerial filaments.
- Pick up the coverslip from the plate and place it so that the side with the cell material is facing up, on cardstock for ease in manipulation of the coverslip.
- Flood the coverslip with ice-cold methanol (~300 µL for a 20 x 20 mm2 coverslip) and let it completely air dry.
CAUTION: Methanol is a mutagen. Avoid direct contact with skin. - Wash the coverslip three times in Phosphate Buffered Saline (PBS)42 by gently dispensing and removing the solution.
- Add 15 µL of 100 μg/ml Propidium Iodide and/or 10 μg/mL Wheat-Germ-Agglutinin conjugated to fluorescein isothiocyanate (WGA-FITC) made in 50% glycerol to a labeled microscope slide.
NOTE: Propidium iodide stains DNA red to detect chromosomal DNA location while WGA-FITC stains the cell wall green. - Invert coverslip face down onto the drop of fluorescent stain on the glass slide. (Optionally) seal with clear nail polish for preservation of slide. As the dyes are light-sensitive, incubate for 15 minutes in a darkened or dimly lit room.
- Observe the slides under a 100X objective lens using a phase-contrast or DIC microscope equipped with epifluorescence excitation cubes for propidium iodide ('Texas Red' Filter Set or 535/617 nm excitation/emission maxima) and FITC (FITC filter set or 490/525 nm excitation/emission maxima).
- Place the slide on the stage and focus using the coarse and fine adjustment knobs.
- Observe the fluorescence as shown elsewhere43.
- Use a free image analysis software package that can help in the identification of spores with lower fluorescence intensity44.
Representative Results
Initial phenotyping experiments are necessary for characterizing new species and strains and can be used as a complimentary approach to the phylogenetics and DNA-DNA hybridization experiments that are used for characterizing new species. Streptomyces mutants resulting from random mutagenesis methods such as chemical, UV, or transposon mutagenesis are typically identified through direct, visual screens on agar plates. Colonies of Streptomyces are examined for changes in phenotype in comparison to the wild-type, parental strain. For example, a lighter colored aerial mycelium may indicate a lower level of gray pigment caused by a defect in sporulation, or the lack of a fuzzy appearance is indicative of a block in aerial mycelium formation (Figure 1). Many streptomycetes produce pigmented antibiotics in vegetative mycelium or surrounding agar. S. coelicolor produces two pigmented antibiotics as well as two non-pigmented ones. Actinorhodin is a blue-pigmented antibiotic and undecylprodigiosin is a red-pigmented antibiotic45. Strains that have undergone initial visual colony screens are then propagated by streaking for single colonies.
Following visual identification of potentially interesting mutants, strains are subjected to microscopic examination. Phase-contrast microscopy is especially suited to examining Streptomyces mutants for developmental defects, using the wild-type strain as a control (Figure 2). Wild type S. coelicolor colonies typically produce an aerial mycelium by about two days of growth at 30 °C on MS agar, and long chains of spores by three days of growth. The life cycle will progress slightly slower or faster on other types of media. It is imperative that the mutants be analyzed under the same growth conditions as the wild type when drawing conclusions about the developmental defects and delays. Bald mutants may produce spores after prolonged growth on agar media. A class of mutants referred to as white mutants may either be delayed for spore formation, show a reduction in the abundance of spores produced, produce spores with shape and/or size defects, or simply produce lower levels of the mature, gray spore pigment46. Other mutants investigated thus far may be delayed or accelerated in the life cycle progression such as the mutants affected for the accumulation of signaling molecules (e.g., cyclic-di-GMP47).
