A Method for 3D Reconstruction and Virtual Reality Analysis of Glial and Neuronal Cells
Summary
The described pipeline is designed for the segmentation of electron microscopy datasets larger than gigabytes, to extract whole-cell morphologies. Once the cells are reconstructed in 3D, customized software designed around individual needs can be used to perform a qualitative and quantitative analysis directly in 3D, also using virtual reality to overcome view occlusion.
Abstract
Serial sectioning and subsequent high-resolution imaging of biological tissue using electron microscopy (EM) allow for the segmentation and reconstruction of high-resolution imaged stacks to reveal ultrastructural patterns that could not be resolved using 2D images. Indeed, the latter might lead to a misinterpretation of morphologies, like in the case of mitochondria; the use of 3D models is, therefore, more and more common and applied to the formulation of morphology-based functional hypotheses. To date, the use of 3D models generated from light or electron image stacks makes qualitative, visual assessments, as well as quantification, more convenient to be performed directly in 3D. As these models are often extremely complex, a virtual reality environment is also important to be set up to overcome occlusion and to take full advantage of the 3D structure. Here, a step-by-step guide from image segmentation to reconstruction and analysis is described in detail.
Introduction
The first proposed model for an electron microscopy setup allowing automated serial section and imaging dates back to 19811; the diffusion of such automated, improved setups to image large samples using EM increased in the last ten years2,3, and works showcasing impressive dense reconstructions or full morphologies immediately followed4,5,6,7,8,9,10.
The production of large datasets came with the need for improved pipelines for image segmentation. Software tools for the manual segmentation of serial sections, such as RECONSTRUCT and TrakEM211,12, were designed for transmission electron microscopy (TEM). As the whole process can be extremely time-consuming, these tools are not appropriate when dealing with the thousands of serial micrographs that can be automatically generated with state-of-the-art, automated EM serial section techniques (3DEM), such as block-face scanning electron microscopy (SBEM)3 or focused ion beam-scanning electron microscopy (FIB-SEM)2. For this reason, scientists put efforts into developing semi-automated tools, as well as fully automated tools, to improve segmentation efficiency. Fully automated tools, based on machine learning13 or state-of-the-art, untrained pixel classification algorithms14, are being improved to be used by a larger community; nevertheless, segmentation is still far from being fully reliable, and many works are still based on manual labor, which is inefficient in terms of segmentation time but still provides complete reliability. Semi-automated tools, such as ilastik15, represent a better compromise, as they provide an immediate readout for the segmentation that can be corrected to some extent, although it does not provide a real proofreading framework, and can be integrated using TrakEM2 in parallel16.
Large-scale segmentation is, to date, mostly limited to connectomics; therefore, computer scientists are most interested in providing frameworks for integrated visualizations of large, annotated datasets and analyze connectivity patterns inferred by the presence of synaptic contacts17,18. Nevertheless, accurate 3D reconstructions can be used for quantitative morphometric analyses, rather than qualitative assessments of the 3D structures. Tools like NeuroMorph19,20 and glycogen analysis10 have been developed to take measurements on the 3D reconstructions for lengths, surface areas, and volumes, and on the distribution of cloud points, completely discarding the original EM stack8,10. Astrocytes represent an interesting case study, because the lack of visual clues or repetitive structural patterns give investigators a hint about the function of individual structural units and consequent lack of an adequate ontology of astrocytic processes21, make it challenging to design analytical tools. One recent attempt was Abstractocyte22, which allows a visual exploration of astrocytic processes and the inference of qualitative relationships between astrocytic processes and neurites.
Nevertheless, the convenience of imaging sectioned tissue under EM comes from the fact that the amount of information hidden in intact brain samples is enormous and interpreting single section images can overcome this issue. The density of structures in the brain is so high that 3D reconstructions of even a few objects visible at once would make it impossible to distinguish them visually. For this reason, we recently proposed the use of virtual reality (VR) as an improved method to observe complex structures. We focus on astrocytes23 to overcome occlusion (which is the blocking of the visibility of an object of interest with a second one, in a 3D space) and ease qualitative assessments of the reconstructions, including proofreading, as well as quantifications of features using count of points in space. We recently combined VR visual exploration with the use of GLAM (glycogen-derived lactate absorption model), a technique to visualize a map of lactate shuttle probability of neurites, by considering glycogen granules as light-emitting bodies23; in particular, we used VR to quantify the light peaks produced by GLAM.
Protocol
1. Image Processing Using Fiji
- Open the image stack by dragging and dropping the native file from the microscope containing the stack, or by dragging and dropping the folder containing the whole image stack into the software window.
