Photobleaching Enables Super-resolution Imaging of the FtsZ Ring in the Cyanobacterium Prochlorococcus
Summary
Here we describe a photobleaching method to reduce the autofluorescence of cyanobacteria. After photobleaching, stochastic optical reconstruction microscopy is used to obtain three-dimensional super-resolution images of the cyanobacterial FtsZ ring.
Abstract
Super-resolution microscopy has been widely used to study protein interactions and subcellular structures in many organisms. In photosynthetic organisms, however, the lateral resolution of super-resolution imaging is only ~100 nm. The low resolution is mainly due to the high autofluorescence background of photosynthetic cells caused by high-intensity lasers that are required for super-resolution imaging, such as stochastic optical reconstruction microscopy (STORM). Here, we describe a photobleaching-assisted STORM method which was developed recently for imaging the marine picocyanobacterium Prochlorococcus. After photobleaching, the autofluorescence of Prochlorococcus is effectively reduced so that STORM can be performed with a lateral resolution of ~10 nm. Using this method, we acquire the in vivo three-dimensional (3-D) organization of the FtsZ protein and characterize four different FtsZ ring morphologies during the cell cycle of Prochlorococcus. The method we describe here might be adopted for the super-resolution imaging of other photosynthetic organisms.
Introduction
Super-resolution microscopies can break the diffraction limit of light and provide images within sub-diffraction resolutions (< 200 nm). They have been widely used in many organisms to study protein localization and subcellular structures. Major super-resolution microscopy methods include structured illumination microscopy (SIM), stimulated emission depletion microscopy (STED), STORM, and photoactivated localization microscopy (PALM). The mechanisms and applications of these super-resolution microscopes have been reviewed elsewhere1,2.
STORM can achieve a resolution as high as 10 nm by spatial separation3,4. For STORM, only one molecule within a diffraction-limited region is activated ("on") and the rest of the molecules are kept inactivated ("off"). By an accumulation of rapid switch-on and -off of single molecules, a "diffraction-unlimited" image can be generated3. Meanwhile, many kinds of organic dyes and fluorescent proteins are applicable in STORM, allowing an easy upgrade from regular fluorescence microscopy to high-resolution microscopy5,6.
STORM has not been widely applied in photosynthetic cells, such as cyanobacteria, algae, and plant cells with chloroplasts7,8, which is due to the fact that STORM requires high laser intensity to drive photoswitching. The high-intensity laser unfavorably excites strong autofluorescence background in photosynthetic cells and interferes with the single-molecule localization in STORM imaging. In order to use STORM to investigate the subcellular structures or protein interactions in photosynthetic cells, we developed a photobleaching protocol to quench the background autofluorescence signals9. In a routine immunofluorescent staining procedure, specimens are exposed to white light of a high intensity during the blocking step, which lowers the autofluorescence of photosynthetic cells to meet the requirements for STORM. Thus, this protocol makes it feasible to investigate pigmented organisms with STORM.
Here, we describe the protocol to use STORM to image the FtsZ ring organization in the unicellular picocyanobacterium Prochlorococcus. FtsZ is a highly conserved tubulin-like cytoskeletal protein which polymerizes to form a ring structure (the Z ring) around the circumference of a cell10 and is essential for the cell division11. Preserved Prochlorococcus cells are first photobleached to reduce the autofluorescent background and immunostained with a primary anti-FtsZ antibody, and then a secondary anti-Rabbit IgG (H+L) antibody is conjugated with a fluorophore (e.g., Alexa Fluor 750). Eventually, STORM is used to observe the detailed FtsZ ring organizations in Prochlorococcus during different cell cycle stages.
Protocol
1. Sample Preparation and Fixation
- Inoculate 1 mL of axenic Prochlorococcus MED4 to 5 mL of the seawater-based Pro99 medium12. Grow Prochlorococcus cultures at 23 °C under the light with an intensity of 35 µmol photons/m2s. Five days later, collect 1 mL of culture into a 1.5-mL tube.
NOTE: Five days after the inoculation, Prochlorococcus MED4 will reach the late log phase, with approximately 108 cell/mL, which is appropriate for STORM imaging. - Add 100 µL of formaldehyde freshly prepared from 25% paraformaldehyde (PFA) (w/v) and 2 µL of 50% glutaraldehyde into the 1.5-mL tube to fix the culture. Incubate the sample in the dark at room temperature for 20 min.