A simple follow-up to the light microscopy technique described above is fluorescence microscopy using propidium iodide to stain chromosomal DNA nucleoids and fluorescently labeled wheat germ agglutinin to stain the cell wall of sporulation septa. Spores lacking a chromosome will be devoid of the red propidium iodide staining, while spores that contain less DNA than usual may be identified if they appear to have decreased staining (Figure 3). Wheat germ agglutinin may be used to stain the cell wall and differences in cell wall staining patterns (i.e., cell division defects) may be observed. In Figure 4 the wild-type strain of S. venezuelae, a species that is recently being used as another model streptomycete because of its faster life cycle and ability to sporulate in liquid45, has been stained with WGA-FITC to elucidate the ladder-like array of division septa that are typically seen early during sporulation.
The initial phenotyping strategies described here should result in the following: 1) Identification of mutant strains of interest for further study; 2) Acquired knowledge about the newly identified mutant and the potential role of the gene that has been mutated; 3) The formulation of a subsequent series of next experimental steps that can be used to further clarify the role of the gene in question. In the case of a newly identified streptomycete from the environment, the researcher will gain knowledge of the potential new species compared to already characterized species of Streptomyces.
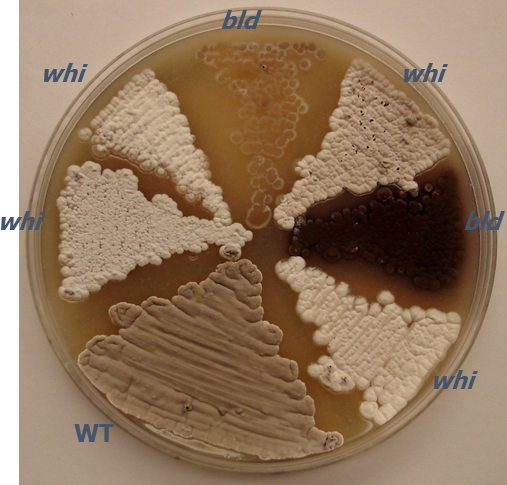
Figure 1: Photograph of a representative agar plate demonstrating macroscopic colony phenotypes of S. coelicolor. The agar plate shows the fuzzy, gray visual appearance of the aerial mycelium for wild-type S. coelicolor strain MT1110 (WT) in comparison to various random transposon insertion mutants with developmental phenotypes. Mutants are either lacking an aerial mycelium [bald (bld)] or an aerial mycelium with reduced spore pigmentation [white (whi)]. Transposon mutants were generated using mini-Tn5 for insertion mutagenesis6,7. The strains were grown on MS agar for 5 days at 30 °C. Petri dish diameter, 100 mm.
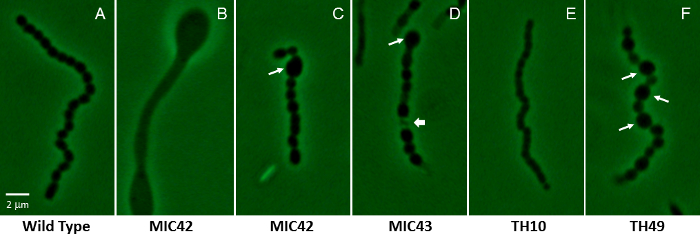
Figure 2: Phase-contrast micrographs showing a representative aerial filament for S. coelicolor white mutant strains containing random transposon insertion mutations. (A) Shown is a representative wild type aerial filament that has undergone synchronous, regularly-spaced cell divisions, producing evenly shaped and sized spores (MT1110). (B–F) The remaining panels show the vastly different microscopic phenotypes of various white mutants that were isolated via random transposon mutagenesis. Panel B shows an aerial filament that has not undergone sporulation, but instead has produced bulging areas within the filament. Long arrows in C, D, and F indicate abnormally large spores that are characteristic of mutants MIC42, MIC43, and TH49. The short arrow in panel D indicates an example of a lysed spore compartment. Panel E shows the partially constricted smaller compartments that are typical of the spore chains produced by mutant TH10. The strains were grown on MS agar for 5 days at 30 °C. The mutants are unrelated to those shown in Figure 1. A wild type spore is approximately 1.1 µm on the long axis. Scale bar = 2 µm. Please click here to view a larger version of this figure.