NOTE: Fiji is able to automatically recognize all the standard image formats, such as .jpg, .tif, and .png, as well as proprietary file formats from microscope providers. While the following protocol has been optimized for image stacks from 3DEM, these steps might be used for light microscopy datasets as well. - Once the stack is opened, go to Image > Properties to make sure the voxel size has been read from the metadata. If not, it can be manually entered.
- Make sure to transform the image into 8-bit. Click on Image > Type and select 8-bit.
- If the original stack was in different sequential files/folders, use Concatenate to merge them in a single stack by selecting Image > Stacks > Tools > Concatenate.
- Save the stack by selecting File > Save as. It will be saved as a single .tif file and can be used as a backup and for further processing.
- If the original stack is acquired as different tiles, apply stitching within TrakEM2.
- Create a new TrakEM2 project using New > TrakEM2 (blank).
- Within the viewport graphical user interface (GUI), import the stacks by clicking the right mouse button > Import > Import stack, and import all the tiles opened in the Fiji main GUI. Make sure to resize the canvas size to fit the entire stack by right-clicking > Display > Resize Canvas/Layer Set, and choose the y/x pixel size depending on the final size of the montage.
NOTE: For instance, if the montage should be finalized using four tiles of 4,096 x 4,096 pixels, the canvas size should be at least 8,192 x 8,192 pixels. Consider make it slightly bigger, and crop it later. - Superimpose the common portions of individual tiles by dragging and dropping them on the layer set. Use the top left slider to modify the transparency to help with the superimposition.
- Montage and realign the stacks by clicking the right mouse button > Align, and then select one of the options (see note below).
NOTE: SBEM or FIB-SEM are usually already well aligned, but minor misalignments might occur. Align layers is the automated pipeline for z alignment; Montage multiple layers is the automated pipeline for aligning tiles on each z stack, for the entire volume; Align multilayer mosaic combines the two previous options. This operation is time-consuming and subject to the random-access memory (RAM) of the machine. Consider using a high-end computer and let it run for hours/days, depending on the size of the stack. - Finally, export the image stacks and save them by using mouse right click > Make flat image, and then make sure to select the first to the last image in the drop-down menus Start and End.
- Using TrakEM2 embedded functions, segment structures of interest if needed.
- In the TrakEM2's GUI, right-click on Anything under the Template window and select Add new child > Area_list.
- Drag and drop Anything on top of the folder under Project Objects, and one Anything will appear there.
- Drag and drop the Area list from the template to the Anything located under Project Objects.
- On the image stack viewport, select the Z space with the cursor. The area list will appear, with a unique ID number. Select the brush tool on top (bottom right) and use the mouse to segment a structure by filling its cytosol, over the whole z stack.
- Export the segmented mask to be used in ilastik as seed for carving by right-clicking either on the Area list object on the Z space list, or on the mask in the viewport, and select Export > Area lists as labels (tif).
- Depending on the resolution needed for further reconstructions, if possible, reduce the pixel size of the image stack by downsampling, considering the memory requirements of the software that will be used for segmentation and reconstruction. Use Image > Adjust > Size.
NOTE: ilastik handles stacks of up to 500 pixels on xy well. Also, downsampling will reduce the resolution of the stack; therefore, take into account the minimum size at which the object still appears recognizable and, thus, can be segmented. - To enhance contrast and help the segmentation, the Unsharp mask filter can be applied to make membranes crisper. Use Process > Filter > Unsharp mask.
- Export the image stack as single images for further processing in segmentation software, (e.g., ilastik), using File > Save as… > Image sequence and choose .tif format.
2. Segmentation (Semi-automated) and Reconstruction Using ilastik 1.3.2
- In the main ilastik GUI, select the Carving module.
- Load the image stack using Add New > Add a single 3D/4D volume from sequence.
- Select Whole directory and choose the folder containing the image stack saved as single files. At the bottom of the new window, where options for loading the images are present, make sure to keep Z selected.
- For the following steps, all operations and buttons can be found on the left side of the main software GUI. Under the Preprocessing tab, use the standard options already checked. Use bright lines (ridge filters) and keep the filter scale at 1.600. This parameter can be modified afterward.
- Once the preprocessing is finished, select the next page in the drop-down menu of the Labeling module. One Object and one Background are present by default.
- Select the object seed by clicking on it, and draw a line on top of the structure of interest; then, select the background seed and draw one or multiple lines outside the object to be reconstructed. Then, click on Segment, and wait.
