Single-cell Screening Method for the Selection and Recovery of Antibodies with Desired Specificities from Enriched Human Memory B Cell Populations
Summary
The BSelex method to identify and recover individual antigen-specific antibodies from human peripheral blood mononuclear cells combines flow cytometry with single cell PCR and cloning.
Abstract
The human antibody repertoire represents a largely untapped source of potential therapeutic antibodies and useful biomarkers. While current computational methods, such as next generation sequencing (NGS), yield enormous sets of data on the antibody repertoire at the sequence level, functional data is required to identify which sequences are relevant for a particular antigen or set of antigens. Here, we describe a method to identify and recover individual antigen-specific antibodies from peripheral blood mononuclear cells (PBMCs) from a human blood donor. This method utilizes an initial enrichment of mature B cells and requires a combination of phenotypic cell markers and fluorescently-labeled protein to isolate IgG memory B cells via flow cytometry. The heavy and light chain variable regions are then cloned and re-screened. Although limited to the memory B cell compartment, this method takes advantage of flow cytometry to interrogate millions of B cells and returns paired heavy and light chain sequences from a single cell in a format ready for expression and confirmation of specificity. Antibodies recovered with this method can be considered for therapeutic potential, but can also link specificity and function with bioinformatic approaches to assess the B cell repertoire within individuals.
Introduction
Antibodies are a growing class of therapeutic molecules, and the existing B cell repertoire in any human is a potential source of such antibodies. When recovered from a human donor, they require no adaptation or "humanization", steps that are required for antibodies generated in other animal systems. Several methods exist for the identification and isolation of human antibodies, including B cell activation and proliferation1, immortalization via EBV transformation2,3, and generation of hybridoma cell lines4,5. However, all of these methods require extensive cell culture to screen and recover antigen-specific antibodies. Information about the human antibody repertoire has been greatly expanded with the development of next generation sequencing (NGS) technology, allowing for the identification of massive amounts of individual sequences present in donor samples. However, because NGS yields an agnostic view of all sequences present, it does not allow for the identification and isolation of antigen-specific antibodies, especially in the case of rare or low frequency antibodies.
The purpose of the "BSelex" method is to identify antigen-specific antibodies from circulating peripheral blood mononuclear cells in human donors, and isolate and recover the sequences of these antibodies for further analysis. This method utilizes flow cytometry and cell sorting to take advantage of the B cell receptor (BCR) expressed on the surface of memory B cells. Millions of B cells can be screened for antigen-specificity via flow cytometry before the more low-throughput molecular biology methods are initiated. Paired heavy and light chain identification is not possible in most NGS methods, which analyze cell sequences in bulk. In the method we describe here, cells are isolated individually, and paired recovery of both heavy and light chain sequences is possible, which allows direct cloning and expression of the full IgG.
Protocol
The use of samples from human volunteers followed protocols approved by The Scripps Research Institute Institutional Review Board. Informed consent was obtained from the donors prior to the blood donation.
1. Reagent Preparation
- Peripheral blood mononuclear cells (PBMCs)
- Recover peripheral blood mononuclear cells from normal human donors by Ficoll-Plaque Plus isolation. Cryopreserve at 50 million cells/mL in 90% fetal bovine serum (FBS) and 10% dimethyl sulfoxide (DMSO). Freeze the cells at -80 °C and transfer to liquid nitrogen for long term storage.
- Labeling target antigen peptides for sorting
- Generate or obtain peptides specific to the target protein (up to 70 residues long) with a terminal biotin.
- Label 4 nmol of each individual biotinylated peptide with streptavidin covalently attached to PE (R-phycoerythrin) or APC (allophycocyanin) in separate tubes. Prepare using a molar ratio of 9:1 peptide to streptavidin (SA), with a final concentration of roughly 3.2 µM for SA-PE and 5.7 µM SA-APC. Prepare Biotin tetramers as a negative control.
- Incubate each peptide mix overnight, in the dark, at 4 °C with slow mixing.
- Remove unbound fluorophore by passing labeled peptides through microcolumns containing polyacrylamide beads.
- Store peptide tetramers up to 2 months at 4 °C.
2. Cell Sorting
- At least 1 h up to 16 h before the CD22+ isolation, thaw donor PBMCs and allow to rest at 37 °C.
- Remove vials of frozen PBMCs from the freezer and immediately transfer to a 37 °C water bath. When the vials are almost thawed, transfer to the work area.