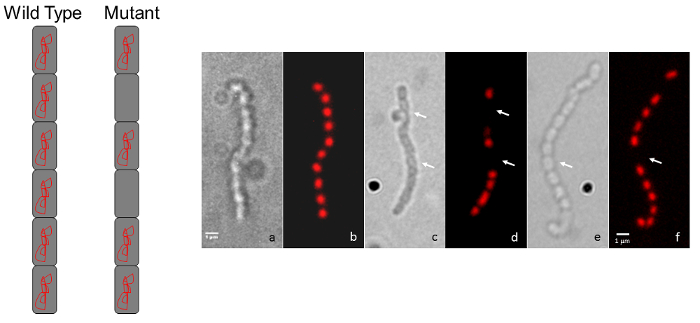
Figure 3: Representative micrographs showing S. coelicolor mutant chromosome segregation phenotypes. (A) A diagram representation shows the typical wild-type, regularly-spaced staining appearance for a chain of spores (every spore contains chromosomal DNA) compared to the intermittent staining pattern for a mutant that displays a chromosome segregation defect. (B) Each pair of panels shows the same spore chain observed by Differential Interference Contrast (DIC) image on the left and a propidium iodide-stained fluorescence image is shown on the right. The wild-type phenotype (a, b) is compared to two random transposon insertion mutants (c, d and e, f). Arrows indicate spores devoid of DNA. The strains were grown on MS agar for 5 days at 30 °C. The mutants shown are unrelated to those in Figure 1 and Figure 2. Scale bar = 1 µm. Please click here to view a larger version of this figure.
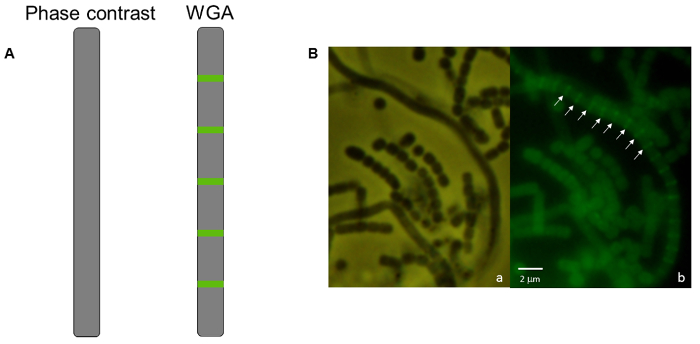
Figure 4: Fluorescence micrograph of S. venezuelae wild-type strain stained with WGA-FITC. (A) A diagram of an aerial filament appears smooth with no indentations and no visible signs of sporulation using phase-contrast microscopy in comparison with the ladder-like array of cell wall staining that indicates an early stage of sporulation has begun in that same filament using WGA-FITC under fluorescence microscopy. (B) Micrographs of wild-type S. venezuelae. (a) Smooth aerial filaments are present among chains of spores. (b) Within the mycelium shown in panel a, one aerial filament at an early stage in development possesses a ladder-like array of cell wall deposition. Arrows indicate the regularly-spaced formation of cross-walls stained by WGA-FITC that develop synchronously within a single aerial filament as it undergoes developmentally-associated sporulation. The strain was grown on MYM(Maltose, Yeast extract, Malt extract) MYM agar for 38 h at 30 °C. Scale bar = 2 µm. Please click here to view a larger version of this figure.
Discussion
Here we present protocols for beginning Streptomyces researchers to initiate studies by including the steps needed to propagate strains and prepare stocks for long-term storage. We then describe the protocols for visual and microscopic characterization of Streptomyces strains. Some typical initial steps in phenotyping developmental mutants are: 1) visual examination of the mutant colonies compared to wild type colonies on agar medium; 2) phase-contrast microscopy; and 3) fluorescence microscopy of sporogenic aerial hyphae. Based upon the phenotype displayed in these three steps, a variety of techniques may be employed to further discern the phenotype of a particular strain.