NOTE: The sample was kept in the dark for fixation because glutaraldehyde is light sensitive. - Spin down the sample at 13,500 x g for 1 min; then, remove the supernatant and resuspend the cells in 100 µL of the Pro99 medium12. Store the sample at 4 °C until immunostaining.
2. Precoating of the Coverslip with Polystyrene Beads
NOTE: Polystyrene beads are considered as the fiducial marker for drift correction.
- Vortex the original vial of polystyrene beads and prepare a 1:20,000 dilution with 50% ethanol as working slurry.
- Turn on the hot plate, set the temperature to 120 °C, and place coverslips onto the hot plate.
- Load 100 µL of working slurry onto each coverslip. Incubate the coverslip on the hot plate for 10 min and, then, carefully transfer the coverslips to Petri dishes for storage at room temperature.
NOTE: The side with the beads attached to it should face up, and the cells should be attached on the same side. The beads are immobilized on the coverslips and used for correcting the sample-drifting in real-time during STORM imaging. After immobilizing the beads, the coverslips cannot be flamed.
3. Coating of Poly-L-lysine onto the Bead-coated Coverslip
NOTE: This is done for the immobilization of cyanobacterial cells.
- Add 100 µL of poly-L-lysine (1 mg/mL) to the center of the coverslip with the bead-coated side facing up. Let it sit for 30 min at room temperature.
- Use a pipette to carefully aspire the unattached poly-L-lysine and transfer it to a 1.5-mL tube. Save the poly-L-lysine at 4 °C for reuse.
- Rinse the coverslip with 10 mL of ultra-filtered water in a 60-mm Petri dish and then briefly dry the coverslip on a paper towel.
- Store the coverslip in a Petri dish (100 mm) for later usage. To prevent the attachment of the coverslip to the dish, line the Petri dish with a layer of parafilm.
NOTE: The coverslip, precoated with beads and poly-L-lysine, can be stored at room temperature for at least half a year. Therefore, preparing a batch of coverslips in advance is highly recommended.
4. Immobilization of Cells on the Coverslip
- Transfer a precoated coverslip to a new Petri dish with parafilm at the bottom, which will be referred to as the staining dish henceforth. Load 100 µL of the fixed sample on the coverslip and let it sit for 30 min to allow cell attachment.
- Transfer the coverslip to a well of a 12-well plate, which will be referred to as the washing dish henceforth. Add 1 mL of phosphate-buffered saline (PBS) (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4) to the well to wash the coverslip. Remove the PBS buffer and add a new PBS buffer to wash the coverslip again.
5. Permeabilization of Cyanobacterial Cells
- Remove the PBS buffer using a pipette. Add 1 mL of freshly prepared permeabilization buffer to the well of the washing dish, which contains 0.05% non-ionic detergent-100 (v/v), 10 mM EDTA, 10 mM Tris (pH 8.0), and 0.2 mg/mL lysozyme.
- Incubate the washing dish at 37 °C for 20 min. Then, remove the permeabilization buffer.
- Add 1 mL of PBS buffer and place the washing dish on a shaker which gently shakes the washing dish for 5 min. Remove the PBS buffer. Repeat this step 3x to wash the coverslip.
6. Photobleaching of the Chlorophyll Pigments in a Blocking Step
- Transfer the coverslip to the staining dish. Add 50 µL of blocking buffer, which contains 0.2% (v/v) non-ionic detergent-100 and 3% (v/v) goat serum, on the top of the coverslip.
- Place the whole staining plate on ice and move it under the xenon light source for photobleaching for at least 60 min, with a light intensity at 1,800 µmol photons/m2·s.
- After photobleaching and blocking, carefully remove the blocking buffer by adsorbing it with the edge of a paper towel.
NOTE: The intensity of xenon light is so high that a pair of proper sunglasses is needed for eye protection. Meanwhile, wrap the light source and the plate using aluminum foil to avoid the leaking of light, which may cause eye damage. Check the coverslip every 15 min to make sure that the coverslip remains moist. If the coverslip is dry, add 50 µL of blocking buffer.
7. Antibody Binding
- Dissolve anti-Anabaena FtsZ antibody in 200 µL of water according to the manufacturer’s instructions.
- Dilute 1 µL of anti-FtsZ antibody into 99 µL of blocking buffer. Add 50 µL of diluted primary antibody on the coverslip and incubate it at room temperature for 30 min.