- Transfer the contents of each vial to a 50 mL tube containing prewarmed medium (keeping the donors separate). Add medium to a final volume of 50 mL. Gently mix the contents.
- Centrifuge at 370 x g for 6 min at room temperature. Discard the supernatant, resuspend the cells in 15 mL of RPMI complete medium and perform a cell count.
- Adjust cell concentration to 2.0 x 107 cells/mL with RPMI complete medium, and transfer cells to a T75 flask or larger and incubate at 37 °C for at least 1 h and up to 16 h to allow recovery time for thawed cells.
- Collect the cells (pooling if desired) and centrifuge at 370 x g for 6 min at 4 °C. Discard supernatant and resuspend the cells in 5 mL of ice cold MACS buffer (PBS, pH 7.6 containing 0.5% BSA and 2 mM EDTA), bring to 50 mL with MACS buffer and perform a cell count.
- Centrifuge the cells at 400 x g for 7 min at 4 °C.
- Isolate the CD22+ B cells by positive selection via capture on CD22 microbeads. Use MACS buffer for all washes. Count and centrifuge 400 x g for 7 min at 4 °C. The expected recovery is 5-10% of the total PBMC population.
- Resuspend cells at 40 million per mL FACS buffer (Tris buffer, pH 8.0 containing 0.5% BSA and 2 mM EDTA) and proceed with cell staining of the donors individually or as a pool.
- Stain cells for memory B cell sorting
- Remove an aliquot of cells for cytometer set up and compensation for each of the labeling fluorophores used. Include unstained cells as a control.
- Calculate the final staining volume for the number of CD22+ cells obtained (staining 2 million per 100 μL).
- Add extracellular markers for B cells (CD19-PerCP-Cy5.5), IgG (IgG-FITC), and memory compartment (CD27-PECy7) as per manufacturers' dilution recommendations, and mix gently. Aliquot 107 cells into a 1.5 mL tube for the negative control and transfer the remaining cells to a 15 mL tube.
- Add the dual labeled biotin-SA tetramers to the negative control tube at a final concentration of 36 nM each for PE and APC multiplied by the number of peptides in the sort tube and bring the volume to 0.5 mL final with FACS buffer (e.g., 10 peptides x 36 nM=360 nM).
- Add the peptide tetramers to the sort tube at 36 nM each for PE and APC and bring the cells to 2×106 per 100 μL with TBS Buffer. Incubate at 4 °C in the dark and with gentle rotation for 30-60 min.
- Wash 2x at 4 °C, 400 x g, 7 min, removing an aliquot to count before the last wash. Filter cells using 5 mL filter cap tubes. Add DAPI (4',6-diamidino-2-phenylindole) stain to 0.3 µM final concentration just prior to sorting as a marker of cell membrane integrity.
- Single cell sorting via flow cytometry
- Prepare 48 wells of 96-well PCR plates for sorting.
- Prepare a master mix: (2 μL of 10x RT buffer, 0.5 μL of RNase inhibitor, 7.5 μL of sterile PCR-grade water) per sample. Aliquot 10 μL per well, cover, and store at 4 °C until ready for sort.
- Run the negative control on the cell sorter, analyzing the entire sample.
- Set flow cytometry gates to isolate the appropriate cell populations for single cell sorting.
- Plot FSC area vs SSC area and set Gate R1 to isolate lymphocytes.
- Plot FSC height vs FSC width and set Gate R3 to exclude FSC doublets.
- Plot SSC height vs SSC width and set Gate R4 to exclude SSC doublets.
- Plot SSC height vs DAPI and set Gate R5 to isolate live cells (DAPI–).
- Plot IgG vs CD19 and set Gate R6 to isolate IgG+ B cells (CD19+ IgG+).
- Plot IgG vs CD27 and set Gate R7 to select memory B cells (CD27high).
- Plot Antigen-PE (Ag-PE) vs antigen-APC (Ag-APC) and set Gate R8 as an Ag-PE+/Ag-APC+ quadrant with a low number of events in the double positive gate.
- Set flow cytometry gates to isolate the appropriate cell populations for single cell sorting.
- Collect an equal number of memory cells from the antigen-positive (Ag+) sample for determination of signal to noise.
- Sort single cells having the phenotype CD19+ CD27+ Ag-PE+ Ag-APC+ (Gate R8) into prepared 96-well PCR plates.
