Preparation and Application of a New Bacterial Biosensor for the Presumptive Detection of Gunshot Residue
Summary
A protocol is presented using synthetic biology techniques to synthesize a set of bacterial biosensors for the analysis of gunshot residue, and to test the functioning of the devices for their intended use using fluorescence spectroscopy.
Abstract
MicRoboCop is a biosensor that has been designed for a unique application in forensic chemistry. MicRoboCop is a system made up of three devices that, when used together, can indicate the presence of gunshot residue (GSR) by producing a fluorescence signal in the presence of three key analytes (antimony, lead, and organic components of GSR). The protocol describes the synthesis of the biosensors using Escherichia coli (E. coli), and the analytical chemistry methods used to evaluate the selectivity and sensitivity of the sensors. The functioning of the system is demonstrated by using GSR collected from the inside of a spent cartridge casing. Once prepared, the biosensors can be stored until needed and can be used as a test for these key analytes. A positive response from all three analytes provides a presumptive positive test for GSR, while each individual device has applications for detecting the analytes in other samples (e.g., a detector for lead contamination in drinking water). The main limitation of the system is the time required for a positive signal; future work may involve studying different organisms to optimize the response time.
Introduction
A biosensor is any analytical device that uses biological components (such as proteins, nucleic acids, or whole organisms) that produce a response that can be used for the detection of a chemical substance or analyte. As an example, the coal mining industry used a biosensor for much of the 20th century to detect the presence of toxic mine gases: the canary in the coal mine1. The biological organism's (canary's) response (death or distress) to a chemical analyte (carbon monoxide) was observed by the miners in order to protect the workers. In a more modern and sophisticated example, bacteria can be altered using synthetic biology techniques to respond to the presence of a certain chemical analyte by exhibiting a specific response, such as the expression of a fluorescent protein.
Synthetic biology is a broad term that refers to the construction of biological devices and systems that do not exist naturally, or the re-design of existing biological systems for a specific purpose2. Synthetic biology is distinguished from genetic engineering by a standard methodology and the existence of standardized parts (standard synthetic biology genetic elements) that can be used to synthesize devices and systems. A part is introduced into the genome of a device, an organism such as a bacterium, to express a certain trait that will serve as an indication of function. For example, in many synthetic devices, the expression of a fluorescent protein is introduced into a single celled organism as a reporter protein. Multiple devices can be combined into a system. The genomes of microorganisms such as bacteria are easy to manipulate in this manner. Numerous examples of biosensors specific to a wide range of chemical analytes have been reported in the literature over the last decade3,4.
In this work, the MicRoboCop system is presented as an example of a biosensor designed using synthetic biology techniques with novel applications in forensic and environmental chemistry. MicRoboCop is a system of three separate devices that, when combined, will allow Escherichia coli to express red fluorescent protein (RFP) in the presence of gunshot residue (GSR) that has been collected from a person’s hands or a surface. Each of the three devices responds to a specific chemical analyte that is known to be a component of GSR5. The three analytes to which the system responds are I. 2,4,6-trinitrotoluene (TNT) and related compounds, II. lead (in the form of lead ions), and III. antimony (also in the form of ions).
GSR consists of many different chemical substances, but the three usually used to identify a residue as GSR are barium, lead, and antimony5. The standard evidentiary test for the identification of GSR is to use scanning electron microscopy (SEM) with energy dispersive X-ray fluorescence (EDX)5. SEM-EDX allows analysts to identify the unique morphology and the elemental components of GSR. Presently, there are few widely used binary presumptive tests available. One recently published presumptive test uses ion-mobility spectroscopy (IMS), which is specialized equipment that might not be available in many labs6. There are also a few color “spot” tests that can be used, though they are typically used for distance determination or for GSR identification on bullet holes and wounds5. Additionally, there has been some limited attention in the literature to electrochemical tests for GSR that employ voltammetric analysis, which has the advantage of potentially being field portable, or anodic stripping voltammetry, which is an extremely sensitive method for metallic elements7. There is very little mention in the literature of biosensors designed specifically for the purpose of detecting GSR, though some biosensors for other forensic applications have been published8.
The biological elements for each device in the MicRoboCop system, and the plasmid construction, are illustrated in Figure 1. The curved arrow in Figure 1b represents the promoter region that is activated in the presence of the analyte, the oval is the ribosomal binding site that allows translation of the reporter protein, the gray box labeled RFP is the gene that expresses red fluorescent protein, and the red octagon is the transcription termination site. All three devices will be used together as a system to detect GSR. Each device with a specific promoter (SbRFP, PbRFP, and TNT-RFP) will be incubated with the sample that is being tested and fluorescence of RFP will be measured. RFP will only be expressed if the appropriate chemical analyte is present and activates the promoter region. Three devices that respond to some of the chemical substances present in GSR have been designed and are presented in this work.