- Transfer the coverslip to the washing dish. Add 1 mL of PBS buffer and place the washing dish on a shake. Gently shake the washing dish for 5 min. Repeat this step 3x to wash the coverslip.
- Transfer the coverslip to the staining dish. Dilute 1 µL of a secondary antibody into 500 µL of blocking buffer. Add 50 µL of diluted secondary antibody onto the coverslip and incubate it for 30 min in the dark at room temperature.
- Transfer the coverslip to the washing dish. Wash it with 1 mL of PBS buffer on a shaker. Gently shake the washing dish for 5 min. Wrap the washing dish with aluminum paper to make it light-proof. Repeat this step 3x.
- Add 500 µL of formaldehyde freshly prepared from 4% paraformaldehyde (PFA) (w/v) to the well. Incubate the coverslip for 15 min in the dark at room temperature.
- Remove the formaldehyde and repeat the washing step 3x.
- Store the coverslip in PBS buffer at 4 °C in the dark until imaging.
8. Preparation of the STORM Imaging Buffer
- Prepare 1 mL of imaging buffer according to Table 1, immediately before the STORM imaging.
NOTE: As the shelf life of the imaging buffer is about 2 h, prepare it fresh, right before usage. Avoid vortexing after the addition of glucose oxidase and catalase.
CAUTION: Methyl viologen is poisonous material. Please handle it with particular care. Cyclooctatetraene is classified as a carcinogen Cat. 1 chemical. Please avoid inhalation and skin contact and make the cyclooctatetraene stock solution and imaging buffer in a chemical hood.
9. Image Acquisition of STORM Data
- Load the coverslip in the loading chamber (Figure 1). Load the freshly prepared imaging buffer gently into the chamber to avoid washing off the cyanobacterial cells. Place a rectangular coverslip on the top of the imaging buffer to prevent its reaction with oxygen in the air. Avoid trapping air bubbles underneath the coverslip.
- Turn on the camera, the LED light, and the laser. Open STORM software for image acquisition, centroid position determination, and sample drifting correction13.
- Add half a drop of immersion oil on top of the lens. Load the chamber and make sure the lens of the objective makes contact with the coverslip.
- Examine the signal with a 750-nm laser.
NOTE: Always include a second antibody-minus-staining negative control to ensure that the autofluorescence is diminished. - Identify a sample area that contains both cells and fiducial markers. Start the software for sample drifting correction.
NOTE: In general, the ideal sample area contains 10 – 30 cells. Meanwhile, the cells should be separated well to avoid any overlapping cells. - Acquire one wide-field image as a reference, with the camera electron multiplication (EM) gain at 300 and an exposure time of 30 ms (Figure 2A).
- Increase the 750-nm excitation laser intensity to a higher power, approximately 4.5 kW/cm2. Once the fluorophores have transitioned into a sparse blinking pattern, acquire one super-resolution image by collecting 10,000 frames at 33 Hz (Figure 2B).
NOTE: If the fluorophores are not isolated, wait for a couple of minutes until more fluorophores are switched to the dark state and the isolated single molecules flicker sequentially. The number of frames described here is suitable for our sample and needs to be optimized for other targeted structures.
10. Reconstruction of Super-resolution Images from Raw Data
- Use a plugin called QuickPALM (Table of Materials)14 in ImageJ to reconstruct a 3-D color super-resolution image.
NOTE: A preliminary chromatic image (Figure 2C) along with a color-coded scale bar (Figure 2C) will appear. The color represents the depth on the z-axis. - To demonstrate the 3-D structure in a video, construct a stack of super-resolution images with z-axis sectioned every 10 nm using QuickPALM14. Adjust the brightness of the stacks and duplicate the stack of one target cell. Use a plugin called 3D Viewer (Table of Materials)15 in ImageJ to generate the 3-D image of the cell. Record the rotation of the 3-D structure for demonstration purposes.
Representative Results
STORM achieves super-resolution imaging by activating individual photoswitchable fluorophores stochastically. The location of every fluorophore is recorded and a super-resolution image is then constructed based on these locations4. Therefore, the precision of the fluorophore location is important for the super-resolution image reconstruction. The absorption spectra of Prochlorococcus peak at 447 nm and 680 nm,and Prochlorococcus has a minimum absorption at wavelengths above 700 nm16. However, Prochlorococcus MED4 cells still emitted high autofluorescence when exposed to an extremely high intensity of the 750-nm laser (Figure 3A), which is required for STORM imaging. Thus, the high autofluorescence background of Prochlorococcus cells heavily interferes with super-resolution imaging.