NOTE: Depending on the power of the computer and the size of the stack, segmentation could take from a few seconds to hours. Once it's done, a semitransparent mask highlighting the segmentation should appear on top of the segmented structure.- Scroll through the stack to check on the segmentation. The segmentation might not be accurate and not follow the structure of interest accurately or spill out from it. Correct any spillover by placing a background seed on the spilled segmentation, and add an object seed over the nonreconstructed segment of the object of interest.
- If the segmentation is still not correct, try to modify with the Bias parameter, which will increase or decrease the amount of uncertain classified pixels as accepted. Its value is 0.95 by default; decrease it to limit any spillover (usually to not less than 0.9) or increase if the segmentation is too conservative (up to 1).
- Another possibility is to go back to step 2.4 (by clicking on Preprocessing) and to modify the size of the filter; increasing the value (e.g., to 2) will minimize the salt-and-pepper noise-like effects but will also make membranes more blurred and smaller details harder to detect. This might limit spillover.
- Reiterate as much as needed, as long as all desired objects have been segmented. Once an object is finished, click on Save current object, below Segment. Two new seeds will appear, to start the segmentation of a new object.
- Extract surface meshes right away as .obj files by clicking on Export all meshes.
- If further proofreading is needed, segmentation can be extracted as binary masks. Select the mask to export it by clicking Browse objects; then, right-click on Segmentation > Export and, then, under Output file info, select tif sequence as format.
3. Proofreading/Segmentation (Manual) in TrakEM2 (Fiji)
- Load the image stack and create a new TrekEM2 project as per step 1.6.1.
- Import the masks that need proofreading using the option Import labels as arealists, available in the same import menu used to import the image stack.
- Select the arealist imported in the Z space and use the Brush tool to review the segmentation.
- Visualize the model in 3D by right-clicking on Area list > Show in 3D. In the following window asking for a resample number, a higher value will generate a lower resolution mesh.
NOTE: Depending on the size of the object, consider using a value between 4 and 6, which usually gives the best compromise between resolution and morphological detail. - Export the 3D mesh as wavefront .obj by choosing from the menu File > Export surfaces > Wavefront.
4. 3D Analysis
- Open Blender. For the following steps, make sure to install the NeuroMorph toolkit (available at available at https://neuromorph.epfl.ch/index.html) and the glycogen analysis toolkit (available at https://github.com/daniJb/glyco-analysis).
- Import the objects using the NeuroMorph batch import by clicking Import Objects under the Scene menu, to import multiple objects at once. Make sure to activate Use remesh and Smooth shading.
NOTE: It is not recommended to click on Finalize remesh if there is uncertainty about the initial size of the object. Import tree depth at value 7 (default) is usually good to maintain an acceptable resolution and morphology of the object. - Select the object of interest from the outliner and modify the octree depth of the remesh function under the Modifiers menu iteratively to minimize the number of vertices and to avoid losing details in resolution and correct morphology. When changing the octree depth, the mesh on the main GUI will change accordingly. Once finished, click on Apply to finalize the process.
NOTE: Values around 4 are usually good for small objects (such as postsynaptic densities), values around 7 for larger ones (such as long axons or dendrites), and values around 8 or 9 for complete cellular morphologies where details of different resolutions should be maintained. - Use the image superimposition tool Image stack interactions on the left panel, under the NeuroMorph menu, to load the image stack. Make sure to enter the physical size of the image stack for x, y, and x (in microns or nanometers, depending on the units of the mesh) and select the path of the stack by clicking on Source_Z. X and Y are orthogonal planes; they are optional and will be loaded only if the user inserts a valid path.
- Then, select a mesh on the viewport by right-clicking on it, enter the edit mode by pressing Tab, select one (or more) vertices using mouse right click, and finally, click on Show image at vertex. One (or more) cut plane(s) with the micrograph will appear superimposed on top of the mesh.
- Select the cut plane by right-clicking on it, press Ctrl + Y, and scroll over the 3D model using the mouse scroll. This can also be used as proofreading method.
- Use Measurement tools for quantifications of surface areas, volumes, and lengths. These operations are documented in more details by Jorstad et al.19 and on the NeuroMorph website24.
- Use the Glycogen analysis tool for quantifying glycogen proximity toward spines and boutons. Operations are documented in more details in a previous publication10 and in the glycogen analysis repository25.
- Import the objects using the NeuroMorph batch import by clicking Import Objects under the Scene menu, to import multiple objects at once. Make sure to activate Use remesh and Smooth shading.
- Run GLAM23. The code, executable, and some test files are available in the GLAM repository26.
- Prepare two geometry files in .ply format: one source file containing glycogen granules and one target file containing the morphology surfaces.