- Cover plates with aluminum tape pads, centrifuge for 1 min at 400 x g, and store at -80 °C for future cloning.
- Prepare 48 wells of 96-well PCR plates for sorting.
3. Single Cell Cloning
- Reverse transcription reactions
- Remove the single B cell sort plate from 80 °C. Thaw on ice for 2 min. Spin the plate for 2 min at 3,300 x g to pool contents in the bottom of the wells before opening.
- Prepare a dNTP/buffer master mix with 10% non-ionic detergent and oligo(dT) as the reverse primer. Add enzyme mix to each well containing the single cell and DO NOT MIX.
- Incubate 5 min at 65 °C. Add enzyme master mix containing 100 mM DTT, RNase inhibitor, and reverse transcriptase enzyme to bring total volume to 20 µL. Incubate 10 min at 50 °C, then 10 min at 80 °C.
- Transfer to ice prior to starting PCR steps
- Multi-step nested PCR reactions
- Use separate nested PCR reactions are used for heavy chain (HC) and light chain (LC) amplification.
- For the first round of PCR amplification (Step I), use a pool of forward primers and a single reverse primer to amplify the HC and LC variable regions in separate PCR reactions (Figure 2). By design, the pool of forward primers will amplify a large percentage of germline leader (L) sequences and the reverse primer is specific to the downstream constant regions of each chain, including both the kappa (κ) and lambda (λ) for light chains16.
- Use 2.5 µL of cDNA product as template for each PCR (Step I) reaction. Add primers to the final reaction concentration of 1 µM for each. Double the 10x polymerase buffer to 2x concentration, bring the final volume to 25 µL per reaction. Run HC PCR reactions using [94 °C/4 min; 94 °C /15 s, 55 °C /20 s, 68 °C/60 s; 68 °C/3 min] for 50 cycles. Run LC PCR reactions using [98 °C/4 min; 98 °C /15 s, 72 °C /20 s, 72 °C/60 s; 72 °C/3 min] for 50 cycles.
- Use 2.5 µL of the Step I product as template for each (Step II) PCR reaction. Add the pool of forward primers specific to the HC and LC (κ and λ) framework 1 region and a pool of reverse primers specific to the junction region of each antibody chain. Double the 10x polymerase buffer to 2x concentration, and run PCR reactions 50 cycles each using the same parameters as Step I.
- Run the completed PCR reactions on 1% agarose gel to visualize positive amplification hits. Recover paired amplicons (both heavy and light chain from the same cell) and isolate via gel extraction. Determine DNA fragment concentration via OD260 for accurate ligation mix calculations.
- Combine recovered heavy and light chain fragments with a linker fragment and the expression vector backbone using a 4-fragment ligation reaction using a commercial kit.
- Transform ligation reactions into chemically competent bacteria using a commercial kit. Once plated onto antibiotic plates, add 4 mL of growth media (with antibiotic) to the remaining transformation culture, and incubate 37 °C overnight at 250 rpm. This is the "ligation mix culture."
- Prepare miniprep DNA from the overnight ligation mix cultures using a commercial kit, and determine the resulting plasmid DNA concentration.
4. ELISA Screen for Confirmation of Antigen-specificity
- Transfect 10 µg of miniprep DNA with cationic lipid-based reagent into 10 mL of suspension 293 cells, and incubate for 3-4 days at 37 °C (8% CO2) while rotating at 125 rpm. For harvest, centrifuge culture 10 min at 1,000 x g and recover clarified media.
- Measure the IgG concentration in the supernatants via affinity to Protein A.
- Test each IgG (in supernatant) at 20 µg/mL by enzyme-linked adsorbent assay (ELISA) against the individual peptides used for the sort, captured on a streptavidin plate or actin as a negative control. Use a goat anti-human peroxidase secondary antibody against human IgG Fab to detect recombinant clones. After subtraction of background, OD > 0.5 is defined as screen positive.
- Confirm screen positive hits by performing an additional ELISA, using dilutions of IgG supernatant to generate a concentration curve starting at 20 µg per mL, and plotting against OD for each antigen showing reactivity in the initial screen ELISA.