In order to utilize STORM to investigate protein organizations in Prochlorococcus, we developed a photobleaching method. After exposure to a white light of high intensity for 30 min, the autofluorescence of Prochlorococcus MED4 cells decreased (Figure 3B), although several cells with autofluorescence were still detected. We further elongated the photobleaching time to 60 min and found that the majority of the cells lost their autofluorescence (Figure 3C). These results indicate that the photobleaching method we developed here can greatly decrease the autofluorescence in photosynthetic organisms.
After photobleaching, we were able to use STORM to visualize the cell division protein FtsZ in Prochlorococcus MED4 cells. The cyanobacterium Prochlorococcus is the smallest and the most abundant photosynthetic organism on earth17. The diameter of a Prochlorococcus cell is only 500 – 700 nm18. With such a small cell size, it is impossible to visualize the FtsZ ring organization in Prochlorococcus cells using conventional wide-field fluorescence microscopy (Figure 2A). However, STORM has a spatial resolution of 9.6 nm in the xy-plane and 41.6 nm in the z-axis. Using STORM, we were able to reveal a detailed morphology of the FtsZ ring in Prochlorococcus (Figures 2B and 4). By rotating 3-D STORM images, we identified four different types of FtsZ ring morphologies: clusters (Figure 4A, Movie S1), an incomplete ring (Figure 4B, Movie S2), a complete ring (Figure 4C, Movie S3), and double rings (Figure 4D, Movie S4). Gaps were observed in the complete FtsZ rings of Prochlorococcus (Figure 4C, Movie S3), which is similar to that found in Caulobacter cresentus19. These four types of FtsZ ring morphologies showed the assembly process of the FtsZ ring during the cell cycle of Prochlorococcus, and this study helped us to understand the role of the FtsZ ring during the cell division of Prochlorococcus9.
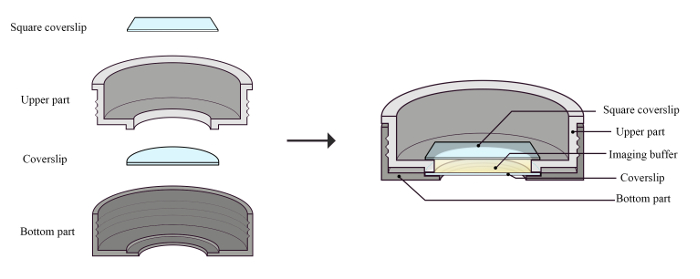
Figure 1: Components of the loading chamber and the assembled chamber with the coverslips and imaging buffer for STORM imaging. The coverslip with the sample was placed on the groove of the bottom part first. The upper part was then carefully screwed on to the bottom part. The imaging buffer was added in the loading chamber and then carefully covered with a square coverslip. Bubbles should be avoided to minimize any movement that may influence the accuracy of the measurement. Please click here to view a larger version of this figure.