- Prepare three .ascii colormap files (each line containing t_norm(0..1) R(0..255) G(0..255) B(0..255)) for representing local absorption values, peak values, and average absorption values.
- Execute the C++ script GLAM, with the geometry files (step 4.2.1) and colormap files (step 4.2.2), by setting the influence radius (in microns), the maximum expected absorption value, and a normalized threshold for clustering the absorption peaks. For information about other parameters, execute the script with the -help option.
- Export the results as .ply or .obj files: target objects color-mapped with GLAM values, absorption peak markers represented as color-coded spheres, and target objects color-coded with respect to the mean absorption value.
- Open VR Data Interact. VR Data Interact-executable code is available in a public repository27.
- Import meshes to be visualized, following the instructions in the VR menu. Make sure to import the .obj files computed in step 4.2.1, in case a GLAM analysis is needed.
Representative Results
By using the procedure presented above, we show results on two image stacks of different sizes, to demonstrate how the flexibility of the tools makes it possible to scale up the procedure to larger datasets. In this instance, the two 3DEM datasets are (i) P14 rat, somatosensory cortex, layer VI, 100 µm x 100 µm x 76.4 µm4 and (ii) P60 rat, hippocampus CA1, 7.07 µm x 6.75 µm x 4.73 µm10.
The preprocessing steps (Figure 1) can be performed the same way for both datasets, simply taking into account that a larger dataset such as the first stack, which is 25 GB, requires more performing hardware to handle large data visualization and processing. The second stack is only 1 GB, with a perfectly isotropic voxel size.
The size of the data might not be directly related to the field of view (FOV), rather than to the resolution of the stack itself, which depends on the maximum pixel size of the sensor of the microscope, and the magnification of the stack. In any case, logically, larger FOVs are likely to occupy more physical space compared to smaller FOVs, if acquired at the same resolution.
Once the image stack is imported, as indicated in section 1 of the protocol, in Fiji software (Figure 1A), a scientific release of ImageJ12, one important point is to make sure that the image format is 8-bit (Figure 1B). This because many acquisition software different microscopy companies producers generate their proprietary file format in 16-bit to store metadata relevant to information about the acquiring process (i.e., pixel size, thickness, current/voltage of the electron beam, chamber pressure) together with the image stack. Such adjustments allow scientists to save memory, as the extra 8-bit containing metadata does not affect the images. The second important parameter to check is the voxel size, which then allows the reconstruction following the segmentation to be performed on the correct scale (micrometers or nanometers; Figure 1B).
Stacks might need realignment and/or stitching if they have been acquired using tiling; these operations can be performed within TrakEM2 (Figure 2A), although regarding realignment, automated 3DEM techniques like FIB-SEM or 3View are usually well realigned.
One last step requires filtering and, possibly, downsampling of the stack, depending on which objects need to be reconstructed and whether downsampling affects the recognition of the reconstructable features. For instance, for the larger stack (of the somatosensory cortex of P14 rats), it was not possible to compromise the resolution for the benefit of the reconstruction efficiency, while for the smaller stack (of the hippocampus CA1 of P60 rats), it was possible to do so because the resolution was far above what was needed for the smallest objects to be reconstructed. Finally, the use of the unsharp mask enhances the difference between the membranes and the background, making it favorable for the reconstructions of software like ilastik which uses gradients to pre-evaluate borders.
Following the image processing, reconstruction can be performed either manually using TrakEM2, or semi-automatically using ilastik (Figure 2C). A dataset like the smaller one listed here (ii), that can be downsampled to fit into memory, can be fully segmented using ilastik (Figure 2B) to produce a dense reconstruction. In the case of the first dataset listed here (i), we have managed to load and preprocess the entire dataset with a Linux workstation with 500 GB of RAM. The sparse segmentation of 16 full morphologies was obtained with a hybrid pipeline, by extracting rough segmentation that was manually proofread using TrakEM2.
The 3D analysis of features like surface areas, volumes, or the distribution of intracellular glycogen can be performed within a Blender environment (Figure 3) using custom codes, such as NeuroMorph19 or glycogen analysis10.
In case of datasets containing glycogen granules as well, analysis on their distribution can be inferred using GLAM, a C++ code that would generate colormaps with influence area directly on the mesh.
Finally, such complex datasets can be visualized and analyzed using VR, which has been proven to be useful for the analysis of datasets with a particular occluded view (Figure 4). For instance, peaks inferred from GLAM maps were readily inferred visually from dendrites in the second dataset discussed here.