The promoters used in the three MicRoboCop devices are an arsenic and antimony sensitive promoter, SbRFP9,10, a lead sensitive promoter, PbRFP11,12 and a TNT sensitive promoter, TNT-RFP13. Because a search in the literature revealed no promoter designed to respond to barium, the TNT promoter was selected instead since this promoter is sensitive to a number of structurally related compounds (in particular, 2,4-dinitrotoluene and dinitrobenzene) that are known to be a part of the organic compounds left behind in GSR. This promoter has successfully been used to specifically detect minute quantities of TNT and 2,4-dinitrotoluene (2,4-DNT) in buried land mines13. Using the three devices together as a system, a positive test for GSR will produce fluorescence in all three devices. A fluorescence signal in only one or two devices will indicate another environmental source of the analyte(s) or in the case of the TNT promoter, activation by a compound that is not an organic compound left behind in GSR. By using all three devices together, the possibility of a false positive results due to environmental sources is minimized. Lead-free ammunition, which is gaining in popularity, still represents only about 5% of ammunition sales in the United States; hence, false negative results due to the absence of lead may be a possibility but there is still utility in a sensor that uses lead as a marker for GSR14. In addition to this specific forensic application, each device can be used separately for purposes of detecting environmental contaminants.
The protocols presented include the synthetic biology techniques used to create the devices (sensor bacteria) and the analytical techniques to check the function of the devices and analyze the fluorescence signals obtained. The protocol also includes collection of forensic evidence in the form of hand wiping to collect GSR from the hands of a suspect or swabbing to collect GSR from a surface. Results from the lead sensor device are presented as example results, along with a demonstration of a positive test for GSR using a spent cartridge casing.
Protocol
NOTE: Synthesis of E. coli expressing RFP is presented.
1. Preparation of plasmid DNA from E. coli
- Thaw E.coli containing a plasmid with an RFP gene and ampicillin resistance gene and grow the E.coli on Luria Broth (LB) agar plates containing 100 µg/mL ampicillin at 37 °C for 24 h. For example, use the J10060 plasmid from the registry of standard biological parts used for synthetic biology (see Table of Materials). The J10060 plasmid includes a gene for RFP (red fluorescent protein) under the control of a pBad promoter region and an ampicillin resistance gene. Alternatively, transform E.coli (refer to step 3.2) with the plasmid prior to growth on the LB agar plates.
- Follow a standard miniprep protocol (see Table of Materials) to isolate DNA from 1 mL of an E. coli culture that contains the J10060 plasmid. The purpose of the following protocol is to remove the pBad promoter and replace it with the desired promoter for the device.
- Following the plasmid miniprep, store DNA in the freezer until ready for digestion.
2. Restriction enzyme digestion
- Set up the following reaction in a microcentrifuge tube for EcoRI and NheI digestion: 10 μL of J10060 plasmid DNA (isolated in step 1), 8 μL of water, and 1 μL each of EcoRI and NheI enzymes pre-mixed with 1 μL of buffer (see Table of Materials).
- For the promoter DNA, set up the following reaction in a microcentrifuge tube for EcoRI and NheI digestion: 10 μL of annealed promoter DNA sequences (8 μL of water, and 1 μL each of EcoRI and NheI enzymes pre-mixed with 1 μL of buffer.
- For Sb-, Pb-, or TNT-RFP (see Table of Materials), dissolve the oligonucleotides in buffer (30 mM HEPES, pH 7.5; 100 mM potassium acetate), incubate in equal molar concentrations, heat to 94 °C for 2 min, and gradually cool at room temperature).
- Mix the samples by pipetting gently up and down with the pipette set to 10 μL.
- Incubate for 30 min at 37 °C.
- Heat inactivate the enzymes at 80 °C for 5 min.
- Store the digested DNA in the freezer until ready for the next step.
3. Ligation and transformation
- Ligation
- Using the plasmid and promoter DNA that were double digested with EcoRI and NheI in step 2, set up the reaction mixture shown in Table 1 in a microcentrifuge tube on ice; add the T4 DNA Ligase last.
NOTE: Table 1 shows a ligation using a molar ratio of 1:3 vector to insert for the indicated DNA sizes. - Gently mix the reaction by pipetting up and down and microcentrifuge briefly.
- Incubate at room temperature for 10 min.
- Heat inactivate at 65 °C for 10 min.