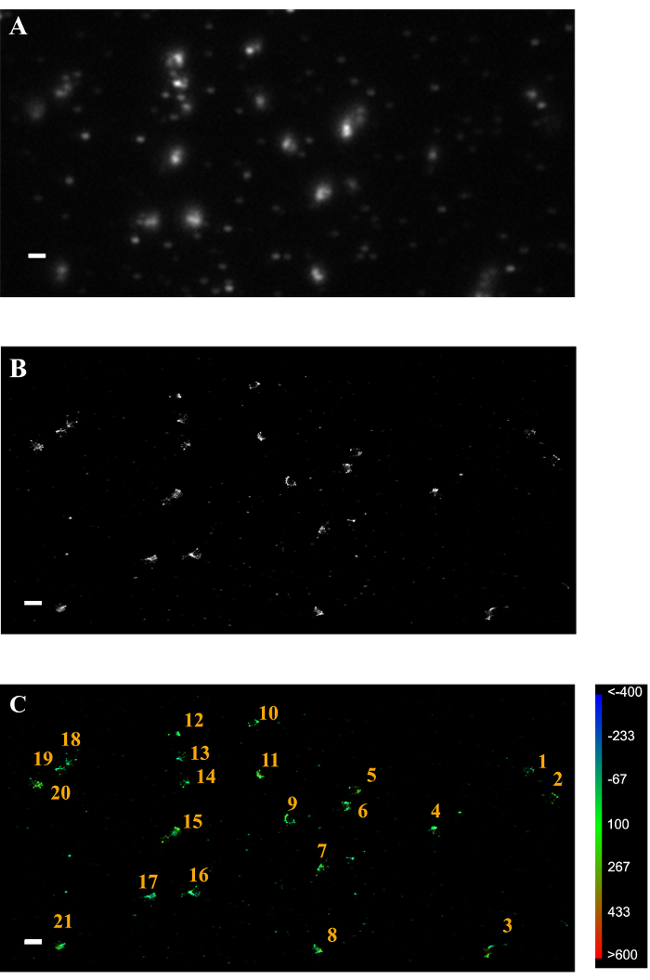
Figure 2: Representative wide-field, 2-D STORM, and 3-D color STORM images of FtsZ in Prochlorococcus MED4. The images were taken from the same field of view using regular (A) wide-field microscopy, (B) 2-D STORM, and (C) 3-D color STORM. The colors in panel C indicate the depth of the fluorescent signals on the z-axis. The numbers in panel C indicate the cells of interest. The scale bars = 1 µm. Please click here to view a larger version of this figure.
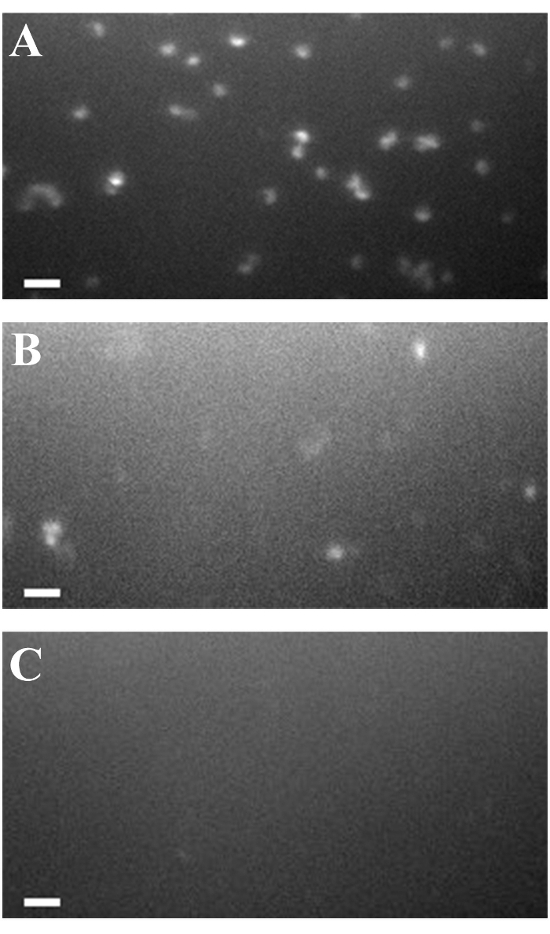
Figure 3: Photobleaching of Prochlorococcus MED4. Fixed Prochlorococcus MED4 cells were (A) not photobleached, or they were photobleached for (B) 30 min and (C) 60 min. The XD-300 xenon light was used at an intensity of 1,800 µmol/m2·s. Cells were excited by a 750-nm laser at the same intensity as was used for STORM imaging. The autofluorescences were imaged with the same filters as used in STORM to make sure the presence of minimal autofluorescence to affect STORM. The scale bars = 2 µm. Please click here to view a larger version of this figure.
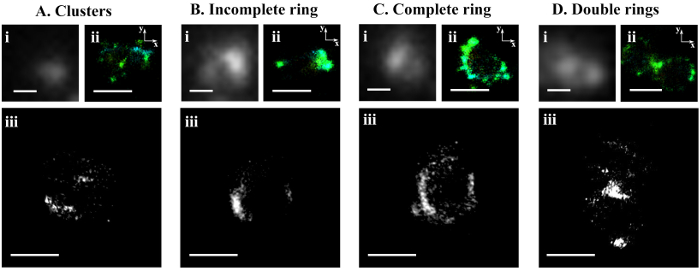
Figure 4: Representative STORM images of four FtsZ ring morphologies. Four different FtsZ ring morphologies were observed in Prochlorococcus: (A) clusters, (B) an incomplete ring, (C) a complete ring, and (D) double rings. For the same cell, images of (i) wide-field fluorescence microscopy, (ii) STORM on the xy-plane, and (iii) STORM after rotation were shown. The 3-D movies corresponding to the cells in panels A, B, C, and D are shown in Movies S1, S2, S3, and S4, respectively. The scale bars = 500 nm. Please click here to view a larger version of this figure.