Representative Results
This method covers a multi-step process to isolate antigen-specific antibodies from human donors. In the representative data shown here, cells were incubated with a pool of fluorescently-labeled peptides representing several different domains of the tau protein, including phosphorylated peptides to mimic putative phosphorylation sites. These peptides were used as "bait" to identify cells that are reactive with tau epitopes(s) of interest. In preparation for sorting, a panel of fluorescently-labeled phenotypic markers were used to identify different cell populations within the enriched B cell population. A series of cytometry gates were devised to isolate the target memory B cells (Figure 1). Lymphocytes were isolated based on their cell size and granularity using forward scatter (FSC) and side scatter (SSC) plots in flow cytometry6,7. Following exclusion of multiple cells ("doublets") and dead cells, phenotypic markers allowed the segregation of IgG+ memory B cells via IgG, CD19 (B cell) and CD27 (memory). In this approach, the CD27 marker does not distribute into two discrete populations, so the top 45% of CD27+ expressing cells are included for the final gate. Finally, cells double-positive for both APC and PE fluorophores distribute into the upper right quadrant of the gating graph, indicating reactivity to both labeled versions of peptides. The cells that fell within the drawn gate were isolated and sorted into individual wells of a 96-well plate. The use of antigen with two different labels increases the signal-to-noise ratio and reduces the number of false positives in the subsequent molecular biology process. The sorted cells represented about 0.1% of the memory B cells in the final gate, and 0.001% of the starting cell sample.
The first readout of single cell cloning is confirmation of amplification of the respective heavy and light variable chains (Figure 2). Since paired recovery of both amplicons is desired, the PCR reactions are evaluated side-by-side on agarose gel, and matched pairs are excised from the gel and DNA fragments extracted. Typical efficiency of amplification is 30-50% heavy chain and 50-70% light (kappa) chain. Recovery of paired amplicons is usually between 25-40% efficiency. These efficiencies vary between donor pools, and the representative data (Figure 3) is an example of very efficient amplification from 24 single cells (42% paired recovery). Following IgG cloning, the IgG expression vector is transfected into human embryonic kidney (HEK-293) suspension cells, which are used to maximize expression. The use of serum-free medium reduces contaminating proteins from the recombinant antibody prep, and helps minimize noise in subsequent binding assays. The recovered antibodies are screened against the original panel of tau peptides and scored for reactivity by ELISA (Figure 4). An initial threshold of OD = 0.5 above background is used to indicate positive antigen reactivity, and β-actin protein is used as a control for non-specific binding. If the flow cytometry and sorting steps use a mixed pool of peptides, the screening ELISA is the first step of deconvoluting specific reactivities of the recovered IgGs. Three of the 10 IgGs assayed demonstrated reactivity against phosphorylated CBTAU22.1 peptide, and one was reactive to non-phosphorylated CBTAU27.1 (Figure 4A). Additional confirmation was completed using a concentration curve of the same recombinant antibody samples against the peptides identified in the initial screen, and an additional peptide as a negative control (Figure 4B). For each positive hit, an individual plasmid clone was isolated from the transformed pool and reconfirmed by the same ELISA method. Only Clone 34 is shown the data presented, and while reactivity to non-phosphorylated CBTAU27.1 was confirmed, lower affinity binding was also observed with the phosphorylated 27.1 peptide (Figure 4C).
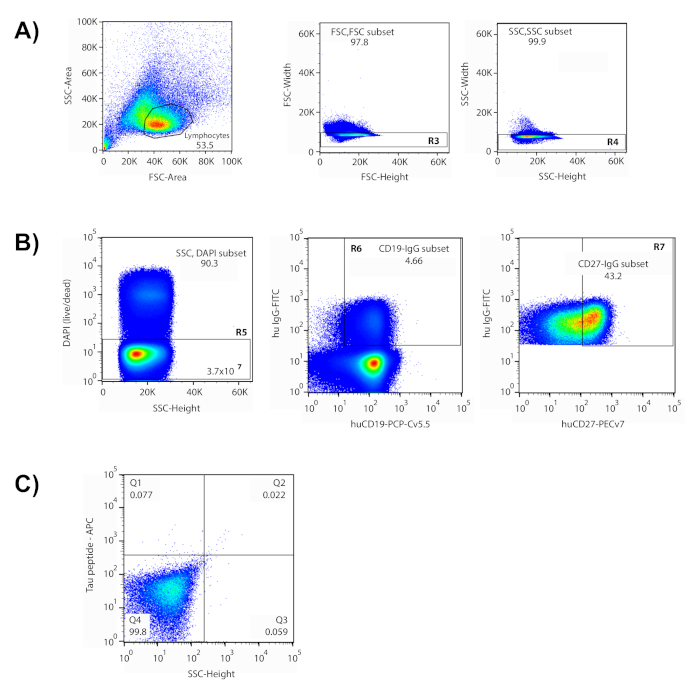
Figure 1: Isolation of antigen-reactive single cells via flow cytometry. (A) Shown is a representative plot where the lymphocyte population was contained within the drawn gate. (B) Gates based upon forward and side scatter (FSC-height v FSC-width (R3); SSC- height v SSC- width(R4)) were used to exclude doublets. Only cells within drawn gates were evaluated in subsequent plots. DAPI+ cells were considered dead and excluded (R5). In this experiment, 3.7 x 107 "live" cells were interrogated. The majority of live cells were B cells (CD19+), and IgG+ cells (4.66%) were isolated from this population (R6). The top 43.2% of CD27+-expressing memory cells were included in the selection gate (R7). (C) Quadrant 2 (Q2) contained cells reactive to both labeled antigens (peptide-APC v peptide-PE), and these cells were sorted and recovered individually into a 96-well plate. Please click here to view a larger version of this figure.