Initial phenotyping experiments are commonly used to characterize new species, identify mutants of interest, partially characterize mutants, and begin to discern the typical role of a particular gene based on the phenotype of an identified mutant. The methods described here have already been used in university teaching laboratories to identify and characterize a wide variety of Streptomyces mutants, including those with defects in cell division48,49,50,51,52,53,54, sporulation55,56, aerial mycelium formation57,58, antibiotic production59, second messenger signaling47, and chromosome segregation60. These techniques are the vital first steps to determining the phenotype of mutants in general and reveal a large amount of important information about the roles of genes of interest. The methods may easily be extended to other species of Streptomyces and have already been used to describe strains of S. griseus, S. venezuelae, S. scabies, and many other streptomycetes. The video protocols described here are expected to serve as an important resource for new researchers entering fields of Streptomyces research, such as in the area of drug discovery. This includes the new instructors who are working to combat the antibiotic resistance crisis and educating the countless numbers of new undergraduate student researchers joining the crowdsourcing efforts of the Small World Initiative.
The techniques described here can be easily adapted to college and high school classroom use in addition to research laboratories, using the modifications described in the video and text. Students in a first year microscopy module at a small liberal arts college were able to streak strains, take digital photographs of strains grown on agar media, and perform phase-contrast and fluorescence microscopy, which culminated in the submission of a portfolio of multi-paneled figures at the end of the 3 week module, representing 15 h of in-lab work. Approximately 160 first year students were responsible for the initial phenotyping of 320 novel transposon mutants. Undergraduate research students at three institutions participated in the initial phenotyping of additional mutants and the subsequent characterization of many of the strains. The comprehensive data obtained in a relatively short period of time, illustrate the value of the protocols described here. Hundreds of additional mutants have been stored as glycerol mycelial stocks for future characterization.
Following the initial experiments described here, a variety of methods may be employed to extend the quality of information pertaining to strains of interest. If the mutation is unknown, a genotyping method should be employed to determine the type and/or location of the mutation. For example, random transposon mutagenesis of the wild-type chromosome6,7,8,9,10,11,12,13 results in colonies that should undergo initial phenotypic screens such as those described above. Then the location of the transposon should be identified using a technique such as inverse polymerase chain reaction (iPCR)61. Determining the genotype of newly discovered mutants is an important step following initial characterization.
Some commonly used advanced methods for subsequent phenotyping analysis that may be mentioned in the classroom or explored through further research include green fluorescent protein (GFP) tagging to determine localization patterns for the protein of interest, gene expression analyses such as real time quantitative PCR (qPCR) and global gene expression patterns of wild-type versus mutant via RNA Sequencing (RNA-seq). Phenotyping skills are also required for genetic complementation analysis. In a complementation experiment, the wild-type copy of a gene is introduced into a mutated strain to determine whether the newly added allele can compensate for the loss-of-function of the mutated allele. Comparing the phenotype of the complemented strain to that of both the original mutant and the parental, wild-type strain is required.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge Otterbein University for Undergraduate Student Research Fellowships and Student Research Fund Awards to GVK and SGK; and the Otterbein Professor Gilbert E. Mills Memorial Endowed Sabbatical Program Award and The Department of Biology and Earth Science Faculty Research and Scholarship Endowed Award to JAB. The Bert and Jane Horn Endowed Student Research Fund in the Sciences was awarded to GVK and SGK. The authors would also like to gratefully acknowledge Duquesne University funded Undergraduate Research Program Fellowships for GVK and SGK. Former Juniata College undergraduate research students, Ryan Johnson and Lindsey Draper contributed the microscopy images for Figures 2 and 3, respectively.