| Chemicals | Stock solution | Final concentration |
| glucose | N/A | 10% (w/v) |
| Tris-Cl (pH 8.0) | 0.5 M | 50 mM |
| Ascorbic acid | 100 mM | 1 mM |
| Methyl viologen | 100 mM | 1 mM |
| Cyclooctatetraene | 200 mM | 2 mM |
| TCEP | 0.5 M | 25 mM |
| Glucose oxidase | 56 mg/ml | 0.56 mg/ml |
| Catalase | 4 mg/ml | 40 µg/ml |
Table 1: Recipe for the imaging buffer.
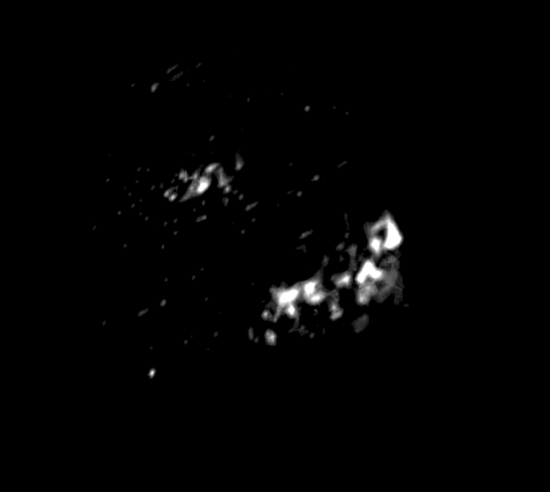
Movie S1: Clusters of FtsZ proteins observed in Prochlorococcus MED4 cells. Please click here to view this video. (Right-click to download.)
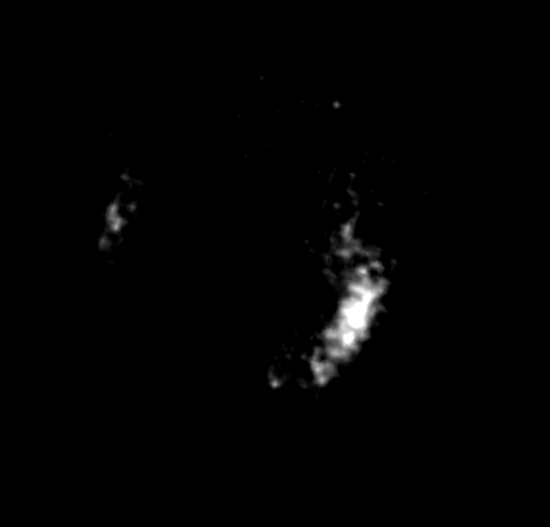
Movie S2: An incomplete ring of FtsZ proteins observed in Prochlorococcus MED4 cells. Please click here to view this video. (Right-click to download.)
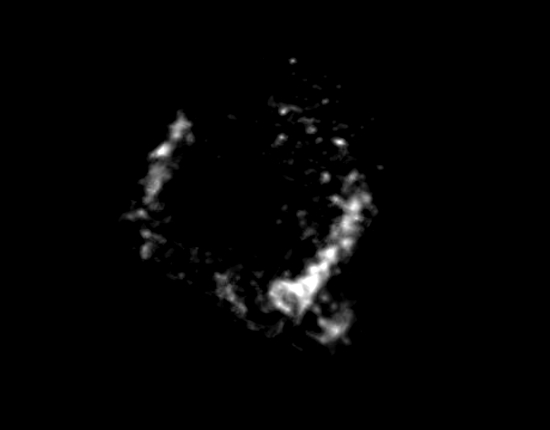
Movie S3: A complete ring of FtsZ proteins observed in Prochlorococcus MED4 cells. Please click here to view this video. (Right-click to download.)

Movie S4: A double-ring of FtsZ proteins observed in Prochlorococcus MED4 cells. Please click here to view this video. (Right-click to download.)