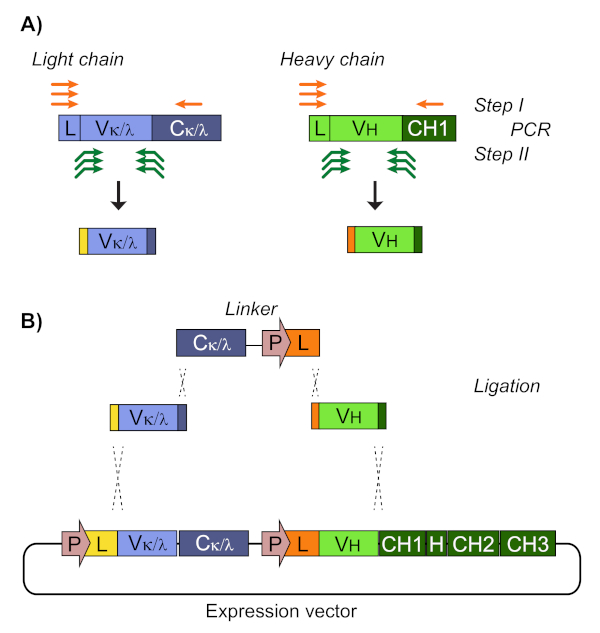
Figure 2: PCR amplification recovers IgG heavy and light variable sequence for cloning. (A) Both heavy (VH) and light (Vk or Vl) chain variable regions are recovered using nested PCR reactions. Step I primers amplify forward from the native leader sequence and reverse from within the constant region. Multiple arrows represent a pool of between 7-12 primers, which are used to ensure broad coverage of possible germlines. Step II primers are nested, specific to the extreme ends of the variable open reading frame, and add sequence to the ends of the amplicons that are homologous to adjacent sequence in the expression vector. (B) The linker consists of the constant light chain (Ck or Cl), followed by the heavy chain promoter and a non-native signal peptide sequence. The amplicons, linker, and plasmid backbone are simultaneously ligated via overlapping homologous sequence generated during the Step II PCR amplification, and isolated as an intact plasmid. The recombinant heavy and light chains in the final expression vector are driven by independent (and identical) CMV promoters, and are translated as separate proteins. Please click here to view a larger version of this figure.
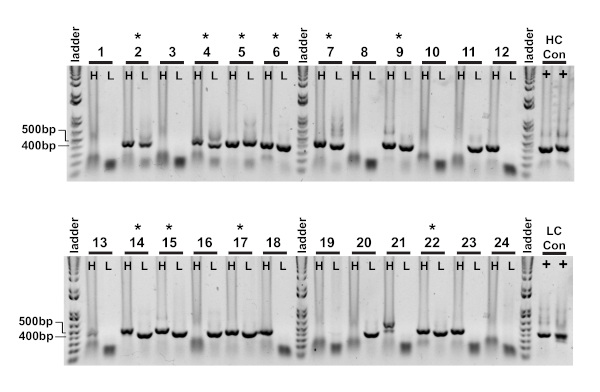
Figure 3: IgG heavy and light variable chain amplicons are recovered from single cells. The nested PCR reactions (Step II) were directly loaded onto 1% agarose gel and visualized. Heavy chain (H) and kappa light chain (L) PCR reactions from the same cell were loaded in adjacent wells so paired amplicons were easily observed. Successful heavy and light chain products are roughly 400 bp and 350 bp respectively. Successful recovery of paired amplicons are denoted by an (*). Please click here to view a larger version of this figure.