- Using the plasmid and promoter DNA that were double digested with EcoRI and NheI in step 2, set up the reaction mixture shown in Table 1 in a microcentrifuge tube on ice; add the T4 DNA Ligase last.
- Transformation
- Thaw a tube with 20 µL of DH5-alpha Competent E. coli cells on ice until the last ice crystals disappear.
- Add 5 µL of plasmid DNA to the cell mixture. Carefully flick the tube 4-5 times to mix the cells and DNA.
- Place the mixture on ice for 2 min.
- Heat shock at exactly 42 °C for exactly 30 s. Do not mix.
- Place on ice for 2 min. Do not mix.
- Pipette 380 µL of room temperature SOC into the mixture. Immediately spread onto an LB agar plate containing ampicillin (100 µg/mL) and incubate overnight at 37 °C.
- Check the plates within 24 h for growth.
- Seal the plates with sealing film and store in the refrigerator until ready for next step.
4. Colony PCR
- Add to a PCR tube (set up 4 reaction tubes) the reaction mixtures shown in Table 2.
- Gently mix the reactions by pipetting up and down.
- Using a yellow pipette tip, scrape a colony (or very small region) of the transformed E. coli. Transfer a swipe of this E. coli onto a new LB/ampicillin/agar plate that has been sectioned off, and then insert the pipette tip into the PCR tube. Shake the pipette tip to mix the E. coli with the PCR mix. Repeat three more times for additional colonies. Transfer the PCR tubes to a PCR machine and begin thermocycling using the program shown in Table 3.
- Run gel electrophoresis using a 2% agarose gel in TAE to determine which colonies have the best ligation into the plasmid and grow those colonies on a new plate.
- Store the plates in the refrigerator until ready for testing. Prepare a liquid culture in Luria broth with 100 µg/mL ampicillin added for chemical testing.
5. DNA Sequencing
- For each sample, add 5 µL of plasmid, 4 µL of the sequencing primer, and 3 µL of deionized water.
- Place this mixture into a tube and send it for DNA sequencing (see Table of Materials).
- Analyze DNA sequence data to compare the expected and observed DNA sequences using DNA sequence analysis software to ensure that there is no mutation and that the genes were correctly inserted.
NOTE: Using E. coli as a chemical sensor is presented below.
6. Preparation of E. coli cultures
- Prepare LB with 100 µg/mL ampicillin for liquid cultures.
- Prepare liquid cultures of the sensor bacteria, the positive control* bacteria and the negative control** bacteria.
NOTE: *Positive control bacteria: E. coli containing a plasmid with the RFP gene under control of a constitutive promoter; plasmid E1010 from the registry of standard biological parts used for synthetic biology (see Table of Materials) was used in this work.
**Negative control bacteria: E. coli containing a plasmid with the RFP gene under control of a different promoter, such as the pBad promoter (plasmid J10060 from the registry of standard biological parts used for synthetic biology (see Table of Materials)) or a plasmid that does not have the RFP gene. - Place the cultures into a shaking incubator at 37 °C and 220 rpm for a minimum of 8 h, maximum of 18 h. Cloudy broth indicates bacterial growth.
7. Titrating E. coli to check function of device
NOTE: Once the sensors have been titrated to check function, this step does not need to be repeated. A positive control in the form of addition of lead, antimony, and 2,4-DNT or 1,3-dinitrobenzene (1,3-DNB) can check the function of the devices for each use without the need for the full titration.
- Prepare a stock solution of the analyte(s) of interest at a concentration of 10 ppm in water. If solubility is an issue, use a 50/50 water/methanol mixture.
- Using Table 4 as a guide, label the appropriate number of sterile culture tubes and place 2 mL of the cultured broth (from protocol step 6) into each tube.
NOTE: In order to determine a general analytical range, do at least three different levels of an analyte with the sensor bacteria, one level with the negative control, and one level with the positive control. There should also be one tube of each of the bacteria that has no added metal (another type of negative control). To more accurately determine analytical range and limits of detection, use a larger range of analyte concentrations. - Add analyte stock solution to the tubes containing 2 mL of broth as noted in Table 4, place the snap caps on the culture tube so that they are loose (to allow air flow into the tube), and vortex the culture tube.
- Leaving the snap caps loosely on the culture tubes, place into a shaking incubator at 220 rpm and 37 °C for at least 24 h.
- Remove the tubes from the incubator, snap the caps onto the tubes tightly, and store the tubes in the refrigerator until ready for fluorescence analysis.