Discussion
In this protocol, we described a procedure to significantly reduce the autofluorescence of the cyanobacterium Prochlorococcus (Figure 3C) and, then, immunostain the proteins in the cells, which enabled us to utilize STORM to study the 3-D FtsZ ring morphologies in Prochlorococcus (Figure 4). This protocol might be adopted for super-resolution imaging in other photosynthetic organisms.
Previous studies on photosynthetic organisms mainly used SIM to achieve super-resolution imaging because SIM does not require lasers of high intensity. However, the spatial resolution of SIM is only ~100 nm1, which is much lower than that of STORM. In this study, the spatial resolution of STORM images could reach 9.6 nm in the xy-plane and 41.6 nm in the z-axis9. The improved resolution provided by photobleaching and STORM imaging allows researchers to study photosynthetic organisms in unprecedented detail.
Photobleaching prior to STORM is a critical step that enables researchers to observe the location and dynamics of proteins in autofluorescent organisms. Besides cyanobacteria, we have demonstrated that autofluorescence of the flowering plant Arabidopsis thaliana could also be quenched after 60 min of photobleaching9. The photobleaching has been widely used, for instance for improving the signal-to-noise ratio and the visualization of multiple biomarkers sequentially20. It was also demonstrated that photobleaching before imaging is advantageous when observing an autofluorescence sample under Raman spectroscopy21. Therefore, it might be feasible to apply this photobleaching method to reduce the autofluorescence of other photosynthetic organisms, including eukaryotic algae, vascular plants, and organisms containing proteorhodopsin and bacteriochlorophyll.
Photosynthetic organisms contain different pigments that have different absorption spectra; therefore, organic dyes and fluorescent proteins need to be prudently selected. For example, in this study, a dye with a maximum excitation wavelength of around 750 nm was used. Although two channels (750 nm and 650 nm) are available for the STORM presented here, Prochlorococcus autofluorescent signals could still be observed after being exposed under a 650-nm laser, even after prolonged photobleaching. This is probably because 650 nm is one of the major absorption wavelengths for Prochlorococcus. On the other hand, some Synechococcus and red algae contain phycoerythrin as their pigment22, which has an absorption spectrum from 488 nm to 560 nm and a minimum absorption at 650 nm23. Therefore, it might be feasible to use a 650-nm channel instead of a 750-nm channel to study the proteins within these phycoerythrin-containing autotrophs.
In summary, a proper combination of photobleaching and STORM provides a powerful approach to understand the protein organization in photosynthetic cells in detail, which will further reveal their dynamics and potential function.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors thank Daiying Xu for her technical assistance and comments on the manuscript. This study is supported by grants from the National Natural Science Foundation of China (Project number 41476147) and the Research Grants Council of the Hong Kong Special Administrative Region, China (project numbers 689813 and 16103414).
Materials
| Polystyrene particles | Spherotech | PP-20-10 | 2.0-2.4 µm |
| Coverslip | Marienfeld | 0111580 | 18 mm ∅, Thickness No. 1 |
| Ethanol | Scharlau | ET00021000 | |
| Poly-L-lysine hydrobromide | Sigma-Aldrich | P9155 | mol wt 70,000-150,000 |
| Paraformaldehyde | Sigma-Aldrich | 158127 | |
| Glutaraldehyde solution, 50% | Sigma-Aldrich | 340855 | |
| PBS | Sigma | P3813 | |
| Triton X-100 | Sigma | T8787 | |
| EDTA Disodium Salt, 2-hydrate | Gold biotechnology | E-210-500 | |
| Trizma base | Sigma | T1503 | |
| Lysozyme | Sigma | L6876 | |
| Goat serum | Sigma | G9023 | |
| anti-Anabaena FtsZ antibody | Agrisera | AS07217 | |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody | Life Technologies | A-21039 | conjugated with Alexa Fluor 750 |
| D-Glucose Anhydrous | Fisher Scientific | D16-1 | |
| L-Ascorbic Acid | Sigma-Aldrich | A5960 | |
| Methyl Viologen | Sigma-Aldrich | 856177 | |
| Cyclooctatetraene | Sigma-Aldrich | 138924 | |
| tris(2-carboxyethyl)phosphine (TCEP) | Sigma-Aldrich | 646547 | |
| Glucose Oxidase | Sigma-Aldrich | G2133 | |
| Catalase | Sigma-Aldrich | C9322 | |
| XD-300 Xenon light source | 250 W | ||
| STORM microscope | NBI | SRiS microscope | |
| Rohdea | NBI | SRiS 3.0 | software for imaging acquisition |
| Luna | NBI | SRiS 3.0 | software for drifting correction |
| QuickPALM | https://code.google.com/archive/p/quickpalm/wikis | ||
| 3D Viewer | http://132.187.25.13/ij3d/?page=Home&category=Home |
Riferimenti
- Huang, B., Babcock, H., Zhuang, X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 143 (7), 1047-1058 (2010).