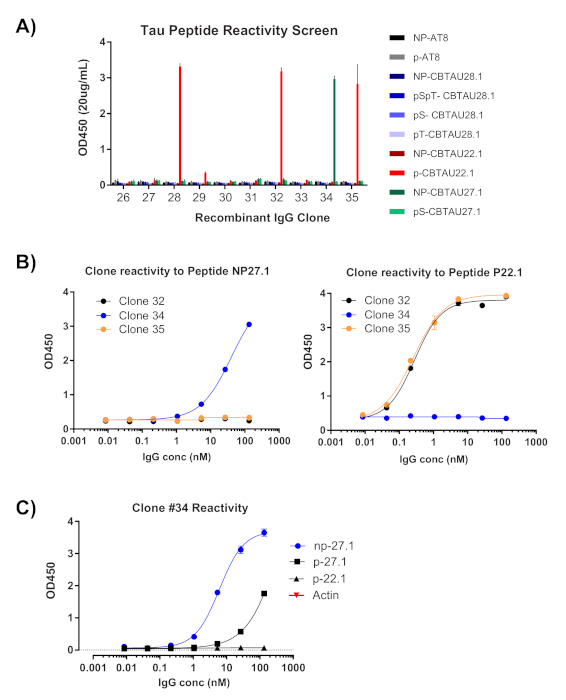
Figure 4: Antigen specificity is confirmed from recombinant IgGs. Plasmids containing the recovered heavy and light chain sequences are transfected into HEK cells for expression. After four days, recombinant antibodies are assessed for reactivity by ELISA. (A) IgG concentration in the clarified media was measured and diluted to 20 µg/mL. The pool of peptides used for sorting were individually assessed using 40 pmol per well in streptavidin plates. All IgGs were tested in duplicate wells. (B) Clones that displayed OD450 measurement >0.5 were re-assessed (Clones #32, #34, #35) using 1:5 dilution steps against the peptides identified in the screen. (C) Once confirmed as a hit, a single plasmid clone was isolated from the clone #34 transformed ligations, transfected, and reconfirmed by ELISA. The same was done for clones #32 and #35 (data not shown). Please click here to view a larger version of this figure.
Discussion
The method presented here combines flow cytometry and single cell cloning, and the methods we describe here we based on methods previously developed by Tiller and colleagues11. Their work describes the recovery and cloning of monoclonal antibodies in order to study the B cell repertoire in humans at the level of individual cells. We have adapted the major components of their process to allow recovery of antigen-specific monoclonal antibodies from a population of memory B cells, including the multi-step amplification primer strategy. The major modification is the addition of labelled antigen "bait". Additional adaptations have been made to the published protocol, including (but not limited to) modifying the cloning vector backbone into a single expression plasmid, additional primer coverage of the germline repertoire (both leader sequence and framework 1), transfection of suspension HEK293 cells for higher expression, and the use of high fidelity polymerases during PCR amplification.
For the methods described here, the most critical steps are near the transition between flow cytometry and single cell cloning. First, the proper placement of sorted single cells into the plate is essential. Setting the drop-delay correctly on the sorter is a key step. Environmental factors, such as low humidity, must also be taken into account, as we have found our recovery efficiencies drop significantly unless a static gun is used on the target sort plates. Following cell placement, plates are centrifuged to ensure cells have contacted the 10 µL of buffer in bottom of the wells. All of these measures are critical for the molecular biology portion to be successful. If the conditions are sub-optimal for the reverse transcription reaction of a single cell, PCR amplification of even β-actin can be difficult. The strength of the method presented is the ability to interrogate millions of cells and only sort the ones that match the antigen reactivity criteria against the antigen of choice. This requires the signal to noise ratio to be as high as possible, which is done by optimizing bait concentrations before sorting. Dual-labeled bait is used to reduce the recovery of false-positive cells, which can occur if one of the fluorophores has high background. Using more than one antigen bait can make the signal:noise optimization more difficult, but the ability to interrogate multiple peptides simultaneously is another benefit of this method.