Materials
| 50% Glycerol | Sigma-Aldrich | G5516 | |
| Immersion Oil (Type DF) | Cragille | 16482 | |
| Lens Paper | Fisherbrand | 11-995 | |
| Sterile Water | GeneMate | G-3250-1L | |
| 100% Ethanol | Sigma-Aldrich | E7023 | |
| Propidium Iodide | Invitrogen | P1304MP | |
| Toothpicks (flat) | Target | 081-22-1957 | |
| Pipette Tips (10 ml) | GeneMate | P-1240-10 | |
| Pipette Tips (200 ml) | GeneMate | P-1240-200Y | |
| Pipette Tips (1250 ml) | GeneMate | P-1240-1250 | |
| Wooden Applicators 6'' | Solon Care | 070809 | |
| Cotton Swabs | Fisherbrand | 23-400-124 | |
| Soy Flour | Bob's Red Mill | 1516C164 | |
| D-Mannitol | Sigma-Aldrich | M4125-1KG | |
| Agar | Sigma-Aldrich | A1296-1KG | |
| Glycerol 100% | VWR amresco life science | 0854-1L | |
| 0.8% NaCl (Saline) | Sigma-Aldrich | SLBB9000V | |
| 1.2 mL freezer tube | NEST | 606101 | |
| Ultra low (-80°C) freezer | SO-LOW | U85-18 | |
| Cryo Safety Gloves | Bel-Art | H13201 | |
| Petri dishes | Sigma-Aldrich | P5856-500EA | |
| Cover slips #1.5 | Thomas Scientific | 64-0721 | |
| Slides | Carolina | 63-2010 | |
| Autoclave | Tuttnauer | 2540E | |
| Phase-Contrast Microscope | Olympus | BX40 | |
| Forceps | Carolina Biological | 624504 | |
| Bunsen Burner | Carolina Biological | 706706 | |
| Methanol | Sigma-Aldrich | 34860-1L-R | |
| PBS (phosphate buffer saline) | Sigma-Aldrich | P4417-50TAB | |
| WGA-FITC | Biotium | 29022-1 | |
| Clear nail polish | OPI | 22001009000 | |
| Image J | NIH | Free Software | |
| 100 mL beaker | Pyrex USA | 1000-T | |
| 1 L beaker | Carolina Biological | 721253 | |
| 1 L flask | Fisherbrand | S63274 | |
| 250 mL flask | Pyrex USA | 4980 | |
| 100 mL graduated cylinder | Carolina Biological | 721788 | |
| 500 mL graduated cylinder | Carolina Biological | 721792 | |
| Stir Bar | Fisher Scientific | 22-271825 | |
| Centrifuge | Eppendorf | 5810R | |
| Camera for Microscope | Olympus | DP72 | |
| Nitrile Examination Gloves (Med) | Bio Excell | 71011002 | |
| Vortex Mixer | Carolina Biological | 701077 |
Riferimenti
- Barka, E. A., et al. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80 (1), 1-43 (2015).
- Bentley, S. D., et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 417 (2), 141-147 (2002).
- Redenbach, M., et al. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21 (1), 77-96 (1996).
- McCormick, J. R., Flardh, K. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36 (1), 206-231 (2012).
- Flardh, K., Buttner, M. J. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7 (1), 36-49 (2009).
- Bennett, J. A. . Molecular Genetic analysis of division and development in Streptomyces coelicolor. , (2007).
- Hull, T. D., et al. Cyclic Di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J. Bacteriol. 194 (17), 4642-4651 (2012).
- Petzke, L., Luzhetskyy, A. In vivo Tn5-based transposon mutagenesis of streptomycetes. Appl. Microbiol. Biotechnol. 83 (5), 979-986 (2009).
- Xu, Z., et al. Large-Scale Transposition Mutagenesis of Streptomyces coelicolor Identifies Hundreds of Genes Influencing Antibiotic Biosynthesis. Appl. Environ. Microbiol. 83 (6), (2017).
- Fernandez-Martinez, L. T., et al. A transposon insertion single-gene knockout library and new ordered cosmid library for the model organism Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek. 99 (3), 515-522 (2011).