8. Using E. coli as chemical sensor for GSR
- Using an ethanol-based wipe designed for removing lead (see Table of Materials), wipe all surfaces of the hands, including between the fingers. Use a separate wipe for each hand. Store the wipes in an appropriately labelled sealable baggie until analysis.
- For surfaces to be tested: use an alcohol-based wipe for large surfaces or a cotton swab moistened with ethanol for small surfaces.
NOTE: To demonstrate the sensors’ response to GSR, the inside of a spent .40 caliber cartridge casing was swabbed with an ethanol moistened cotton swab. - Wearing clean gloves and using scissors that have been cleaned with alcohol, cut an approximately 1 cm2 section out of the center of the wipe.
- Place the cut piece of the hand wipe or the cotton swab directly into a culture tube that contains 2 mL of the sensor bacteria, ensuring that it is submerged in the broth.
- Proceed as described above in steps 7.4 – 7.5.
9. Fluorescence analysis using portable spectrometer (see Table of Materials)
- Use a vortex mixer to shake the tubes.
- Prepare the spectrometer to collect fluorescence emission at the appropriate wavelength for your RFP variant with an excitation wavelength of 500 nm.
- Use the Luria broth to record a blank spectrum.
- Carefully transfer each supernatant to a low volume cuvette and collect the emission intensity.
10. Fluorescence analysis using 96-well plate reader (see Table of Materials)
- Use a vortex mixer to shake the tubes.
- Transfer 200 mL of the broth to a well in the well plate. Record which samples went into each well of the plate.
- Set up the fluorimeter to collect the emission intensity at the appropriate wavelength for the RFP variant.
11. Data analysis
- Using the signal obtained from all negative controls (RFP negative bacteria or sensor bacteria with no analyte added), calculate an average fluorescence signal for the background.
- Subtract the average background signal from each fluorescence signal obtained for the sensor bacteria to get a background corrected fluorescence intensity.
- Estimate the signal-to-noise (S/N) ratio by dividing the background corrected fluorescence intensity by the average background signal. If the S/N ratio is greater than 3, the test is positive.
Representative Results
Fluorescence spectra for the RFP variant used in this work are shown in Figure 2. These data are from the PbRFP device as it responds to lead and the TNT-RFP device as it responds to two analytes, 2,4-DNT and 1,3-DNB. This figure shows the spectrum of a negative control (no analyte added), and the spectra at two different levels of analyte added. The maximum fluorescence signal for the RFP variant used was observed at 575 nm (excitation wavelength 500 nm). The data in Figure 3 are representative of a single titration experiment (hence no error bars are included) of the PbRFP device, titrated as in step 7 of protocol. Figure 3a shows data collected from the portable spectrometer, while Figure 3b shows data collected from the fluorimeter (from the same set of solutions). There is a general trend of increasing fluorescence as the concentration of metal increases. It is worth noting that at high concentrations, greater than about 800 ppb, the response drops off due to the toxicity of the metal at such a high concentration. This maximum response level may vary depending on the analyte used. Our previous work with the SbRFP showed that the bacteria could tolerate higher levels (at least up to 1,000 ppb) of arsenic and antimony10. Literature on levels of these analytes collected from hand swabs indicates that these levels of lead and antimony are consistent with what might be collected from a hand swab15. Additionally, the results presented in Figure 4 demonstrate that the bacteria can tolerate the amounts of analytes present in a cartridge case swab without cell death, which will be significantly higher than what is collected from a hand swab.
Using the calculated S/N values for these data, the lowest detectable level of lead was 12 ppb (detectable as defined by an S/N greater than 3). In contrast, the S/N for the portable spectrometer data is only 2 at the highest level tested. However, the trend of increasing fluorescence with increasing analyte concentration is still clearly noted.
Figure 4a shows a positive test for GSR. To obtain this result, ethanol swabs were collected from the inside of a spent .40 caliber cartridge casing and added to the three sensor bacteria, as in step 8 of the protocol. This figure also shows a positive control (bacteria that constitutively expresses RFP) and a negative control in the form of the SbRFP device with no analyte added. The cartridge case swabs were used as proof-of-principle results. In future work, hand swabs will be collected from persons who are known to have fired a gun to show that the sensors are responsive to hand swabs as well.
| COMPONENT | 20 μL REACTION |
| 10X T4 DNA Ligase Buffer | 2 μL |
| Plasmid DNA (3 kb) | 3 μL |
| Promoter DNA (0.7kb) | 10 μL |
| Nuclease-free water | 4 μL |
| T4 DNA Ligase | 1 μL |
Table 1. Reaction mixture for ligation, protocol step 3.1.1.