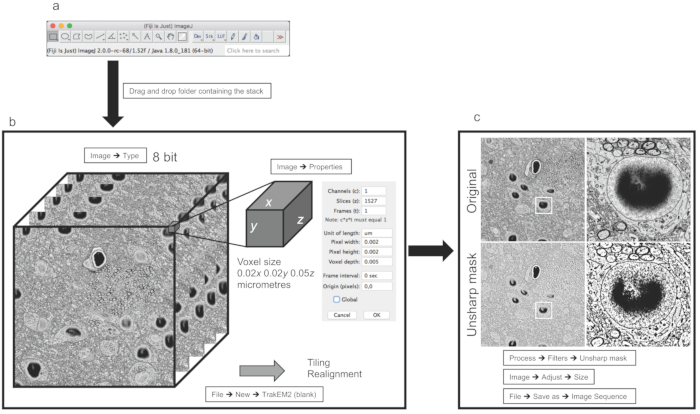
Figure 1: Image processing and preparation for image segmentation. (a) Main Fiji GUI. (b) Example of stacked images from dataset (i) discussed in the representative results. The panel on the right shows properties allowing the user to set the voxel size. (c) Example of a filer and resize operation applied to a single image. The panels on the right show magnifications from the center of the image. Please click here to view a larger version of this figure.
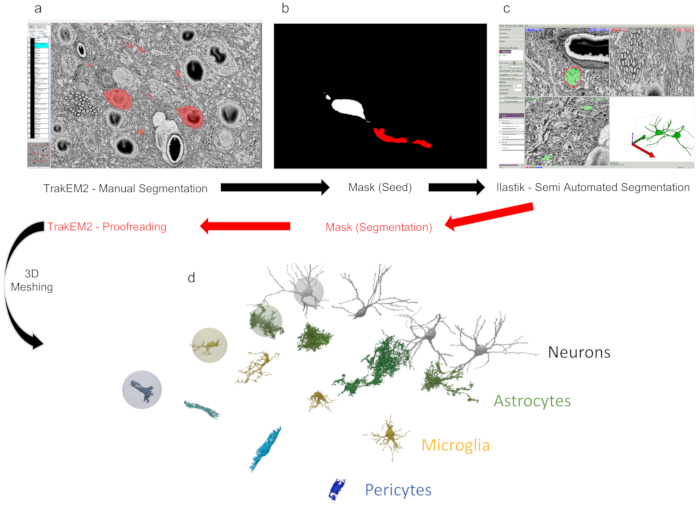
Figure 2: Segmentation and reconstruction using TrakEM2 and ilastik. (a) TrakEM2 GUI with objects manually segmented (in red). (b) The exported mask from panel a can be used as input (seed) for (c) semi-automated segmentation (carving). From ilastik, masks (red) can be further exported to TrakEM2 for manual proofreading. (d) Masks can be then exported as 3D triangular meshes to reveal reconstructed structures. In this example, four neurons, astrocytes, microglia, and pericytes from dataset (i) (discussed in the representative results) were reconstructed using this process. Please click here to view a larger version of this figure.
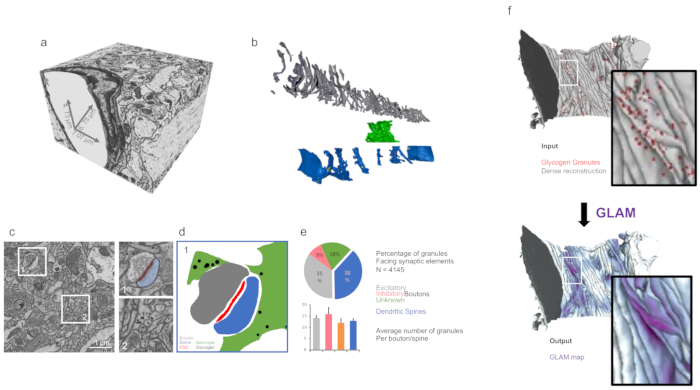
Figure 3: 3D analysis of reconstructed morphologies using customized tools. (a) Isotropic imaged volume from FIB-SEM dataset (ii) (as discussed in the representative results). (b) Dense reconstruction from panel a. Grey = axons; green = astrocytic process; blue = dendrites. (c) Micrograph showing examples of targets for quantifications such as synapses (dataset (i)) and astrocytic glycogen granules (dataset (ii)) in the right magnifications. (d) Mask from panel c showing the distribution of glycogen granules around synapses. (e) Quantification of glycogen distribution from the dataset from panel c, using the glycogen analysis toolbox from Blender. The error bars indicate standard errors. N = 4,145 glycogen granules. (f) A graphic illustration of the input and output visualization of GLAM. Please click here to view a larger version of this figure.