- Gehring, A. M., Nodwell, J. R., Beverley, S. M., Losick, R. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc. Natl. Acad. Sci. U.S.A. 97 (17), 9642-9647 (2000).
- Bishop, A., Fielding, S., Dyson, P., Herron, P. Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 14 (5), 893-900 (2004).
- Bilyk, B., Weber, S., Myronovskyi, M., Bilyk, O., Petzke, L., Luzhetskyy, A. In vivo random mutagenesis of streptomycetes using mariner-based transposon Himar1. Appl. Microbiol. Biotechnol. 97 (1), 351-359 (2013).
- Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., Hopwood, D. A. . Practical Streptomyces Genetics. , (2000).
- Gust, B., Kieser, T., Chater, K. F. . REDIRECT technology: PCR-targeting system in Streptomyces coelicolor. , (2002).
- Gust, B., Chandra, G., Jakimowicz, D., Yuqing, T., Bruton, C. J., Chater, K. F. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54, 107-128 (2004).
- Tong, Y., Charusanti, P., Zhang, L., Weber, T., Lee, S. Y. CRISPR-Cas9 based engineering of Actinomycetal genomes. ACS Synth. Biol. 4 (9), 1020-1029 (2015).
- Huang, H., Zheng, G., Jiang, W., Hu, H., Lu, Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta BiochimBiophys. Sin.(Shanghai). 47 (4), 231-243 (2015).
- Cobb, R. E., Wang, Y., Zhao, H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 4 (6), 723-728 (2015).
- Carey, M. F., Peterson, C. L., Smale, S. T. PCR-mediated site-directed mutagenesis. Cold Spring Harb Protoc. 2013 (8), 738-742 (2013).
- Brown, E. D., Wright, G. D. Antibacterial drug discovery in the resistance era. Nature. 529 (7586), 336-343 (2016).
- Dodds, D. R. Antibiotic resistance: A current epilogue. Biochem. Pharmacol. 134, 139-146 (2017).
- Caruso, J. P., Israel, N., Rowland, K., Lovelace, M. J., Saunders, M. J. Citizen Science: The Small World Initiative improved lecture grades and California critical thinking skills test scores of nonscience major students at Florida Atlantic University. J. Microbiol. Biol. Educ. 17 (1), 156-162 (2016).
- Davis, E., et al. Antibiotic discovery throughout the Small World Initiative: A molecular strategy to identify biosynthetic gene clusters involved in antagonistic activity. Microbiology Open. 6 (3), (2017).
- van Keulen, G., Dyson, P. J. Production of specialized metabolites by Streptomyces coelicolor A3(2). Adv. Appl. Microbiol. 89, 217-266 (2014).
- Aigle, B., et al. Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol. 41 (2), 251-263 (2014).
- Antoraz, S., Santamaria, R. I., Diaz, M., Sanz, D., Rodriguez, H. Toward a new focus in antibiotic and drug discovery from the Streptomyces arsenal. Front. Microbiol. 6, 461 (2015).
- Onaka, H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J. Antibiot.(Tokyo). 70 (8), 865-870 (2017).
- Chen, J., Wu, Q., Hawas, U. W., Wang, H. Genetic regulation and manipulation for natural product discovery. Appl. Microbiol. Biotechnol. 100 (7), 2953-2965 (2016).
- Katz, L., Baltz, R. H. Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 43 (2-3), 155-176 (2016).
- Liu, G., Chater, K. F., Chandra, G., Niu, G., Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77 (1), 112-143 (2013).
- den Hengst, C. D., Tran, N. T., Bibb, M. J., Chandra, G., Leskiw, B. K., Buttner, M. J. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78 (2), 361-379 (2010).
- Tran, N. T., Den Hengst, C. D., Gomez-Escribano, J. P., Buttner, M. J. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 193 (12), 3100-3108 (2011).