| Component | 25 μL reaction |
| 10 µM Forward Primer | 0.5 µL |
| 10 µM Reverse Primer | 0.5 µL |
| OneTaq 2X Master Mix | 12.5 µL |
| Nuclease-free water | 11 µL |
Table 2. Reaction mixtures for colony PCR, protocol step 4.1.
| STEP | TEMP | TIME |
| Initial Denaturation | 94 °C | 30 s |
| 30 Cycles | 94 °C | 30 s |
| 55 °C | 45 s | |
| 68 °C | 60 s | |
| Final Extension | 68 °C | 5 min |
| Hold | 4 °C |
Table 3. PCR thermocycling parameters for protocol step 4.3.
| Tube ID | Bacteria | Concentration of analyte solution added (ppm) | Metal added | Volume of analyte solution added to 2,000 µL broth | [analyte], ppb |
| 1 | PbRFP | 10 | Pb | 2.5 | 12 |
| 2 | PbRFP | 10 | Pb | 75 | 361 |
| 3 | PbRFP | 10 | Pb | 150 | 698 |
| 4 | PbRFP | 0 | none | 0 | 0 |
| 5 | RFP neg | 10 | Pb | 10 | 50 |
| 6 | RFP pos | 10 | Pb | 10 | 50 |
Table 4. General experiment set up for titration of biosensors, protocol step 7.2.
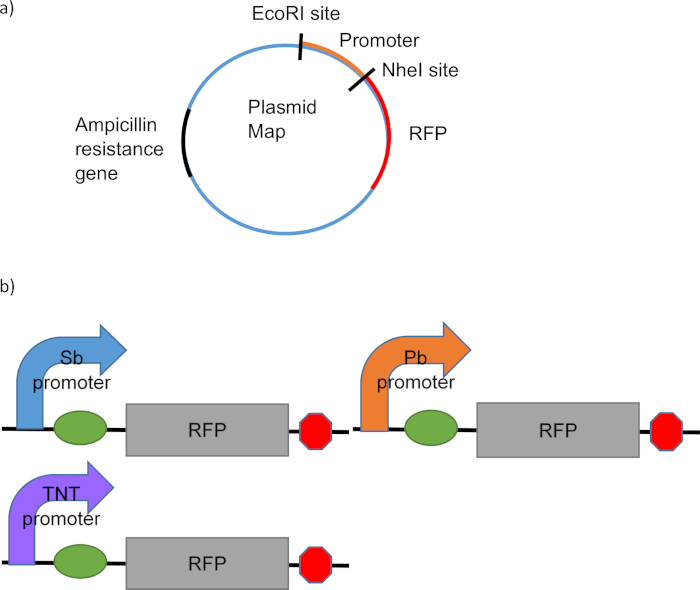
Figure 1. Biological elements of the MicRoboCop devices. (a) Diagram of the general device for MicRoboCop in a plasmid with an ampicillin resistance gene. (b) Diagram of each device that is combined to create the MicRoboCop system. Please click here to view a larger version of this figure.
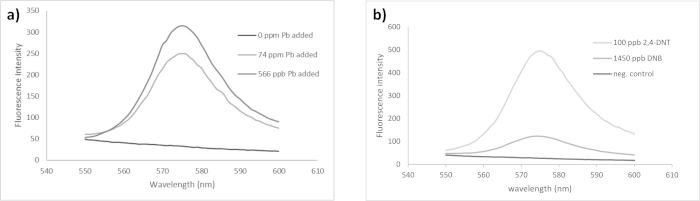
Figure 2. Fluorescence spectra of PbRFP and TNT-RFP bacteria in the presence and absence of analyte. Data collected on fluorimeter. (a) Fluorescence spectra of PbRFP bacteria in the presence and absence of analyte (Pb). (b) Fluorescence spectra of TNT-RFP bacteria in the presence and absence of two analytes (2,4-DNT and 1,3-DNB). Please click here to view a larger version of this figure.
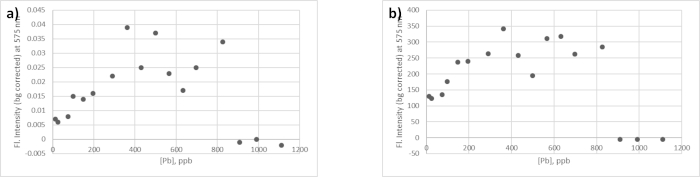
Figure 3. Comparison of the portable spectrometer system and fluorimeter for detection of the fluorescence spectra of PbRFP bacteria in the presence and absence of analyte (lead). (a) Lead titration data for PbRFP sensor bacteria collected on portable spectrometer system. (b) Lead titration data (same samples) for PbRFP sensor bacteria collected on fluorimeter. Please click here to view a larger version of this figure.