When recovery efficiencies are low, a set of nested β-actin primers are used to confirm that template cDNA is present. The drawback to this approach is that it is difficult to determine whether there was no cell present in the well or the reverse transcription reaction failed, making troubleshooting difficult. Occasionally the opposite will occur and the efficiencies are higher than expected. The typical recovery for heavy chain is between 30-45% and light chains are higher, between 40-60%. Each PCR reaction (Step I & II) uses 50 cycles to amplify from a single cell. The high number of total cycles also makes this method susceptible to contamination. Sequence analysis of amplicons can be used to determine whether a contaminant has been introduced, or an unusually high recovery rate has been legitimately achieved.
The utility of this method to recover native human antibodies against an antigen of choice has several significant limitations. First, only soluble proteins that can be labeled may be used as "bait" for the flow cytometry. For the sorts presented in the representative results, we used a series of synthesized overlapping peptides from tau protein, since using the whole protein proved difficult. The use of linear peptides is likely not optimal, since it may limit the identification of antibodies against non-linear or structural epitopes. However, we were able to identify several unique antibodies against tau and these are currently undergoing further evaluation8,9. Another limitation is the low throughput of the molecular biology recovery and cloning of IgGs. The initial sorting strategy allows for the screening of millions of cells, but the subsequent molecular biology processing consists of multiple stages. Ongoing optimization efforts to incorporate newer technologies, such as Gibson Assembly10 have streamlined several steps, but the bottleneck remains cloning individual heavy and light chain pairs.
The "BSelex" method utilizes the BCR expressed on the surface of memory B cells to identify cells that display antigen reactivity, and then recover these individual cells via flow cytometry11,12. Due to this reliance on the BCR, the method is restricted to the memory B cell compartment, and does not capture antibody secreting cells (ASCs) such as plasmablasts. However, this approach may be advantageous to recovering a broader repertoire of antigen-specific immunoglobins when compared to ASCs. Influenza vaccination studies demonstrate that while reactive ASCs can be dominated by a small number of expanded B cell clones, the antigen-specific memory B cell population is rarely clonal12. The T cell receptor (TCR) on the surface of T cells possesses similarities to the B cell receptor in gene arrangement and recombination to maximize diversity. Single cell approaches similar to what is described in the method presented here have been developed for assessing the T cell repertoire, including recovery and single cell cloning of alpha and beta chains13. However, TCR recognition requires peptides to be presented by MHC molecules, adding significant complexity to the labeled bait approach to identify peptide-specific T cells. Unlike B cells, T cells do not undergo affinity maturation of the entire variable region, so identification of the short CDR3 region and some flanking sequence is all that is required for identification and reconstitution. Finally, this method describes the identification of IgG molecules using flow cytometry, but it is also possible to use alternative phenotypic markers to identify B cells of different isotypes.
Currently, there are methods being developed to connect the enormous amount of data collected from next generation sequencing with functional analysis of antigen specificity, but these methods are still being refined. Despite its limitations, the method described here has been used to identify antibodies with potential therapeutic value for both infectious and non-infectious diseases8,14,15, and represents a reliable approach to recover relevant antigen-specific IgGs from humans, without extensive manipulation of B cells or extensive cell culture.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Lucy Chammas, Martha Costa, Julie Kim, Nancy Heredia, and Jeremy Macedo for the extensive testing of many method modifications and refinement of the current BSelex platform.
Materials
| MoFlo Astrio EQ, Cell Sorter | MoFlo | cell sorting | |
| Biotinylated peptides | New England Peptide | Cell sorting "bait" | |
| Micro Bio-spin P30 gel column | BioRad | #7326223 | |
| Streptavidin (R-PE) | Thermo Scientific | SA10041 | |