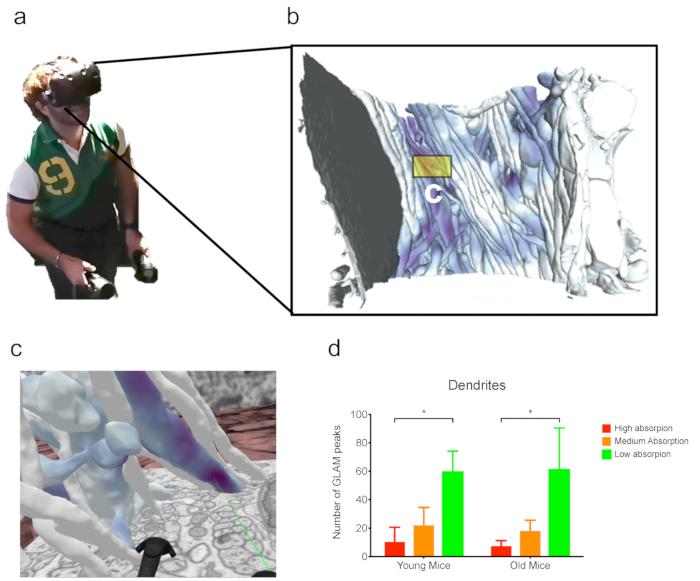
Figure 4: Analysis in VR. (a) A user wearing a VR headset while working on (b) the dense reconstruction from FIB-SEM dataset (ii) (as discussed in the representative results). (c) Immersive VR scene from a subset of neurites from panel b. The green laser is pointing to a GLAM peak. (d) Example of an analysis from GLAM peak counts in VR. N = 3 mice per each bar. Analysis of FIB-SEM from a previous publication28. The error bars indicate the standard errors; *p < 0.1, one-way ANOVA. Please click here to view a larger version of this figure.
Discussion
The method presented here is a useful step-by-step guide for the segmentation and 3D reconstruction of a multiscale EM dataset, whether they come from high-resolution imaging techniques, like FIB-SEM, or other automated serial sectioning and imaging techniques. FIB-SEM has the advantage of potentially reaching perfect isotropy in voxel size by cutting sections as thin as 5 nm using a focused ion beam, its FOV might be limited to 15-20 µm because of side artifacts, which are possibly due to the deposition of the cut tissue if the FOV exceeds this value. Such artifacts can be avoided by using other techniques, such as SBEM, which uses a diamond knife to cut serial sections inside the microscope chamber. In this latter case, the z resolution can be around 20 nm at best (usually, 50 nm), but the FOV might be larger, although the pixel resolution should be compromised for a vast region of interest. One solution to overcome such limitations (magnification vs. FOV) is to divide the region of interest in tiles and acquire each of them at a higher resolution. We have shown here results from both an SBEM stack-dataset (i) in the representative results-and a FIB-SEM stack-dataset (ii) in the representative results.
As the generation of larger and larger datasets is becoming increasingly more common, efforts in creating tools for pixel classification and automated image segmentation are multiplying; nevertheless, to date, no software has proven reliability comparable to that of human proofreading, which is therefore still necessary, no matter how time-consuming it is. In general, smaller datasets that can be downsampled, like in the case of dataset (ii), can be densely reconstructed by a single, expert user in a week, including proofreading time.
The protocol presented here involves the use of three software programs in particular: Fiji (version 2.0.0-rc-65/1.65b), ilastik (version 1.3.2 rc2), and Blender (2.79), which are all open-source and multi-platform programs and downloadable for free. Fiji is a release of ImageJ, powered by plugins for biological image analysis. It has a robust software architecture and is suggested as it is a common platform for life scientists and includes TrakEM2, one of the first and most widely used plugins for image segmentation. One issue experienced by many users lately is the transition from Java 6 to Java 8, which is creating compatibility issues; therefore, we suggest refraining from updating to Java 8, if possible, to allow Fiji to work properly. ilastik is a powerful software providing a number of frameworks for pixel classification, each one documented and explained on their website. The carving module used for the semi-automated segmentation of EM stacks is convenient as it saves much time, allowing scientists to reduce the time spent on manual work from months to days for an experienced user, as with a single click an entire neurite can be segmented in seconds. The preprocessing step is very intense from a hardware point of view, and very large datasets, like the SBEM stack presented here, which was 26 GB, require peculiar strategies to fit into memory, considering that one would acquire large dataset because cannot compromise field of view and resolution. Therefore, downsampling might not be an appropriate solution in this case. The latest release of the software can do the preprocessing in a few hours with a powerful Linux workstation, but the segmentation would take minutes, and scrolling through the stack would still be relatively slow. We still use this method for a first, rough segmentation, and proofread it using TrakEM2. Finally, Blender is a 3D modeling software, with a powerful 3D rendering engine, which can be customized with python scripts that can be embedded in the main GUI as add-ons, such as NeuroMorph and glycogen analysis. The flexibility of this software comes with the drawback that, in contrast to Fiji, for instance, it is not designed for the online visualization of large datasets; therefore, visualizing and navigating through large meshes (exceeding 1 GB) might be slow and not efficient. Because of this, it is always advisable to choose techniques that reduce mesh complexity but are careful not to disrupt the original morphology of the structure of interest. The remesh function comes in handy and is an embedded feature of the NeuroMorph batch import tool. An issue with this function is that, depending on the number of vertices of the original mesh, the octree depth value, which is related to the final resolution, should be modified accordingly. Small objects can be remeshed with a small octree depth (e.g. 4), but the same value might disrupt the morphology of larger objects, which needs bigger values (6 at best, to 8 or even 9 for a very big mesh, such as a full cell). It is advisable to make this process iterative and test the different octree depths if the size of the object is not clear.