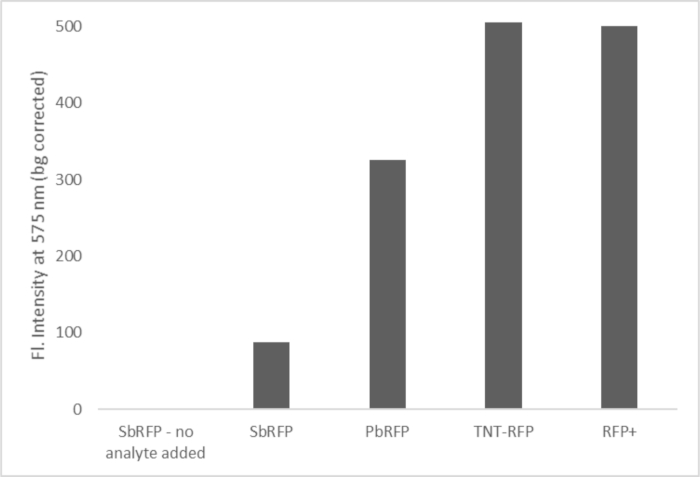
Figure 4. Ethanol swabs taken from the inside of a .40 caliber spent pistol cartridge to show the response of the three devices to GSR. S/N for all signals was greater than 3, indicating a positive test for GSR. Please click here to view a larger version of this figure.
Discussion
Modifications and troubleshooting
The experiment described in Table 4 can be modified in any way appropriate to the sensors that have been designed. The most important aspect of a chemical sensor is to evaluate its sensitivity and specificity. It is beneficial to ensure that a wide range of concentrations of the analyte is analyzed to determine the useful analytical range of the sensor. It is also worth determining a maximum level of analyte for the cells. Because the analytes used in this study are toxic metals (Pb and Sb) or organic compounds in a methanol solution (for the TNT derivatives), there is an upper level at which cell death due to the toxicity of the analyte or solution will occur (generally higher than 500 – 1,000 ppb for the experiments conducted thus far).
Limitations of the technique
The results presented in this work are qualitative in nature but are meant to demonstrate the quantitative capabilities of RFP modified E. coli. The sensitivity of the sensor can vary significantly between cultured batches depending on the density of the cells in the broth. If quantitative results are required, the cell concentration should be estimated by measuring the optical density of the liquid cultures before analysis. If the optical density of the cultures is determined, then the cells can be diluted appropriately to reduce variability between experiments. As a presumptive test for the desired analytes, however, the qualitative “present/not present” response is acceptable for the applications discussed here. The life span of the cells on the agar plate should also be noted – previous work has indicated that the plates can be stored in the refrigerator for up to 2 weeks, but the devices do not work very well towards the end of that time frame and beyond.
Another consideration is the choice of equipment used to analyze the fluorescence signal. Using a research grade spectrophotometer with a 96-well plate reader allows selection of exact excitation and emission wavelengths, which can increase sensitivity. Using this system, the results of up to 96 experiments can be collected simultaneously. RFP fluorescence may also be analyzed using a portable spectrometer system. Portable instruments typically allow selected excitation bands, which may or may not coincide with the excitation maxima of the RFP variant being used. However, as long as the excitation wavelength is within a reasonable range of the excitation maxima, the portable instrument will generally be serviceable (though with a loss in sensitivity). The cost of the portable systems is significantly less than the research grade spectrophotometer, and portability may certainly be an advantage. Based on the potential application of the bacteria, the analyst can decide whether or not the additional cost and loss of portability with the spectrophotometer system is justified.
Significance with respect to existing methods
The three-part MicRoboCop system described in this work is intended to be used as a qualitative, presumptive test for the presence of GSR. Currently, the “gold standard” evidentiary test for GSR requires expert analysis by SEM-EDX. SEM-EDX equipment is expensive and typically operated by highly specialized analysts. Additionally, GSR evidence is highly variable in forensic casework and many variables contribute to the deposition of GSR on hands and surfaces16. A presumptive test for GSR may be useful to investigators as providing probable cause for a search of person or property. When compared to electrochemical tests or tests such as ion mobility spectroscopy, this method offers simple, readily available instrumentation to which most analytical laboratories should have access.