| Streptavidin (APC) | Thermo Scientific | S32362 | |
| CD22 MicroBeads, human | Miltenyi | #130-046-401 | |
| LS columns | Miltenyi | #130-042-401 | |
| Pre separation filters | Miltenyi | #130-041-407 | |
| PerCp-Cy5.5 mouse anti-human CD19 | BD Biosciences | BD#340951 | |
| PE-Cy7 mouse anti-human CD27 | BD Biosciences | #560609 | |
| FITC mouse anti-human IgG | BD Biosciences | #560952 | |
| DAPI | Life Technologies | #D21490 | |
| RNaseOUT | Life Technologies | #10777-019 | |
| Bovine serum albumin, Fraction V | Sigma | #A4503 | |
| RPMI media | Hyclone | #SH30096.01 | |
| HI FBS (Fetal bovine serum) | Invitrogen | #10082147 | |
| Mastercycler Gradient | Eppendorf | Model #6325 | PCR machine |
| PCR 96-well plates | Phenix | MPS-500 | |
| PCR Plate mats | Phenix | SMX-PCR96 | |
| Advantage UltraPure PCR Deoxynucleotide Mix | Clontech | #639125 | |
| Superscript IV First-strand synthesis system | Invitrogen | #18091050 | |
| Phusion High Fidelity Polymerase | Thermo Scientific | F-531-L | |
| Platinum polymerase | Invitrogen | #10966108 | |
| Nuclease-free water | QIAGEN | #129114 | |
| Oligonucleotide primers | IDT | assorted | Primers for all PCR steps |
| Gel Extraction Kit | QIAGEN | #28706 | |
| PCR Purification kit | QIAGEN | #28106 | |
| Miniprep Kit | QIAGEN | #27106 | |
| NEBuilder HiFi DNA Assembly Cloning Kit | New England Biolabs | E5520S | Gibson assembly |
| Expi293 Expression Media | Invitrogen | #A14351-01 | |
| ExpiFectamine 293 Transfection Kit | Invitrogen | #A14525 | |
| Opti-MEM Media | Invitrogen | #31985070 | |
| CO2 incubator (Multitron) | |||
| Octet RED384 System | Pall/ Forte Bio | ||
| Pierce Streptavidin Coated High Binding Capacity Clear 96-well Plates | Thermo Scientific | #15500 | |
| Corning Costar 96-well Polystyrene Plate High Binding Half Area | Corning | #3690 | |
| Goat Anti-human IgG Fab Secondary Antibody | Jackson Labs | #109-036-097 | |
| Goat Anti-mouse HRP Secondary Antibody | Jackson Labs | #115-035-072 | |
| SureBlueTM TMB Microwell Peroxidase Substrate (1-Component) | KPL | #52-00-03 | |
| TMB Stop Solution | KPL | #50-85-06 |
Riferimenti
- Su, K. Y., Watanabe, A., Yeh, C. H., Kelsoe, G., Kuraoka, M. Efficient Culture of Human Naive and Memory B Cells for Use as APCs. Journal of Immunology. 197 (10), 4163-4176 (2016).
- Corti, D., Lanzavecchia, A. Broadly neutralizing antiviral antibodies. Annual Review of Immunology. 31, 705-742 (2013).
- Steinitz, M., Klein, G., Koskimies, S., Makel, O. EB virus-induced B lymphocyte cell lines producing specific antibody. Nature. 269 (5627), 420-422 (1977).
- Bloom, A. D., Nakamura, F. T. Establishment of a tetraploid, immunoglobulin-producing cell line from the hybridization of two human lymphocyte lines. Proceedings of the National Academy of Sciences of the United States of America. 71 (7), 2689-2692 (1974).
- Olsson, L., Kaplan, H. S. Human-human hybridomas producing monoclonal antibodies of predefined antigenic specificity. Proceedings of the National Academy of Sciences of the United States of America. 77 (9), 5429-5431 (1980).
- Lechner, J., et al. Alterations in Circulating Immune Cells in Neovascular Age-Related Macular Degeneration. Scientific Reports. 5, 16754 (2015).
- Loken, M. R., Brosnan, J. M., Bach, B. A., Ault, K. A. Establishing optimal lymphocyte gates for immunophenotyping by flow cytometry. Cytometry. 11 (4), 453-459 (1990).
- Pascual, G., et al. Immunological memory to hyperphosphorylated tau in asymptomatic individuals. Acta Neuropathologica. 133 (5), 767-783 (2017).
- van Ameijde, J., et al. Enhancement of therapeutic potential of a naturally occurring human antibody targeting a phosphorylated Ser(422) containing epitope on pathological tau. Acta Neuropathologica Communications. 6 (1), 59 (2018).
- Gibson, D. G., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6 (5), 343-345 (2009).
- Tiller, T., et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. Journal of Immunological Methods. 329 (1-2), 112-124 (2008).
- Wrammert, J., et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 453 (7195), 667-671 (2008).
- Han, A., Glanville, J., Hansmann, L., Davis, M. M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nature Biotechnology. 32 (7), 684-692 (2014).
- Apetri, A., et al. A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities. Acta Neuropathologica Communications. 6 (1), 43 (2018).
- Jones, H. G., et al. Structural basis for recognition of the central conserved region of RSV G by neutralizing human antibodies. PLoS Pathogens. 14 (3), 1006935 (2018).
- . . US patent. , (2017).