As mentioned previously, one aspect that should be taken into account is the computational power to be dedicated to reconstruction and analysis, related to the software that is being used. All the operations shown in the representative results of this manuscript were obtained using a MacPro, equipped with an AMD FirePro D500 Graphics card, 64 GB of RAM, and an Intel Xeon E5 CPU with 8 cores. Fiji has a good software architecture for handling large datasets; therefore, it is recommended to use a laptop with a good hardware performance, such as a MacBook Pro with a 2.5 GHz Intel i7 CPU and 16 GB of RAM. ilastik software is more demanding in terms of hardware resources, in particular during the preprocessing step. Although downsampling the image stack is a good trick to limit the hardware requests from the software and allows the user to process a stack with a laptop (typically if it is below 500 pixels in x,y,z), we suggest the use of a high-end computer to run this software smoothly. We use a workstation equipped with an Intel Xeon Gold 6150 CPU with 16 cores and 500 GB of RAM.
When provided with an accurate 3D reconstruction, scientists can discard the original micrographs and work directly on the 3D models to extract useful morphometric data to compare cells of the same type, as well as different types of cells, and take advantage of VR for qualitative and quantitative assessments of the morphologies. In particular, the use of the latter has proven to be beneficial in the case of analyses of dense or complex morphologies that present visual occlusion (i.e., the blockage of view of an object of interest in the 3D space by a second one placed between the observer and the first object), making it difficult to represent and analyze them in 3D. In the example presented, an experienced user took about 4 nonconsecutive hours to observe the datasets and count the objects. The time spent on VR analysis might vary as aspects like VR sickness (which can, to some extent, be related to car sickness) can have a negative impact on the user experience; in this case, the user might prefer other analysis tools and limit their time dedicated to VR.
Finally, all these steps can be applied to other microscopy and non-EM techniques that generate image stacks. EM generates images that are, in general, challenging to handle and segment, compared with, for instance, fluorescence microscopy, where something comparable to a binary mask (signal versus a black background), that in principle can be readily rendered in 3D for further processing, often needs to be dealt with.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the King Abdullah University of Science and Technology (KAUST) Competitive Research Grants (CRG) grant "KAUST-BBP Alliance for Integrative Modelling of Brain Energy Metabolism" to P.J.M.
Materials
| Fiji | Open Source | 2.0.0-rc-65/1.65b | Open Source image processing editor www.fiji.sc |
| iLastik | Open Source | 1.3.2 rc2 | Image Segmentation tool www.ilastik.org |
| Blender | Blender Foundation | 2.79 | Open Source 3D Modeling software www.blender.org |
| HTC Vive Headset | HTC | Vive / Vive Pro | Virtual Reality (VR) Head monted headset www.vive.com |
| Neuromorph | Open Source | — | Collection of Blender Addons for 3D Analysis neuromorph.epfl.ch |
| Glycogen Analysis | Open Source | — | Blender addon for analysis of Glycogen https://github.com/daniJb/glyco-analysis |
| GLAM | Open Source | — | C++ Code For generating GLAM Maps https://github.com/magus74/GLAM |
Riferimenti
- Leighton, S. B. SEM images of block faces, cut by a miniature microtome within the SEM – a technical note. Scanning Electron Microscopy. , 73-76 (1981).
- Knott, G., Marchman, H., Wall, D., Lich, B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. Journal of Neuroscience. 28, 2959-2964 (2008).
- Denk, W., Horstmann, H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLOS Biology. 2, e329 (2004).