Other applications
The devices described in this manuscript are designed to be combined into a three-part system for the presumptive identification of GSR. However, each device in the MicRoboCop system (SbRFP, PbRFP, and TNT-RFP) can also be used individually to detect chemical contamination in food, water, or environmental samples. Previous work has shown that the TNT-RFP device can be used as an in situ sensor for land mines13,17. Results presented here and in our previous work10 have shown that the SbRFP and PbRFP devices can detect concentrations low enough to rival more expensive and sophisticated equipment such as inductively coupled plasma atomic emission spectroscopy (ICP-AES) and atomic absorption spectroscopy (AAS). The SbRFP sensor is sensitive to arsenic as well as antimony. These devices may provide a low-cost option for analysis of toxic heavy metal contamination.
The synthetic biology protocol for preparing the E. coli presented here is applicable to any system that uses standard synthetic biology genetic parts to synthesize E. coli that express RFP. The analytical method is applicable to any system that expresses RFP, and so can be used to analyze any bacterial biosensor system that has been created using synthetic biology methods.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors wish to acknowledge the students at Longwood University in BIOL 324 (Genetics) and the students in CHEM 403 (Advanced Chemical Laboratory Problem Solving) who were involved in the initial preparation and testing of the antimony and lead biosensors. The idea for MicRoboCop was conceived at the GCAT SynBIO workshop (summer 2014), which is funded by NSF and Howard Hughes Medical Institute and hosted by the University of Maryland Baltimore County. The authors also acknowledge funding received from Longwood University’s Cook-Cole College of Arts and Sciences and the GCAT SynBio Alumni Grant.
Materials
| 1,3-dinitrobenzene, 97% | Aldrich | D194255-25G | |
| 2,4-dinitrotoluene, 97% | Aldrich | 101397-5G | |
| Agar | Fisher Scientific | BP1423-500 | |
| Ampicillin | Fisher Scientific | BP1760-5 | |
| Antimony, Reference Standard Solution (1000ppm ±1%/Certified) | Fisher Scientific | SA450-100 | Standard in dilute HNO3 |
| Cut Smart Buffer | New England BioLabs | B7204S | |
| Duplex Buffer | Integrated DNA Technologies | 11-01-03-00 | |
| EcoRI-HF Restriction Enzyme | New England BioLabs | R3101S | |
| Ethanol, HPLC grade, denatured | Acros Organics | AC611050040 | Solvents do not need to be HPLC grade, ACS or reagent grade will work. |
| Eurofins Genomics SimpleSeq DNA Sequencing Kits | Eurofins Genomics | SimpleSeq Kit Standard | |
| Forward primer for colony PCR | Integrated DNA Technologies | 5’- GCCGCTTGAATTCGTCATATAT-3’ | |
| Forward primer for DNA sequencing | Integrated DNA Technologies | 5’- GTAAAACGACGGCCAGTG-3’ | |
| IBI Science High Speed Plasmid Mini-kit | IBI Scientific | IB47101 | |
| LB Broth, Miller | Fisher Scientific | BP1426-500 | |
| Lead, Reference Standard Solution (1000ppm ±1%/Certified) | Fisher Scientific | SL21-100 | Standard in dilute HNO3 |
| LeadOff Disposable Cleaning and Decon Wipes | Hygenall | 45NRCN | Sold in canisters or individually wrapped, any alcohol based wipe will work. |
| Methanol, HPLC grade | Fisher Scientific | A452-4 | Solvents do not need to be HPLC grade, ACS or reagent grade will work. |
| NEB 5-alpha Competent E. coli cells | New England BioLabs | C2987I | |
| NheI-HF Restriction Enzyme | New England BioLabs | R3131S | |
| Nuclease free water | New England BioLabs | B1500S | |
| OneTaq 2X Master Mix with Standard Buffer | New England BioLabs | M0482S | |
| Plasmids from the registry of standard biological parts used for synthetic biology | Registry of Standard Biological Parts | http://parts.igem.org/Main_Page | |
| Promoter Sequences | Integrated DNA Technologies | Sb promoter: 5’-GCATGAATTCAGTCAT ATATGTTTTTGACTTATCCGCTTCGAAGAGAG AGACACTACCTGCAACAATCGCTAGCGCAT-3’ 3’-CGTACTTAAGCTCACTATATACAAAAACT GAATAGGCGAAGCTTCTCTCTCTGTGATGGAC GTTGTTAGCGATCGCGTA-5’ Pb promoter: 5’-GCATGAATTCGTCTTG ACTCTATAGTAACTAAGGGTGTATAATCGGCA ACGCGAGCTAGCGCAT-3’ 3’-CGTACTTAAGCAGAACTGAGATATCATTG ATCTCCCACATCTTAGCCGTTGCGCTGCGATCGCGTA-5’ TNT promoter: 5’GCATTCTAGATCAATT TATTTGAACAAGGCGGTCAATTCTCTTCGATT TTATCTCTCGTAAAAAAACGTGATACTCATCA CATCGACGAAACAACGTCACTTATACAAAAAT CACCTGCGAGAGATTAATTGAATTCGCAT3’ 3’CGTAAGATCTAGTTAAATAAACTTGTTCCG CCAGTTAAGAGAAGCTAAAATAGAGAGCATTT TTTTGCACTATGAGTAGTGTAGCTGCTTTGTT GCAGTGAATATGTTTTTAGTGGACGCTCTCTA ATTAACTTAAGCGTA5’ |
|
| Reverse primer for colony PCR | Integrated DNA Technologies | 5’- GCCGCTTGAATTCGTCTAGACT- 3’ | |
| Reverse primer for DNA sequencing | Integrated DNA Technologies | 5’- GGAAACAGCTATGACCATG-3’ | |
| T4 DNA Ligase | New England BioLabs | M0202S |
Riferimenti
- Eschner, K. "The Story of the Real Canary in the Coal Mine.". The Smithsonian Magazine. , (2016).