- Toomre, D., Bewersdorf, J. A New Wave of Cellular Imaging. Annual Review of Cell and Developmental Biology. 26 (1), 285-314 (2010).
- Rust, M. J., Bates, M., Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods. 3 (10), 793-795 (2006).
- Huang, B., Wang, W., Bates, M., Zhuang, X. Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science. 319, 810-813 (2008).
- Dempsey, G. T., Vaughan, J. C., Chen, K. H., Bates, M., Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nature Methods Methods. 8 (12), 1027-1036 (2012).
- Linde, S., et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nature Protocols. 6 (7), 991-1009 (2011).
- Komis, G., Šamajová, O., Ovečka, M., Šamaj, J. Super-resolution Microscopy in Plant Cell Imaging. Trends in Plant Science. 20 (12), 834-843 (2015).
- Schubert, V. Super-resolution Microscopy – Applications in Plant Cell Research. Frontiers in Plant Science. 8, 531 (2017).
- Liu, R., et al. Three-Dimensional Superresolution Imaging of the FtsZ Ring during Cell Division of the Cyanobacterium Prochlorococcus. mbio. 8 (6), e00657 (2017).
- Adams, D. W., Errington, J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Reviews Microbiology. 7, 642-653 (2009).
- Boer, P. A. Advances in understanding E. coli cell fission. Current Opinion in Microbiology. 13 (6), 730-737 (2010).
- Moore, L. R., Post, A. F., Rocap, G., Chisholm, S. W. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnology and Oceanography. 47 (4), 989-996 (2002).
- Zhao, T., et al. A user-friendly two-color super-resolution localization microscope. Optics Express. 23 (2), 1879 (2015).
- Henriques, R., Lelek, M., Fornasiero, E. F., Valtorta, F., Zimmer, C., Mhlanga, M. M. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nature Methods. 7 (5), 339-340 (2010).
- Schmid, B., Schindelin, J., Cardona, A., Longair, M., Heisenberg, M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 11, 274 (2010).
- Ting, C. S., Rocap, G., King, J., Chisholm, S. W. Cyanobacterial photosynthesis in the oceans: The origins and significance of divergent light-harvesting strategies. Trends in Microbiology. 10 (3), 134-142 (2002).
- Biller, S. J., Berube, P. M., Lindell, D., Chisholm, S. W. Prochlorococcus: The structure and function of collective diversity. Nature Reviews Microbiology. 13 (1), 13-27 (2015).
- Morel, A., Ahn, Y. -. H., Partensky, F., Vaulot, D., Claustre, H. Prochlorococcus and Synechococcus – a Comparative-Study of Their Optical-Properties in Relation To Their Size and Pigmentation. Journal of Marine Research. 51, 617-649 (1993).
- Holden, S. J., Pengo, T., Meibom, K. L., Fernandez, C., Collier, J., Manley, S. High throughput 3D super-resolution microscopy reveals Caulobacter crescentusin vivo Z-ring organization. Proceedings of the National Academy of Sciences. 111 (12), 4566-4571 (2014).
- Zhao, M., Wei, B., Chiu, D. T. Imaging multiple biomarkers in captured rare cells by sequential immunostaining and photobleaching. Methods. 64 (2), 108-113 (2013).
- Golcuk, K., Mandair, G. S., Callender, A. F., Sahar, N., Kohn, D. H., Morris, M. D. Is photobleaching necessary for Raman imaging of bone tissue using a green laser?. Biochimica et Biophysica Acta. 1758 (7), 868-873 (2006).
- Haxo, F. T., Blinks, L. R. Photosynthetic Action Spectra of Marine Algae. The Journal of General Physiology. 33 (4), 389-422 (1950).
- Bryant, D. A. Phycoerythrocyanin and Phycoerythrin: Properties and Occurrence in Cyanobacteria. Journal of General Microbiology. 128 (4), 835-844 (1982).