- Coggan, J. S., et al. A Process for Digitizing and Simulating Biologically Realistic Oligocellular Networks Demonstrated for the Neuro-Glio-Vascular Ensemble. Frontiers in Neuroscience. 12, (2018).
- Tomassy, G. S., et al. Distinct Profiles of Myelin Distribution Along Single Axons of Pyramidal Neurons in the Neocortex. Science. 344, 319-324 (2014).
- Wanner, A. A., Genoud, C., Masudi, T., Siksou, L., Friedrich, R. W. Dense EM-based reconstruction of the interglomerular projectome in the zebrafish olfactory bulb. Nature Neuroscience. 19, 816-825 (2016).
- Kasthuri, N., et al. Saturated Reconstruction of a Volume of Neocortex. Cell. 162, 648-661 (2015).
- Calì, C., et al. The effects of aging on neuropil structure in mouse somatosensory cortex—A 3D electron microscopy analysis of layer 1. PLOS ONE. 13, e0198131 (2018).
- Calì, C., Agus, M., Gagnon, N., Hadwiger, M., Magistretti, P. J. Visual Analysis of Glycogen Derived Lactate Absorption in Dense and Sparse Surface Reconstructions of Rodent Brain Structures. , (2017).
- Calì, C., et al. Three-dimensional immersive virtual reality for studying cellular compartments in 3D models from EM preparations of neural tissues. Journal of Comparative Neurology. 524, 23-38 (2016).
- Fiala, J. C. Reconstruct: a free editor for serial section microscopy. Journal of Microscopy. 218, 52-61 (2005).
- Cardona, A., et al. TrakEM2 software for neural circuit reconstruction. PLOS ONE. 7, e38011 (2012).
- Januszewski, M., et al. High-precision automated reconstruction of neurons with flood-filling networks. Nature Methods. 1, (2018).
- Shahbazi, A., et al. Flexible Learning-Free Segmentation and Reconstruction of Neural Volumes. Scientific Reports. 8, 14247 (2018).
- Sommer, C., Straehle, C., Köthe, U., Hamprecht, F. A. Ilastik: Interactive learning and segmentation toolkit. , (2011).
- Holst, G., Berg, S., Kare, K., Magistretti, P., Calì, C. Adding large EM stack support. , (2016).
- Beyer, J., et al. ConnectomeExplorer: query-guided visual analysis of large volumetric neuroscience data. IEEE Transactions on Visualization and Computer Graphics. 19, 2868-2877 (2013).
- Beyer, J., et al. Culling for Extreme-Scale Segmentation Volumes: A Hybrid Deterministic and Probabilistic Approach. IEEE Transactions on Visualization and Computer. , (2018).
- Jorstad, A., et al. NeuroMorph: A Toolset for the Morphometric Analysis and Visualization of 3D Models Derived from Electron Microscopy Image Stacks. Neuroinformatics. 13, 83-92 (2015).
- Jorstad, A., Blanc, J., Knott, G. NeuroMorph: A Software Toolset for 3D Analysis of Neurite Morphology and Connectivity. Frontiers in Neuroanatomy. 12, 59 (2018).
- Calì, C. Astroglial anatomy in the times of connectomics. Journal of Translational Neuroscience. 2, 31-40 (2017).
- Mohammed, H., et al. Abstractocyte: A Visual Tool for Exploring Nanoscale Astroglial Cells. IEEE Transactions on Visualization and Computer Graphics. , (2017).
- Agus, M., et al. GLAM: Glycogen-derived Lactate Absorption Map for visual analysis of dense and sparse surface reconstructions of rodent brain structures on desktop systems and virtual environments. Computers & Graphics. 74, 85-98 (2018).
- École Polytechnique Fédérale de Lausanne. . NeuroMorph Analysis and Visualization Toolset. , (2018).
- . GitHub – daniJb/glycol-analysis: Glycogen_analysis.py is a python-blender API based script that performs analysis on a reconstructed module of glycogen data Available from: https://github.com/daniJb/glyco-analysis (2018)
- . GitHub – magus74/GLAM: Glycogen Lactate Absorption Model Available from: https://github.com/magus74/GLAM (2019)
- Agus, M., et al. GLAM: Glycogen-derived Lactate Absorption Map for visual analysis of dense and sparse surface reconstructions of rodent brain structures on desktop systems and virtual environments. Dryad Digital Repository. , (2018).
- Calì, C., et al. Data from: The effects of aging on neuropil structure in mouse somatosensory cortex—A 3D electron microscopy analysis of layer 1. Dryad Digital Repository. , (2018).