- Roda, A., et al. Progress in chemical luminescence-based biosensors: A critical review. Biosensors & Bioelectronics. 76, 164-179 (2016).
- He, W., Yuan, S., Zhong, W. H., Siddikee, M. A., Dai, C. C. Application of genetically engineered microbial whole-cell biosensors for combined chemosensing. Applied Microbiology and Biotechnology. 100 (3), 1109-1119 (2016).
- Dalby, O., Butler, D., Birkett, J. W. Analysis of Gunshot Residue and Associated Materials-A Review. Journal of Forensic Sciences. 55 (4), 924-943 (2010).
- Bell, S., Seitzinger, L. From binary presumptive assays to probabilistic assessments: Differentiation of shooters from non-shooters using IMS, OGSR, neural networks, and likelihood ratios. Forensic Science International. 263, 176-185 (2016).
- O'Mahony, A. M., Wang, J. Electrochemical Detection of Gunshot Residue for Forensic Analysis: A Review. Electroanalysis. 25 (6), 1341-1358 (2013).
- Vigneshvar, S., Sudhakumari, C. C., Senthilkumaran, B., Prakash, H. Recent Advances in Biosensor Technology for Potential Applications – An Overview. Frontiers in Bioengineering and Biotechnology. 4, 9 (2016).
- Fernandez, M., Morel, B., Ramos, J. L., Krell, T. Paralogous Regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a Basis for Arsenic Biosensor Development. Applied and Environmental Microbiology. 82 (14), 4133-4144 (2016).
- Porter, S. E. G., Barber, A. E., Colella, O. K., Roach, T. D. Using Biological Organisms as Chemical Sensors: The MicRoboCop Project. Journal of Chemical Education. 95 (8), 1392-1397 (2018).
- Borremans, B., Hobman, J. L., Provoost, A., Brown, N. L., Van der Lelie, D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. Journal of Bacteriology. 183 (19), 5651-5658 (2001).
- Hobman, J. L., Julian, D. J., Brown, N. L. Cysteine coordination of Pb(II) is involved in the PbrR-dependent activation of the lead-resistance promoter, PpbrA, from Cupriavidus metallidurans CH34. Bmc Microbiology. 12, (2012).
- Yagur-Kroll, S., Amiel, E., Rosen, R., Belkin, S. Detection of 2,4-dinitrotoluene and 2,4,6-trinitrotoluene by an Escherichia coli bioreporter: performance enhancement by directed evolution. Applied Microbiology and Biotechnology. 99 (17), 7177-7188 (2015).
- Gorman, M. "Guns in America: The Debate Over Lead Based Bullets.". Newsweek. , (2017).
- Yuksel, B., Ozler-Yigiter, A., Bora, T., Sen, N., Kayaalti, Z. GFAAS Determination of Antimony, Barium, and Lead Levels in Gunshot Residue Swabs: An Application in Forensic Chemistry. Atomic Spectroscopy. 37 (4), 164-169 (2016).
- Blakey, L. S., Sharples, G. P., Chana, K., Birkett, J. W. Fate and Behavior of Gunshot Residue-A Review. Journal of Forensic Sciences. 63 (1), 9-19 (2018).
- Yagur-Kroll, S., et al. Escherichia coli bioreporters for the detection of 2,4-dinitrotoluene and 2,4,6-trinitrotoluene. Applied Microbiology and Biotechnology. 98 (2), 885-895 (2014).