- Anonymous. Lab Safety. Proper Use of Autoclaves. JoVE Science Education Database. , (2017).
- Border, B. G., Rice-Spearman, L. Microwaves in the laboratory: effective decontamination. Clin. Lab. Sci. 12 (3), 156-160 (1999).
- Bhattacharjee, M. K., Delsol, J. K. Does microwave sterilization of growth media involve any non-thermal effect?. J. Microbiol. Methods. 96, 70-72 (2014).
- Sanders, E. R. Aseptic laboratory techniques: plating methods. J. Vis. Exp. (63), e3064 (2012).
- Anonymous, General Laboratory Techniques. An Introduction to the Micropipettor. JoVE Science Education Database. , (2017).
- Baltz, R. H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 43 (2-3), 343-370 (2016).
- Frohlich, V. C. Phase Contrast and Differential Interference Contrast (DIC) Microscopy. J. Vis. Exp. (18), e844 (2008).
- Anonymous, General Laboratory Techniques. Introduction to Light Microscopy. JoVE Science Education Database. , (2017).
- Sambrook, J., Fritsch, E. F., Maniatis, T. . Molecular Cloning: A Laboratory. 3, (1989).
- Anonymous, General Laboratory Techniques. Introduction to Fluorescence Microscopy. JoVE Science Education Database. , (2017).
- Jensen, E. C. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat. Rec.(Hoboken). 296 (3), 378-381 (2013).
- Chater, K. F. Recent advances in understanding Streptomyces. F1000Res. 5, (2016).
- Chater, K. F. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex?. Curr. Opin. Microbiol. 4 (6), 667-673 (2016).
- Tschowri, N. Cyclic dinucleotide-controlled regulatory pathways in Streptomyces species. J. Bacteriol. 198 (1), 47-54 (2016).
- Bennett, J. A., et al. Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J. Bacteriol. 191 (2), 661-664 (2009).
- Mistry, B. V., Del Sol, R., Wright, C., Findlay, K., Dyson, P. FtsW is a dispensable cell division protein required for Z-ring stabilization during sporulation septation in Streptomyces coelicolor. J. Bacteriol. 190 (16), 5555-5566 (2008).
- Bennett, J. A., Aimino, R. M., McCormick, J. R. Streptomyces coelicolor genes ftsL and divIC play a role in cell division but are dispensable for colony formation. J. Bacteriol. 189 (24), 8982-8992 (2007).
- Bennett, J. A., McCormick, J. R. Two new loci affecting cell division identified as suppressors of an ftsQ-null mutation in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 202 (2), 251-256 (2001).
- Dharmatilake, A. J., Kendrick, K. E. Expression of the division-controlling gene ftsZ during growth and sporulation of the filamentous bacterium Streptomyces griseus. Gene. 147 (1), 21-28 (1994).
- McCormick, J. R., Losick, R. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 178 (17), 5295-5301 (1996).
- McCormick, J. R., Su, E. P., Driks, A., Losick, R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14 (2), 243-254 (1994).
- Flärdh, K., Findlay, K. C., Chater, K. F. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology. 145 (9), 2229-2243 (1999).
- van Wezel, G. P., van der Meulen, J., Kawamoto, S., Luiten, R. G., Koerten, H. K., Kraal, B. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182 (20), 5653-5662 (2000).
- Capstick, D. S., Willey, J. M., Buttner, M. J., Elliot, M. A. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol. Microbiol. 64 (3), 602-613 (2007).
- Ma, H., Kendall, K. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176 (12), 3800-3811 (1994).
- Xu, Z., et al. Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl. Environ. Microbiol. 83 (6), (2017).
- Dedrick, R. M., Wildschutte, H., McCormick, J. R. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. J. Bacteriol. 191 (1), 320-332 (2009).
- Pavlopoulos, A. Identification of DNA sequences that flank a known region by inverse PCR. Methods Mol. Biol. 772, 267-275 (2011).

